Abstract
Newcastle disease virus (NDV) is one of the most important viral diseases of birds. Wild birds constitute a natural reservoir of low-virulence viruses, while poultry are the main reservoir of virulent strains. Exchange of virus between these reservoirs represents a risk for both bird populations. Samples from wild and domestic birds collected between 2006 and 2010 in Luxembourg were analyzed for NDV. Three similar avirulent genotype I strains were found in ducks during consecutive years, suggesting that the virus may have survived and spread locally. However, separate introductions cannot be excluded, because no recent complete F gene sequences of genotype I from other European countries are available. Detection of vaccine-like strains in wild waterbirds suggested the spread of vaccine strains, despite the nonvaccination policy in Luxembourg. Among domestic birds, only one chicken was positive for a genotype II strain differing from the LaSota vaccine and exhibiting a so-far-unrecognized fusion protein cleavage site of predicted low virulence. Three genotype VI strains from pigeons were the only virulent strains found. The circulation of NDV in wild and free-ranging domestic birds warrants continuous surveillance because of increased concern that low-virulence wild-bird viruses could become more virulent in domestic populations.
INTRODUCTION
Newcastle disease, caused by virulent strains of the Newcastle disease virus (NDV), is one of the most important viral diseases in poultry, together with highly pathogenic avian influenza. NDV, or avian paramyxovirus type 1, is a negative single-stranded RNA virus belonging to the Avulavirus genus in the Paramyxoviridae family. Based on clinical signs in chickens, several pathotypes have been defined (1). The sequence of the fusion (F) protein cleavage site is considered a major virulence determinant, and it appears that a minimum of three basic amino acids between residues 112 and 116, followed by a phenylalanine at residue 117, are required for virulence (1). However, other factors are also involved in the virulent phenotype, as indicated by strains found in pigeons around the world and in healthy migratory ducks in Alaska that have a virulent cleavage site motif but are not always virulent for chickens in standard pathogenicity tests (2, 3).
Besides its phenotypic heterogeneity, NDV is also genetically diverse; several phylogenetic classes and genotypes (4) or lineages (5) are recognized, and its diversity is still unfolding (4, 6). Class I strains and genotypes I and X of class II are mainly constituted by avirulent strains. Genotype II contains a broad spectrum of strains, but nowadays, mainly avirulent strains are found. The other class II genotypes (III to IX and XI to XV) contain almost exclusively viruses with a virulent cleavage site.
Wild birds constitute a natural reservoir of viruses of low virulence for chickens. In particular, waterbirds may play an important role in NDV epidemiology due to shedding of viral particles into the aquatic environment more favorable for virus stability and due to their potential for long-distance dissemination by migration (7). Although no direct epidemiological link was found, successions of outbreaks in the United Kingdom, Sweden, Denmark, and Finland between 1996 and 2005 were all related at least on the basis of strain similarity, suggesting multiple introductions of viruses from the same pool by wild birds (8). Moreover, isolation of similar viruses in wild birds (a goosander in Finland and a cormorant in Denmark) together with the proximity to water of a significant number of the affected flocks was suggestive of a wild-bird reservoir at least of genotype XIII strains in Western and Northern Europe (9). Another more common threat for poultry arises from pigeon paramyxovirus type 1 (PPMV-1) strains, variants of NDV grouping within genotype VI. PPMV-1 strains exhibit a broad range of pathogenicities for poultry (2), and pathogenicity may increase after serial passages in chickens (2, 10). On several occasions, PPMV-1 strains caused outbreaks in chickens (11–13).
During the last decade, virulent viruses from genotypes VI, VII, and XIII have been detected in wild and domestic birds in several European countries (9), and avirulent strains of class I and class II (genotypes I and II) have also been reported sporadically (http://ec.europa.eu/food/animal/diseases/controlmeasures/avian/crls_proceedings_en.htm). However, in Europe, only a few sequences from wild birds are available, despite their importance for epidemiological surveillance. In this retrospective study, we examined stored samples collected in the framework of avian influenza surveillance in Luxembourg to investigate the presence of avirulent and virulent NDV in wild and domestic birds. All viruses found had a predicted low virulence, except for three PPMV-1 strains.
MATERIALS AND METHODS
Sample collection.
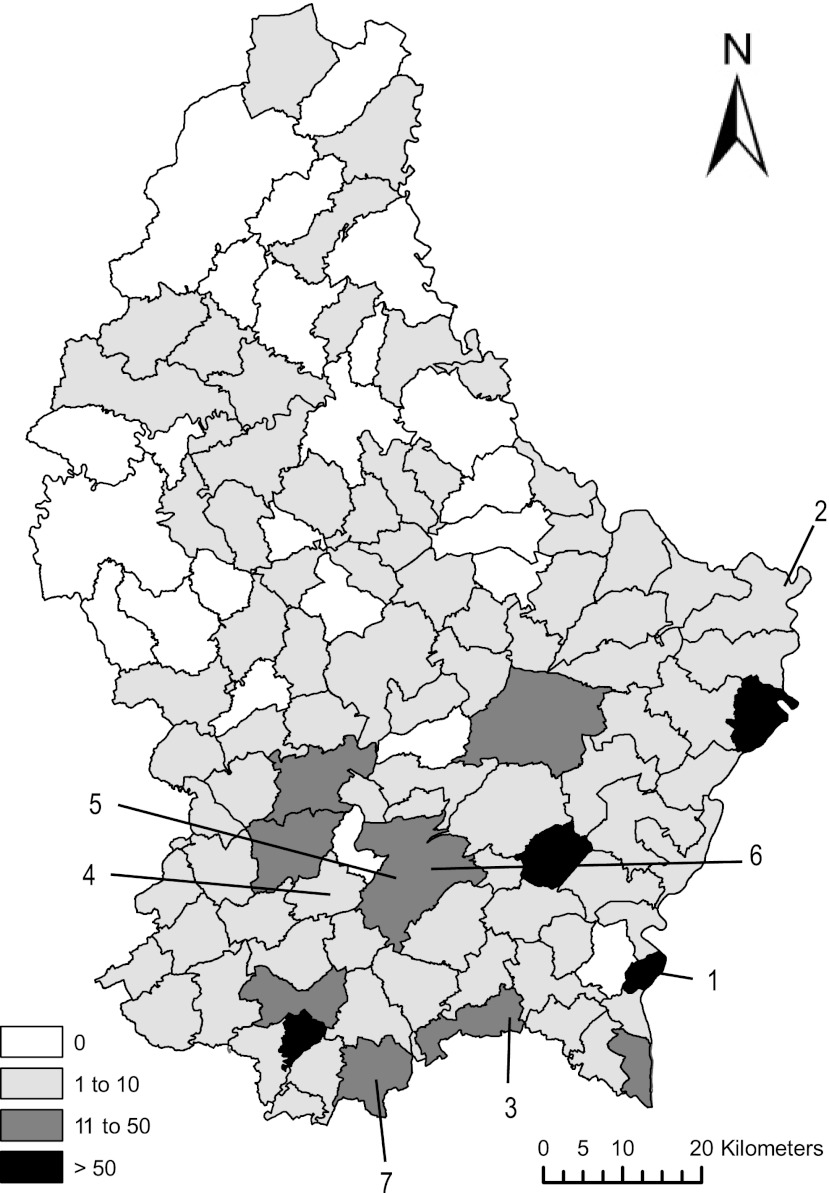
Between January 2006 and July 2010, pooled tracheal and cloacal swabs (n = 576), cloacal swabs (n = 196), tracheal swabs (n = 22), and fresh feces (n = 337) were collected during active and passive surveillance for avian influenza virus. The majority of the samples originated from wild birds (n = 1,003), but a smaller number of domestic birds was also sampled, including chickens (n = 120), a turkey (n = 1), a quail (n = 1), a peafowl (n = 1), and pheasants (n = 2) (see Table 2). Samples from healthy passerines were mainly collected with mist nests during migration surveys at three locations: Nospelt, Ubersyren, and Schifflange. Wild waterfowl, primarily targeted for avian influenza surveillance, were mainly sampled along the Moselle River in Remich and Wasserbillig. Some samples from injured or sick animals, especially from birds of prey, were also collected at a wildlife shelter, while exotic species (n = 3) were sampled at a zoological park and at the international airport. The other bird species were sampled throughout the country but with a bias toward the southern part of the country (Fig. 1).
Table 2.
List of sampled domestic and wild bird species in Luxembourg between 2006 and 2010 tested for class I and class II and genotype classification of positive samples
| Family | No. of positives/no. tested for class II | No. of positives/no. tested for class I | Yearly distributiona |
||
|---|---|---|---|---|---|
| 2006 | 2007 | 2008 | |||
| Phasianidaeb | 1/125 | 0/62 | 1× G-II | ||
| Anatidae | 4/340 | 0/92 | 1× G-II | 2× G-I | 1× G-I |
| Phalacrocoracidae | 1/3 | 0/1 | 1× G-II | ||
| Ardeidae | 0/13 | 0/2 | |||
| Accipitridae | 0/135 | 0/7 | |||
| Falconidae | 0/26 | 0/6 | |||
| Rallidae | 0/52 | 0/4 | |||
| Laridae | 0/8 | ||||
| Columbidae | 3/59 | 0/11 | 2× G-VI | 1× G-VI | |
| Psittacidaec | 0/3 | 0/3 | |||
| Strigidae | 0/12 | 0/1 | |||
| Tytonidae | 0/14 | 0/1 | |||
| Picidae | 0/5 | 0/1 | |||
| Corvidae | 0/20 | 0/3 | |||
| Paridae | 0/5 | 0/2 | |||
| Hirundinidae | 0/9 | 0/9 | |||
| Phylloscopidae | 0/5 | 0/2 | |||
| Acrocephalidae | 0/42 | 0/32 | |||
| Locustellidae | 0/4 | 0/3 | |||
| Sylviidae | 0/95 | 0/37 | |||
| Sturnidae | 0/7 | ||||
| Turdidae | 0/21 | 0/15 | |||
| Muscicapidae | 0/9 | 0/4 | |||
| Passeridae | 0/42 | ||||
| Prunellidae | 0/2 | 0/1 | |||
| Motacillidae | 0/4 | ||||
| Fringillidae | 0/1 | 0/1 | |||
| Emberizidae | 0/4 | 0/3 | |||
| Undetermined | 0/66 | 0/42 | |||
| Total | 9/1,131 | 0/345 | 4/619 | 4/349 | 1/84 |
G-, genotype.
Domestic species including chicken, turkey, quail, peafowl, and pheasant.
Exotic species.
Fig 1.
Geographic distribution of collected samples by municipalities in Luxembourg. The shading corresponds to the number of samples collected per municipality. The numbers indicate the origins of isolates as follows: 1, duck/Luxembourg/26/2006 and pigeon/Luxembourg/119/2006; 2, great cormorant/Luxembourg/2547/2006; 3, chicken/Luxembourg/2871-18/2007; 4, duck/Luxembourg/3785/2007 and duck/Luxembourg/3786/2007; 5, pigeon/Luxembourg/3821-1/2007; 6, mallard/Luxembourg/4178/2008. The strain pigeon/Luxembourg/2657-2/2006 originated from an animal rescued at the animal wildlife shelter in Dudelange (7). Template map © Origine Cadastre: Droits Réservés è l'Etat du Grand-Duché de Luxembourg (2012).
All swab samples were immediately placed in the field in 500 μl of virus transport medium (phosphate-buffered saline [PBS; pH 7.0] with 2,000 U of penicillin/ml, 200 mg of streptomycin/ml, 2,000 U of polymyxin B/ml, 250 mg of gentamicin/ml, 60 mg of ofloxacin/ml, 200 mg of sulfamethoxazole/ml, and 2.5 mg of amphotericin B/ml). All samples were kept refrigerated until delivery to the laboratory. Approximately 100 mg of fecal samples was homogenized in 500 μl of virus transport medium upon arrival at the laboratory. A total of 786 samples were processed immediately and cDNA was kept at −20°C, while 345 samples were stored at −80°C before being processed. The majority of the samples (86%; 968/1,131) were stored at −80°C within 48 h after collection.
Nucleic acid extraction, PCR, and sequencing.
RNA was extracted from 140 μl of medium using a QIAamp viral RNA minikit (Qiagen, Venlo, The Netherlands) or from 50 μl using a MagMAX-96 AI/ND viral RNA isolation kit (Life Technologies, Merelbeke, Belgium) with KingFisher (Thermo Fisher, Waltham, MA). Screening for class I strains was performed only on the 345 samples collected after June 2007 (according to the method of Kim et al. [14]). Class II strain detection by nested PCR (6, 15) and sequencing of positive PCR products (6) were carried out as described previously. Whenever enough material was available, the entire F gene was amplified using several primer combinations in (semi)nested PCR formats (Table 1).
Table 1.
Sequences of primers used to amplify and sequence partial or full F genes
| Primer | Orientation | Sequence (5′–3′) | Localization | Reference |
|---|---|---|---|---|
| FOP1 | Forward | TACACCTCATCCCAGACAGGGTC | F gene | 15 |
| FOP2 | Reverse | AGGCAGGGGAAGTGATTTGTGGC | F gene | 15 |
| FIP1 | Forward | TACTTTGCTCACCCCCCTT | F gene | 15 |
| FIP2 | Reverse | CATCTTCCCAACTGCCACT | F gene | 15 |
| M610 | Forward | CTGTACAATCTTGCGCTCAATGTC | M gene | 16 |
| P1 | Forward | ATGGGCYCCAGAYCTTCTAC | F gene | 17 |
| F581 | Reverse | CTGCCACTGCTAGTTGTGATAATCC | F gene | 16 |
| F-4639f | Forward | TGAYGGCAGGCCTCTT | F gene | This study |
| F-4932f | Forward | CAACCGCTGCACAGATAA | F gene | This study |
| F-4954f | Forward | AGCTGCGGCYCTRATACAA | F gene | This study |
| F-5042f | Forward | GAGGTCACYGACGGATTAT | F gene | This study |
| F-5488f | Forward | TCAGCACTTGTCCCAAA | F gene | This study |
| F-1258-R | Reverse | ACATTGCATGAWTGTCTRTC | F gene | 18 |
| F-5566r | Reverse | CAGTATGAGGTGTCAAGTT | F gene | This study |
| F-5749f | Forward | AGACCCTCCAGGYATCA | F gene | This study |
| F-5888f | Forward | GGCTCAGTGGGGAAT | F gene | This study |
| F-6086f | Forward | GGTACACTTAGCCTGRTHTT | F gene | This study |
| F-6146r | Reverse | CTTYTGTTGCGCCTTT | F gene | This study |
| F-7979-R | Reverse | AGRGCCACYTGCTTRTATA | HN gene | 18 |
Statistical analyses.
Statistical analyses to assess whether differences in sampling or sample processing had an effect on the outcome of the detection tests were performed using the chi-square test with Yates correction in SigmaPlot software.
Sequence analyses.
Kimura distances were calculated according to the Kimura 2-parameter model on partial (240-nucleotide [nt]) or complete (1,662-nt) F gene sequences. The sequence lengths used for distance calculations are mentioned in Results. Phylogenetic relationships were inferred by comparing the Luxembourgish strains with all NDV sequences available on the NCBI website (downloaded in October 2012) after data set curation. Data sets were aligned using ClustalW (19). Trees were calculated with the neighbor-joining method, using the Kimura 2-parameter model and 1,000 bootstrap replicates as implemented in MEGA, version 5.03, software (20). Representative strains were selected based on these preliminary analyses and are displayed in Fig. 2 and 3. The classification nomenclature of Diel et al. (4) was used, and the nucleotide numbering of the F gene sequence was done according to the method of Kho et al. (15).
Fig 2.
Phylogenetic analysis of partial F gene sequences based on nucleotides 332 to 571. Sequences generated in this study are indicated by the circles (strains presented in Fig. 2 and Fig. 3) and squares (strains presented in Fig. 2 only). Previously published sequences are indicated with their accession numbers. Only bootstrap values of ≥50% are shown. The scale corresponds to number of base substitutions per site.
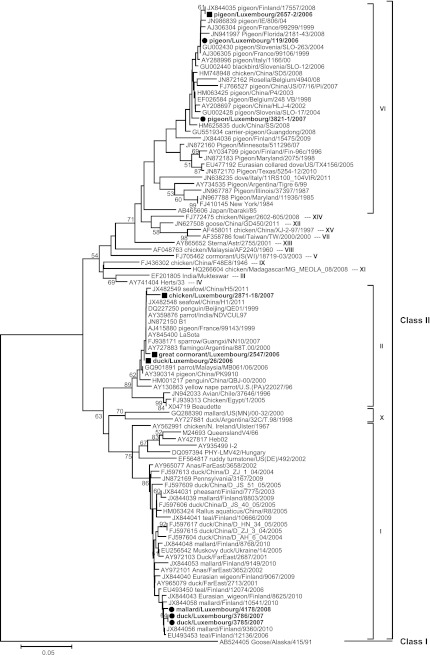
Fig 3.
Phylogenetic analysis of complete F gene sequences (1,662 nt). Symbols are as in Fig. 2. Only bootstrap values of ≥50% are shown. The scale corresponds to number of base substitutions per site.
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank under accession numbers HE972209 to HE972217. The following strain nomenclature was used: host/country/strain number/year.
RESULTS
None of the 345 samples tested for class I strains was positive for these strains. A total of 9 samples out of 1,131 were positive in the PCR detecting class II NDV strains (Table 2), corresponding to an overall prevalence of 0.8% during the period from 2006 to 2010 (4/619 in 2006, 4/349 in 2007, 1/84 in 2008, 0/67 in 2009, and 0/12 in 2010). However, prevalences may be somewhat underestimated, since virus isolation and some reverse transcription-PCR (RT-PCR) protocols may be more sensitive. Statistical analyses revealed that the type of samples, the time between collection and storage (categorized as less or more than 48 h), or the type of material used in the PCR (cDNA stored at −20°C or freshly prepared from original samples stored at −80°C) had no significant effect on the number of positive samples per group (P values = 0.816, 0.847, or 0.583, respectively). The apparent absence of NDV in 2009 and 2010 as well as in the northern part of the country (Fig. 1) was most probably due to a suboptimal surveillance effort rather than a disappearance of NDV in Luxembourg or the existence of regional differences.
Phylogenetic analyses of partial (9 strains) (Fig. 2) and complete (5 strains) (Fig. 3) F gene sequences revealed equal distributions of the samples in three class II genotypes.
Genotype I.
Three similar strains (Kimura distances from 0 to 0.5% [1,662 nt]) from ducks, including one mallard (Anas platyrhynchos), clustered in genotype I. They clustered together with strains from waterfowls from Finland, the Far East, and China (Fig. 2 and 3) and were most closely related to each other based on complete F gene sequences (Fig. 3). They all had a Kimura distance of 0.4% (240 nt) to mallard/Finland/9360/2010. The deduced amino acid sequence of the F protein cleavage site 112GKQGR*L117 (where the asterisk indicates the site of cleavage of the precursor protein, F0, into its subunits, F1 and F2) was typical of avirulent genotype I strains.
Genotype II.
Three samples from a chicken, a great cormorant (Phalacrocorax carbo), and a duck clustered in genotype II, together with the commonly used vaccines LaSota and B1. Kimura distances (240 nt) to the LaSota and B1 vaccines ranged from 0 (duck/Luxembourg/26/06) to 1.3% (chicken/Luxembourg/2871-18/07). Both duck/Luxembourg/26/06 and Great Cormorant/Luxembourg/2547/2006 exhibited a cleavage site typical of avirulent genotype II strains, 112GRQGR*L117, while the chicken/Luxembourg/2871-18/07 strain encoded 112GGQGR*L117 due to a nonsynonymous A-to-G substitution at nucleotide position 380.
Genotype VI.
The last three sequences from pigeons (Columba livia var. domestica) clustered in genotype VI, together with recent isolates mainly found in Columbiformes worldwide. Kimura distances ranged from 0.4% between pigeon/Luxembourg/2657-2/06 and pigeon/Luxembourg/119/06 (240 nt) to 2.6% between pigeon/Luxembourg/119/06 and pigeon/Luxembourg/3821-1/07 (1,662 nt). All genotype VI strains encoded virulent fusion cleavage site motif 112RRQKR*F117, as defined by the World Organization for Animal Health standard (1), and were similar to the cleavage site sequences of other previously described PPMV-1 strains.
DISCUSSION
There is increasing evidence that wild waterbirds are natural carriers of avirulent class I and class II genotype I and X strains (21–25). It was thus not surprising to find five out of nine positive samples from waterbirds, including three avirulent genotype I strains in ducks, in our study. Based on the full F gene sequences (Fig. 3), these genotype I strains formed a monophyletic cluster, which may suggest that they evolved from a recent common ancestor and resulted from a single introduction event in Luxembourg. In this scenario, the detection of similar strains in 2007 and 2008 would indicate that avirulent viruses could be maintained in the local bird population throughout the year. However, their relationship with NDV isolates recently identified in migratory species in Finland (23, 26) could not be further clarified on the basis of the short Finnish sequences. Wild Anatidae, such as Eurasian teals, mallards, and northern shovelers, have different migratory routes, which mainly depend on weather conditions and the availability of food resources. These three species, among others, are also commonly observed at migratory stopovers in Luxembourg. Similarly, the three ducks sampled in August 2007 and in October 2008 in Luxembourg could have arrived in Luxembourg shortly before being sampled. It is therefore possible that genotype I strains have been introduced by migratory species on a single occasion or several occasions. Complete F gene sequences from other European countries, as well as more detailed information on the bird species sampled, would be required to further address this possibility.
The strains of genotype VI found in three Luxembourg pigeons probably originate from separate introduction events, as they do not share a direct common ancestor and are interspersed with strains found in other countries (Fig. 2 and 3). While PPMV-1 strains initially circulated mainly in racing and show pigeons, they are now considered enzootic in feral pigeons and doves in countries such as Germany and Italy (13, 27). Cases in pigeons have been detected almost every year between 2000 and 2009 in the neighboring countries (9), and dissemination to Luxembourg is not unexpected.
While genotype I and X (class II) and class I strains are often detected in waterfowl, vaccine-like strains of genotype II are mainly found in poultry and are usually associated with the use of live vaccines (6, 28, 29). In this respect, our finding of a vaccine-like strain in a great cormorant and a duck in Luxembourg is somewhat unusual. The detection of vaccine strains in nonvaccinated flocks suggested that lentogenic vaccine strains can spread at least within poultry (30). In addition, wild-type virus transmission between wild and domestic birds was already suspected to be at the origin of the similarity of strains found in wild birds and domestic birds in live-bird markets (22, 31) or in flocks with possible contacts with wild birds (7), and of the spill-back of virulent strains into wild birds (29, 32). Therefore, it seems reasonable to expect that vaccine strains may also be exchanged between poultry and wild birds. Similar cases of strains close to LaSota or B1 vaccines in wild birds have been reported elsewhere, including Asia (China, India, and Malaysia), Argentina, and France (Fig. 2). Although NDV vaccination is not allowed in Luxembourg, vaccinated animals are sometimes imported (S. Losch, personal communication). Also, bridge species, such as sparrows, that live in close proximity with domestic birds (29), in particular in regions where backyard chickens are commonly reared, or food or water contamination (11, 33) may have contributed to the transmission of vaccine-like strains to waterbirds in Luxembourg. On the other hand, wild birds may have been infected in other European countries which sometimes allow vaccination. Since great cormorants tend to migrate late during the season, it is difficult to known whether the bird infected with a genotype II strain sampled in April spent the winter locally or was sampled during its northward migration. Unfortunately, the species of the other bird infected with a genotype II strain was not known.
The only strain found in a domestic bird belonged to genotype II, but it did not seem to be directly related to a vaccine strain because of three mutations in a short region of the F gene leading to two amino acid substitutions, one being in the cleavage site. To our knowledge, this particular cleavage site sequence has not been reported before, but this strain is probably not virulent for chickens, as it contains only one basic amino acid between residues 112 and 116 and a leucine at residue 117. Unfortunately, no further information about potential clinical signs in the flock was available.
No virulent NDV strains were found in wild or domestic birds in Luxembourg, except for the three PPMV-1 strains. Although these are normally found in pigeons, PPMV-1 transmission to poultry was reported on a few occasions in Europe during the past decade (9). Even if most cases occurred in small backyard flocks with low biosecurity, the circulation of PPMV-1 in pigeons represents a potential threat for the poultry industry. The presence of avirulent strains in wild birds may also be a risk for poultry. Although excessively rare in the field, virulent strains may develop from low-virulence strains after mutation, as was postulated for outbreaks that occurred in Ireland (34) and Australia (35). This was also demonstrated by serial experimental passages in chickens (36), which may have given minor populations of virulent NDV in field isolates a selective advantage (37). All these scenarios highlight the importance of virological surveillance and preventive measures to reduce the intermingling of wild and domestic birds.
In conclusion, we found avirulent genotype I strains in waterfowls in Luxembourg similar to those circulating in wild migratory birds in Finland, suggesting that these viruses represent typical avirulent strains found in European wild birds and that migratory birds may contribute to their spread. Detection of vaccine-like strains in wild waterbirds suggests the spread of vaccine strains, despite the nonvaccination policy in Luxembourg. Although the three PPMV-1 strains from pigeons were the only virulent strains found in Luxembourg, the presence of NDV in wild and free-ranging domestic birds justifies the need for continuous surveillance in wild and domestic birds.
ACKNOWLEDGMENTS
We thank A. Reye and F. Leenen for technical assistance using ArcGIS and SigmaPlot software, as well as S. Farinelle for sharing her ornithological knowledge. We gratefully acknowledge the Administration des Services Vétérinaries and the Lëtzebuerger Natur-a Vulleschutzliga for their expertise in sample collection. We thank Claudio Afonso, Southeast Poultry Research Laboratory, USDA, for providing positive controls.
We acknowledge the Ministry of Health, the Ministry of Research, and the Centre de Recherche Public-Santé for their generous financial and moral support. Chantal J. Snoeck was supported by an AFR fellowship (TR_PHD BFR08-095) from the Fonds National de la Recherche, Luxembourg.
Footnotes
Published ahead of print 16 November 2012
REFERENCES
- 1. OIE 2012. Chapter 2.3.14. Newcastle disease. In OIE (ed), Manual of diagnostic tests and vaccines for terrestrial animals, 7th ed OIE Organisation Mondiale de la Santé Animale, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.14_NEWCASTLE_DIS.pdf [Google Scholar]
- 2. Collins MS, Strong I, Alexander DJ. 1994. Evaluation of the molecular basis of pathogenicity of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses.” Arch. Virol. 134:403–411 [DOI] [PubMed] [Google Scholar]
- 3. Takakuwa H, Ito T, Takada A, Okazaki K, Kida H. 1998. Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations. Jpn. J. Vet. Res. 45:207–215 [PubMed] [Google Scholar]
- 4. Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL. 2012. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 12:1770–1779 [DOI] [PubMed] [Google Scholar]
- 5. Aldous EW, Mynn JK, Banks J, Alexander DJ. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239–256 [DOI] [PubMed] [Google Scholar]
- 6. Snoeck CJ, Ducatez MF, Owoade AA, Faleke OO, Alkali BR, Tahita MC, Tarnagda Z, Ouedraogo JB, Maikano I, Mbah PO, Kremer JR, Muller CP. 2009. Newcastle disease virus in West Africa: new virulent strains identified in non-commercial farms. Arch. Virol. 154:47–54 [DOI] [PubMed] [Google Scholar]
- 7. Jørgensen PH, Handberg KJ, Ahrens P, Therkildsen OR, Manvell RJ, Alexander DJ. 2004. Strains of avian paramyxovirus type 1 of low pathogenicity for chickens isolated from poultry and wild birds in Denmark. Vet. Rec. 154:497–500 [DOI] [PubMed] [Google Scholar]
- 8. Alexander DJ, Banks J, Collins MS, Manvell RJ, Frost KM, Speidel EC, Aldous EW. 1999. Antigenic and genetic characterisation of Newcastle disease viruses isolated from outbreaks in domestic fowl and turkeys in Great Britain during 1997. Vet. Rec. 145:417–421 [DOI] [PubMed] [Google Scholar]
- 9. Alexander DJ. 2011. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 40:547–558 [DOI] [PubMed] [Google Scholar]
- 10. Kommers GD, King DJ, Seal BS, Brown CC. 2001. Virulence of pigeon-origin Newcastle disease virus isolates for domestic chickens. Avian Dis. 45:906–921 [PubMed] [Google Scholar]
- 11. Alexander DJ, Parsons G, Marshall R. 1984. Infection of fowls with Newcastle disease virus by food contaminated with pigeon faeces. Vet. Rec. 115:601–602 [DOI] [PubMed] [Google Scholar]
- 12. Irvine RM, Aldous EW, Manvell RJ, Cox WJ, Ceeraz V, Fuller CM, Wood AM, Milne JC, Wilson M, Hepple RG, Hurst A, Sharpe CE, Alexander DJ, Brown IH. 2009. Outbreak of Newcastle disease due to pigeon paramyxovirus type 1 in grey partridges (Perdix perdix) in Scotland in October 2006. Vet. Rec. 165:531–535 [DOI] [PubMed] [Google Scholar]
- 13. Werner O, Romer-Oberdorfer A, Kollner B, Manvell RJ, Alexander DJ. 1999. Characterization of avian paramyxovirus type 1 strains isolated in Germany during 1992 to 1996. Avian Pathol. 28:79–88 [DOI] [PubMed] [Google Scholar]
- 14. Kim LM, Suarez DL, Afonso CL. 2008. Detection of a broad range of class I and II Newcastle disease viruses using a multiplex real-time reverse transcription polymerase chain reaction assay. J. Vet. Diagn. Invest. 20:414–425 [DOI] [PubMed] [Google Scholar]
- 15. Kho CL, Mohd-Azmi ML, Arshad SS, Yusoff K. 2000. Performance of an RT-nested PCR ELISA for detection of Newcastle disease virus. J. Virol. Methods 86:71–83 [DOI] [PubMed] [Google Scholar]
- 16. Abolnik C, Horner RF, Bisschop SP, Parker ME, Romito M, Viljoen GJ. 2004. A phylogenetic study of South African Newcastle disease virus strains isolated between 1990 and 2002 suggests epidemiological origins in the Far East. Arch. Virol. 149:603–619 [DOI] [PubMed] [Google Scholar]
- 17. Wang Z, Liu H, Xu J, Bao J, Zheng D, Sun C, Wei R, Song C, Chen J. 2006. Genotyping of Newcastle disease viruses isolated from 2002 to 2004 in China. Ann. N. Y. Acad. Sci. 1081:228–239 [DOI] [PubMed] [Google Scholar]
- 18. Cattoli G, Fusaro A, Monne I, Molia S, Le Menach A, Maregeya B, Nchare A, Bangana I, Maina AG, Koffi JN, Thiam H, Bezeid OE, Salviato A, Nisi R, Terregino C, Capua I. 2010. Emergence of a new genetic lineage of Newcastle disease virus in West and Central Africa—implications for diagnosis and control. Vet. Microbiol. 142:168–176 [DOI] [PubMed] [Google Scholar]
- 19. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jindal N, Chander Y, Chockalingam AK, de Abin M, Redig PT, Goyal SM. 2009. Phylogenetic analysis of Newcastle disease viruses isolated from waterfowl in the upper midwest region of the United States. Virol. J. 6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim LM, King DJ, Curry PE, Suarez DL, Swayne DE, Stallknecht DE, Slemons RD, Pedersen JC, Senne DA, Winker K, Afonso CL. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81:12641–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindh E, Huovilainen A, Ratti O, Ek-Kommonen C, Sironen T, Huhtamo E, Poysa H, Vaheri A, Vapalahti O. 2008. Orthomyxo-, paramyxo- and flavivirus infections in wild waterfowl in Finland. Virol. J. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruenphet S, Jahangir A, Shoham D, Morikawa K, Miyoshi Y, Hanawa E, Okamura M, Nakamura M, Takehara K. 2011. Surveillance and characterization of Newcastle disease viruses isolated from northern pintail (Anas acuta) in Japan during 2006–09. Avian Dis. 55:230–235 [DOI] [PubMed] [Google Scholar]
- 25. Zanetti F, Berinstein A, Pereda A, Taboga O, Carrillo E. 2005. Molecular characterization and phylogenetic analysis of Newcastle disease virus isolates from healthy wild birds. Avian Dis. 49:546–550 [DOI] [PubMed] [Google Scholar]
- 26. Lindh E, Ek-Kommonen C, Väänänen VM, Alasaari J, Vaheri A, Vapalahti O, Huovilainen A. 2012. Molecular epidemiology of outbreak-associated and wild-waterfowl-derived Newcastle disease virus strains in Finland, including a novel class I genotype. J. Clin. Microbiol. 50:3664–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terregino C, Cattoli G, Grossele B, Bertoli E, Tisato E, Capua I. 2003. Characterization of Newcastle disease virus isolates obtained from Eurasian collared doves (Streptopelia decaocto) in Italy. Avian Pathol. 32:63–68 [DOI] [PubMed] [Google Scholar]
- 28. Wu S, Wang W, Yao C, Wang X, Hu S, Cao J, Wu Y, Liu W, Liu X. 2011. Genetic diversity of Newcastle disease viruses isolated from domestic poultry species in Eastern China during 2005–2008. Arch. Virol. 156:253–261 [DOI] [PubMed] [Google Scholar]
- 29. Zhu W, Dong J, Xie Z, Liu Q, Khan MI. 2010. Phylogenetic and pathogenic analysis of Newcastle disease virus isolated from house sparrow (Passer domesticus) living around poultry farm in southern China. Virus Genes 40:231–235 [DOI] [PubMed] [Google Scholar]
- 30. Wehmann O, Herczeg J, Tanyi J, Nagy E, Lomniczi B. 1999. Lentogenic field isolates of Newcastle disease virus isolated in Canada and Hungary are identical with the vaccine type used in the region. Avian Pathol. 28:6–12 [DOI] [PubMed] [Google Scholar]
- 31. Kim BY, Lee DH, Kim MS, Jang JH, Lee YN, Park JK, Yuk SS, Lee JB, Park SY, Choi IS, Song CS. 2012. Exchange of Newcastle disease viruses in Korea: the relatedness of isolates between wild birds, live bird markets, poultry farms and neighboring countries. Infect. Genet. Evol. 12:478–482 [DOI] [PubMed] [Google Scholar]
- 32. Vidanović D, Sekler M, Asanin R, Milic N, Nisavic J, Petrovic T, Savic V. 2011. Characterization of velogenic Newcastle disease viruses isolated from dead wild birds in Serbia during 2007. J. Wildl. Dis. 47:433–441 [DOI] [PubMed] [Google Scholar]
- 33. Alexander DJ. 1995. The epidemiology and control of avian influenza and Newcastle disease. J. Comp. Pathol. 112:105–126 [DOI] [PubMed] [Google Scholar]
- 34. Alexander DJ, Campbell G, Manvell RJ, Collins MS, Parsons G, McNulty MS. 1992. Characterisation of an antigenically unusual virus responsible for two outbreaks of Newcastle disease in the Republic of Ireland in 1990. Vet. Rec. 130:65–68 [DOI] [PubMed] [Google Scholar]
- 35. Gould AR, Kattenbelt JA, Selleck P, Hansson E, Della-Porta A, Westbury HA. 2001. Virulent Newcastle disease in Australia: molecular epidemiological analysis of viruses isolated prior to and during the outbreaks of 1998–2000. Virus Res. 77:51–60 [DOI] [PubMed] [Google Scholar]
- 36. Shengqing Y, Kishida N, Ito H, Kida H, Otsuki K, Kawaoka Y, Ito T. 2002. Generation of velogenic Newcastle disease viruses from a nonpathogenic waterfowl isolate by passaging in chickens. Virology 301:206–211 [DOI] [PubMed] [Google Scholar]
- 37. Kattenbelt JA, Stevens MP, Selleck PW, Gould AR. 2010. Analysis of Newcastle disease virus quasispecies and factors affecting the emergence of virulent virus. Arch. Virol. 155:1607–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]