Abstract
Microarray analyses revealed that the expression of genes for secondary metabolism together with that of primary metabolic genes was induced by chitin in autoclaved soil cultures of Streptomyces coelicolor A3(2). The data also indicated that DasR was involved in the regulation of gene expression for chitin catabolism, secondary metabolism, and stress responses.
TEXT
Streptomycetes are widely distributed in soil and play an important ecological role in the biodegradation of insoluble biomaterials through their broad range of extracellular hydrolytic enzymes (1). Streptomycetes are known to decompose chitin, the second-most-abundant biomass in nature (2), and possess many genes for various chitinases with different enzymatic characteristics (3). Moreover, according to environmental conditions, streptomycetes exhibit morphological differentiation accompanied by the formation of spores and the production of a wide range of secondary metabolites, such as antibiotics. In fact, streptomycetes are important producers of approximately two-thirds of all antibiotics and other valuable secondary metabolites found so far (4).
Our previous study revealed that the expression of chitinase genes of Streptomyces coelicolor A3(2) in soil cultures was significantly higher than that in liquid cultures (5). The expression of chitinase genes was positively regulated by DasR, the pleiotropic GntR regulator of chitin metabolism and antibiotic production (5, 6), the expression level of which in soil cultures was 3-fold higher than that in liquid cultures (5). DasR binds to the conserved sequences, DasR-responsive elements (dre) in the promoter regions of the target genes (7). A genome scan for dre sites revealed that >200 candidate genes are potential targets for DasR. Approximately 40% of these target genes are regulated in chitin metabolism (6). DasR also plays a pivotal role in regulating antibiotic production and differentiation (9). It represses the expression of actII-ORF4 and redZ, which encode transcriptional activators for the production of the blue-pigmented antibiotic actinorhodin (Act) and red-pigmented antibiotic undecylprodigiosin (Red), respectively (6). However, its roles in regulating the transcription of genes involved in these metabolisms in soil have yet to be investigated. The involvement of DasR in the regulation of the soil-specific expression of chitinase genes suggested that DasR may also regulate other functional target genes in soil.
To provide new insights into the cellular response to chitin and the role of DasR in the global gene expression of S. coelicolor A3(2) in soil-like circumstances, genome-wide transcriptional analysis with microarray technology was performed using RNA extracted from the wild type (M145) and dasR-disrupted mutant (YU1) (5) of S. coelicolor A3(2) grown in soil cultures. To clarify the responses of the target strains in soil cultures, it is necessary to remove the orthologue transcripts derived from indigenous streptomycetes. So, we used autoclaved soil for the cultures. The preparation of soil cultures with or without 0.2% (wt/wt) colloidal chitin and RNA extraction from soil was performed as described previously (5). Cultivation was performed at 30°C for 48 h. To obtain enough RNA from soil cultures for microarray analysis, RNA samples were prepared from a total of 16 g of chitin-amended soil cultures or 20 g of control soil cultures. The RNA samples were purified and concentrated using the protocol previously described (5). The RNA samples (10 μg each) extracted from soil cultures were subjected to whole-genome microarrays for S. coelicolor A3(2), which was designed and manufactured by Roche (Roche NimbleGen, Madison, WI). The further steps, i.e., cDNA synthesis, labeling, hybridization, microarray handling, generation of gene expression values, and data evaluation, were performed as described previously (10). The significance of differential expression in two samples was determined using Student's t test, which was corrected for multiple-hypothesis testing using the Benjamini and Hochberg false-discovery rate (FDR) correlation (11). The genes exhibiting P values of <0.05 and fold changes of >2 were judged to be significant. Microarray experiments were performed in triplicate using three independent RNA samples extracted from soil, with two technical replications (six data sets) for each culture.
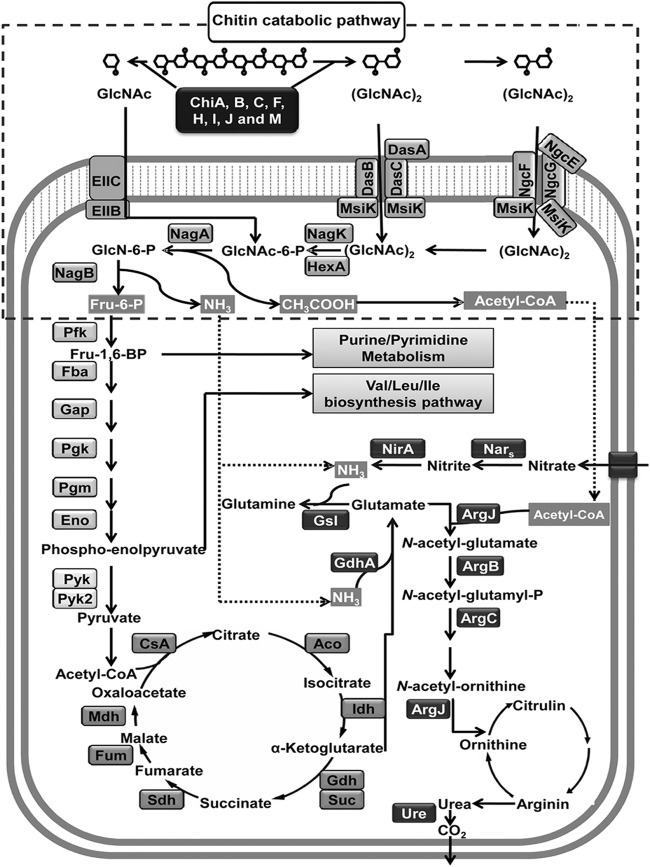
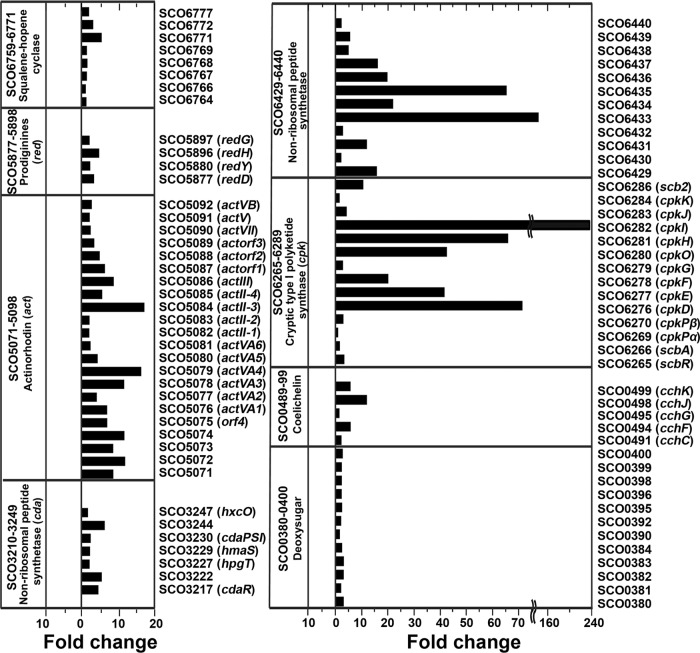
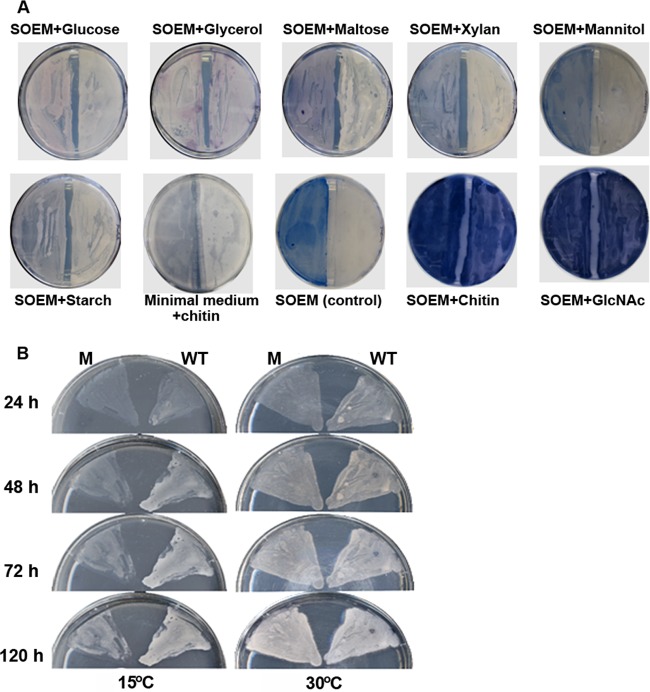
The microarray data indicated that the expression of 675 genes (8.6%) was significantly changed. Of these genes, 437 were upregulated and 238 were downregulated upon the addition of chitin. To characterize the cellular response of S. coelicolor A3(2) to chitin, the differentially expressed genes were categorized according to their cellular roles as annotated by gene ontology and pathway analyses according to the protocols described previously (10) using the DAVID database server (http://david.abcc.ncifcrf.gov/) (see data set S1 in the supplemental material). The addition of chitin to a soil culture of S. coelicolor A3(2) resulted in significant changes in the expression of genes involved in primary metabolism, including major genes for the chitin catabolic, central carbohydrate metabolism, nitrogen and sulfur metabolism, oxidative phosphorylation, and amino acid biosynthesis pathways (see Fig. S1 and data set S1). In the chitin catabolic pathway, eight chitinase genes were dramatically induced in response to chitin. In particular, the induction of chiC and chiH was higher than that of other chitinase genes (see Fig. S1 and data set S1). The results were largely consistent with those obtained with quantitative reverse transcription-PCR (qRT-PCR) in the previous work (5). These facts indicate that the microarray analyses were successful in this study. The taken-up chitin-derived products, namely, N-acetylglucosamine (GlcNAc), taken up by the sugar phosphotransferase system for GlcNAc, and N,N′-diacetylchitobioside (GlcNAc)2, taken up by two specific ABC transporters, dasABC and ngcEFG, are further metabolized through degradation, phosphorylation, deacetylation, and deamination involving the gene products of hexA, nagK, nagA, and nagB, respectively (Fig. 1). The final products of the chitin catabolic pathway comprising the above-named enzymes are fructose 6-phosphate, ammonia, and acetate. Fructose-6-phosphate is a direct or indirect substrate for central metabolic pathways (glycolysis and the tricarboxylic acid [TCA] cycle), purine/pyrimidine metabolism, and the valine/leucine/isoleucine biosynthesis pathway (Fig. 1) (12). Ammonium produced from glucosamine-6-phosphate by NagB could be supplied to the glutamate/glutamine biosynthesis pathway, and acetate produced from glucosamine-6-phosphate by NagA participates in different metabolic pathways, such as the urea cycle, fatty acid biosynthesis, or secondary metabolic pathways (Fig. 1) (13). Upon microarray analysis, eight of 23 gene clusters for secondary metabolites (14) (a total of 100 genes), including the act and red gene clusters, were found to be induced in S. coelicolor A3(2) grown in chitin-amended soil cultures (Fig. 2; also see data set S1). In particular, prominent upregulation of the cryptic type I polyketide synthase gene cluster (cpk) (SCO6265-86) and a putative nonribosomal peptide synthetase (NRPS) gene cluster (SCO6429-40) was observed (Fig. 2). To evaluate the observed differential expression of genes in several secondary metabolic pathways, a two-step qRT-PCR was performed using the specific primers (listed in Table S1). The primers were designed based on probe information used in microarray analysis. One microgram of each RNA sample extracted from the soil cultures was reverse transcribed, and the copy number of the synthesized cDNA in each sample was determined by qRT-PCR as described previously (5). It was confirmed that expression of the examined genes in several secondary metabolic pathways was upregulated in soil cultures in the presence of chitin, without exception (see Fig. S2). The effect of chitin on pigmented antibiotic production by S. coelicolor A3(2) was also confirmed on soil extract medium (SOEM) agar plates. For preparation of SOEM, 200 g of autoclaved soil was suspended in 1,000 ml of double-distilled water. After the preparation was shaken at 200 rpm on an orbital shaker overnight at room temperature, the pH of the filtered supernatant of the soil suspension was adjusted to 7.2 and then used as SOEM. As a result, positive effects of chitin and its monomer (GlcNAc) on antibiotic production by S. coelicolor A3(2) were observed on SOEM agar, whereas no effects on antibiotic overproduction were observed with other carbon sources (Fig. 3A). It has been reported that chitin also stimulates the production of geldanamycin by Streptomyces melanosporofaciens and andrimid by Vibrio coralliilyticus (15, 16). Overexpression of the act, red, and cpk gene clusters has been demonstrated upon the addition of GlcNAc to a minimal medium, but this has not been demonstrated with chitin (6, 17). Chitin induced antibiotic production only in soil cultures or on agar plates containing soil extract. The addition of chitin to the minimal medium had no effect on antibiotic production by wild-type S. coelicolor A3(2) (Fig. 3A) (6). These findings indicate that some water-soluble soil factor(s) may enhance antibiotic production by S. coelicolor A3(2) in the presence of chitin. It would be very advantageous for streptomycetes to produce antibiotics to compete with other microorganisms for nutrients. The enhancement of antibiotic production was repressed by the addition of other carbon sources (Fig. 3A). These data suggest that the enhancement is under the control of carbon catabolite repression (CCR).
Fig 1.
Representation of the S. coelicolor A3(2) chitin metabolism, which was significantly influenced at the transcriptional level during growth in soil. Gene products for which expression was upregulated are shown in the metabolic pathways. Acetyl-CoA, acetyl coenzyme A; EIIC, enzyme IIC; EIIB, enzyme IIB; Pfk, 6-phosphofructokinase; Fba, fructose-bisphosphate aldolase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, phosphopyruvate hydratase; Pyk, pyruvate kinase; CsA, citrate synthase A; Aco, aconitate hydratase; Idh, isocitrate dehydrogenase; Gdh, α-ketoglutarate dehydrogenase; Suc, succinyl-CoA synthetase; Sdh, succinate dehydrogenase; Fum, fumarate hydratase; Mdh, malate dehydrogenase; GsI, glutamin synthase I; ArgJ, N-acetylglutamate synthase J; ArgB, N-acetylglutamate kinase, ArgC, argininosuccinate synthase; Nar, nitrate reductase; Nir, nitrite reductase; Ure, urease.
Fig 2.
Transcriptional responses of the genes involved in the secondary metabolite biosynthetic gene clusters in S. coelicolor A3(2) grown in soil. Upregulated genes for secondary metabolites in S. coelicolor A3(2) grown in chitin-amended soil are shown. The analysis was performed using DAVID bioinformatics resources, which allow online server analysis. The complete sets of differentially expressed genes are listed in data set S1 in the supplemental material.
Fig 3.
(A) Effects of various carbon sources on antibiotic production by S. coelicolor A3(2). The effects of various carbon sources on the production of a pigmented antibiotic were investigated in the dasR mutant (left) and wild-type strains of S. coelicolor A3(2) (right) using SOEM. (B) Growth of the wild-type strain (WT) (right) and dasR mutant (M) (left) on SOEM agar plates with chitin at 30°C and 15°C. Poor growth of the dasR mutant was observed under cold conditions.
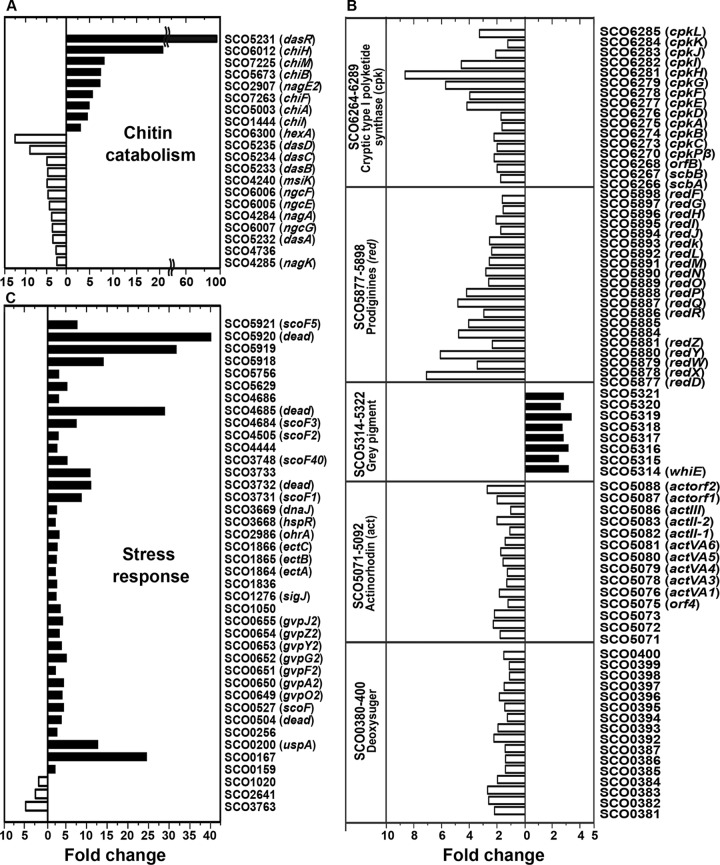
To elucidate the roles of DasR in chitin metabolism and antibiotic production in soil cultures, genome-wide transcription profiles of the wild-type and dasR mutant of S. coelicolor A3(2) were compared using RNA extracted from soil cultures with chitin. The microarray data indicate that there are more DasR target genes than previously reported (6). In total, 679 genes were drastically affected on disruption of dasR (Fig. 4 and see data set S2 in the supplemental material). As reported previously (5), the induction of all chitinase genes decreased in mutant YU1, whereas the transcriptional levels of transporter genes and the genes involved in subsequent intracellular metabolism increased in the mutant (Fig. 4) (S. Rigali et al., European patent application EP1818398). Among secondary metabolic genes, the expression of genes in the Act, Red, and cryptic type I polyketide (Cpk) biosynthesis pathways and the deoxysugar/glycosyltransferase cluster (SCO0381-400) was upregulated in the dasR mutant, whereas transcription of the gray pigment gene cluster for spore formation was downregulated in the mutant (Fig. 4). The results of the microarray analysis were confirmed by qRT-PCR analyses of expression of these gene clusters (see Table S1). The results were largely consistent with those obtained with qRT-PCR (see Fig. S2). It is unclear whether DasR regulates these differentially expressed genes directly or indirectly. DasR might act as a repressor and an activator, like OhrR, which plays dual regulatory roles in the organic hydroperoxide resistance of S. coelicolor A3(2) (18). The overproduction of antibiotics by the mutant was also confirmed on SOEM agar plates; higher Act production was observed in the dasR mutant in the presence and absence of chitin than in the wild-type strain (Fig. 3A). These results are in agreement with the finding that DasR controls the production of Act, Red, and Cpk antibiotics as a repressor through binding to the dre sites of pathway-specific activators of actII-ORF4, redZ, and cpkO, respectively (6, 17). The overproduction of pigmented antibiotics by the dasR mutant on SOEM plates decreased upon the addition of other carbon sources, suggesting that CCR is independent of DasR regulation. It seems that CCR has priority over other means of regulation involved in the production of secondary metabolites.
Fig 4.
Differentially expressed genes in the dasR-disrupted mutant (□) and wild-type strain (■) of S. coelicolor A3(2) grown in soil in the presence of chitin. The differentially expressed genes involved in similar biological processes were categorized according to their functional classification. Of the gene categories, the chitin catabolism genes (A), stress response genes (B), and several gene sets for secondary metabolic pathways (C) are presented. The complete sets of differentially expressed genes are listed in data set S2 in the supplemental material.
The expression level of dasR was upregulated in the wild-type strain in chitin-amended soil (Fig. S1 and see data set S1 in the supplemental material). Under such conditions, act, red, and cpk expression should be repressed by DasR induced by chitin. The same situation was expected for the downstream genes in the chitin catabolic pathway as well as the transporters of chitin degradation products. However, the expression of all these genes was upregulated by chitin in soil cultures to levels similar to that of dasR. This suggests that there is another mechanism(s) that induces the expression of these genes, the effect of which is stronger than the inhibitory effect of DasR on expression. It has been demonstrated that the intermediate product of chitin and GlcNAc metabolism, glucosamine-6-phoshphote (GlcN-6-P), inhibits the binding of DasR to dre (9). In the presence of chitin, an increase in the intracellular level of GlcN-6-P during the degradation of chitin may weaken the repressive activity of DasR toward its targets.
On the other hand, the expression of the putative NRPS gene cluster (SCO6429-40) was not affected by the dasR mutation in soil cultures (see data set S2 in the supplemental material), whereas prominent upregulation of this gene cluster was observed when chitin was added to the soil cultures (Fig. 2). Therefore, another positive regulator is involved in the induction of this gene cluster. Comparison of the expression levels of this gene cluster in soil and liquid cultures indicated that it is induced in the presence of chitin in the soil cultures. No induction was observed in inorganic salts-yeast liquid medium with or without chitin (see Fig. S3). The results suggest that some other factors in soil may trigger the expression of this gene cluster in the presence of chitin. Such an inducer has been identified for the activation of chitinase genes in streptomycetes, it being called “allosamidin.” It strongly promotes the transcription and production of chitinases by various species of Streptomyces in the presence of chitin (19).
The disruption of dasR also dramatically changed the expression levels of a wide range of stress response genes (a total of 46 genes). In particular, the levels of expression of the genes for six cold shock proteins and four DEAD box RNA helicases were significantly reduced in the mutant (Fig. 4). The organization of these genes in the genome of S. coelicolor A3(2) (see Fig. S4 in the supplemental material) and their expression profiles in microarray data suggested that genes for both DEAD box RNA helicases and cold shock proteins formed operons, which were cotranscribed under positive regulation by DasR. The dre in the promoters of two cold shock proteins (ScoF and ScoF2) has been identified, which suggests that DasR is involved in the regulation of the expression of these genes (6). This assumption was confirmed by the observation of the growth of the wild-type and dasR mutant strains on agar plates at a low temperature. The growth of the dasR mutant was delayed on SOEM plates containing 0.2% chitin at 15°C (Fig. 3B). In other microorganisms, disruption of these DEAD box RNA helicases also leads to the formation of a cold-sensitive phenotype (20). Moreover, the transcriptional analysis of one of the gene sets, scoF5 (SCO5921) and DEAD box RNA helicase (SCO5920), by qRT-PCR indicated that their expression was strongly induced after a temperature downshift from 30°C to 10°C in the wild type, while the expression of these genes was partially induced in the mutant (see Fig. S5). The expression of cold shock proteins and DEAD box RNA helicases was also enhanced upon the addition of chitin in S. coelicolor A3(2) grown in soil (see Fig. S1). The qRT-PCR data revealed that DasR plays an important positive role in the expression of cold stress proteins and DEAD box RNA helicases in S. coelicolor A3(2) (see Fig. S5). The expression of dasR is induced by chitin (see Fig. S1 and data set S1), which may affect the expression of these genes. On the other hand, most eukaryotic and bacterial DEAD box proteins and cold shock proteins have now been shown to participate in every aspect of RNA metabolism under non-cold conditions, including RNA unwinding, ribosome biogenesis, translation, RNA decay, and RNA chaperoning (21). The similarity of the amino acid sequences of DEAD box RNA helicases in S. coelicolor A3(2) to that of initiation translation factor eIF4A in Saccharomyces cerevisiae (21) support the idea that DEAD box RNA helicases may be involved in some process in protein synthesis, which is more active in chitin-amended soil cultures than in those without chitin.
The expression of other genes with previously characterized roles in stress responses, including hspR (SCO3668), the dnaJ chaperone (SCO3669) (22), gene set SCO1864-66 (ectoine biosynthesis) (23), gas vesicles (SCO0649-655) (24), and sigma factors sigJ, sigK, and sigM (22), was significantly reduced in the dasR mutant strain of S. coelicolor A3(2) grown in soil, indicating that DasR is also involved in the expression of these genes (Fig. 4).
The addition of chitin to soil cultures of S. coelicolor A3(2) significantly induced not only the expression of chitin catabolic genes but also that of the gene sets for primary metabolism, some of those for secondary metabolism, and the genes related to RNA metabolism (20). The production of antibiotics will be advantageous for cells to compete for food with other microorganisms in the soil environment, and RNA-metabolic proteins will be required for high levels of protein synthesis for the upregulated metabolisms. As chitin is one of the major nutrients in the soil environment, the above-described responses of S. coelicolor A3(2) to chitin are useful for the utilization of chitin for effective competition with other microbes in their original niche, soil. DasR should be recognized as one of the most important regulators for survival in soil, because it is involved in many steps in these responses.
Microarray data accession number.
The microarray details and transcriptomics data are accessible through GEO series accession number GSE39164 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39164).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Yoshiteru Hashimoto for helpful discussions.
This work was supported by KAKENHI (grant no. 21380050) and a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02217-12.
REFERENCES
- 1. Chater KF, Bior S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34:171–198 [DOI] [PubMed] [Google Scholar]
- 2. Metcalfe AC, Krsek M, Gooday GW, Prosser JI, Wellington EMH. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saito A, Fujii T, Miyashita K. 2003. Distribution and evolution of chitinase genes in Streptomyces species: involvement of gene-duplication and domain-deletion. Antonie Van Leeuwenhoek 84:7–16 [DOI] [PubMed] [Google Scholar]
- 4. Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York, NY [Google Scholar]
- 5. Nazari B, Saito A, Kobayashi M, Miyashita K, Wang Y, Fujii T. 2011. High expression levels of chitinase genes in Streptomyces coelicolor A3(2) grown in soil. FEMS Microbiol. Ecol. 77:623–635 [DOI] [PubMed] [Google Scholar]
- 6. Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colson S, Stephan J, Hertrich T, Saito A, van Wezel GP, Titgemeyer F, Rigali S. 2007. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J. Mol. Microbiol. Biotechnol. 12:60–66 [DOI] [PubMed] [Google Scholar]
- 8. Reference deleted.
- 9. Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Müller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237–1251 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Morimoto S, Ogawa N, Fujii T. 2011. A survey of the cellular responses in Pseudomonas putida KT2440 growing in sterilized soil by microarray analysis. FEMS Microbiol. Ecol. 78:220–232 [DOI] [PubMed] [Google Scholar]
- 11. Reimers M. 2010. Making informed choices about microarray data analysis. PLoS Comput. Biol. 6:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swiatek MA, Tenconi E, Rigali S, van Wezel GP. 2012. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in control of development and antibiotic production. J. Bacteriol. 194:1136–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Challis GL, Hopwood DA. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U. S. A. 25:14555–14561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clermont N, Legault G, Lerat S, Beaulieu C. 2010. Effect of biopolymers on geldanamycin production and biocontrol ability of Streptomyces melanosporofaciens strain EF-76. Can. J. Plant Pathol. 32:481–489 [Google Scholar]
- 16. Wietz M, Mansson M, Gram L. 2011. Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ. Microbiol. Rep. 3:559–564 [DOI] [PubMed] [Google Scholar]
- 17. van Wezel GP, McKenzie NL, Nodwell JR. 2009. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458:117–141 [DOI] [PubMed] [Google Scholar]
- 18. Oh SY, Shin JH, Roe JH. 2007. Dual role of OhrR as a repressor and an activator in response to organic hydro-peroxides in Streptomyces coelicolor. J. Bacteriol. 189:6284–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuki S, Nakanishi E, Furihata K, Miyamoto K, Tsujibo H, Watanabe T, Ohnishi Y, Horinouchi S, Nagasawa H, Sakuda S. 2008. Chitinase inhibitor allosamidin promotes chitinase production of Streptomyces generally. Int. J. Biol. Macromol. 43:13–19 [DOI] [PubMed] [Google Scholar]
- 20. Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J. Bacteriol. 188:240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37 [DOI] [PubMed] [Google Scholar]
- 22. Lee EJ, Karoonuthaisiri N, Kim HS, Park JH, Cha CJ, Kao CM, Roe JH. 2005. A master regulator, sigmaB, governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 57:1252–1264 [DOI] [PubMed] [Google Scholar]
- 23. Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E. 2008. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74:7286–7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keulen GV, Hopwood DA, Dijkhuizen L, Sawers RG. 2005. Gas vesicles in actinomycetes: old buoys in novel habitats? Trends Microbiol. 13:350–354 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.