Abstract
Nearly 690 raw surface water samples were collected during a 6-year period from multiple watersheds in the South Nation River basin, Ontario, Canada. Cryptosporidium oocysts in water samples were enumerated, sequenced, and genotyped by detailed phylogenetic analysis. The resulting species and genotypes were assigned to broad, known host and human infection risk classes. Wildlife/unknown, livestock, avian, and human host classes occurred in 21, 13, 3, and <1% of sampled surface waters, respectively. Cryptosporidium andersoni was the most commonly detected livestock species, while muskrat I and II genotypes were the most dominant wildlife genotypes. The presence of Giardia spp., Salmonella spp., Campylobacter spp., and Escherichia coli O157:H7 was evaluated in all water samples. The greatest significant odds ratios (odds of pathogen presence when host class is present/odds of pathogen presence when host class is absent) for Giardia spp., Campylobacter spp., and Salmonella spp. in water were associated, respectively, with livestock (odds ratio of 3.1), avian (4.3), and livestock (9.3) host classes. Classification and regression tree analyses (CART) were used to group generalized host and human infection risk classes on the basis of a broad range of environmental and land use variables while tracking cooccurrence of zoonotic pathogens in these groupings. The occurrence of livestock-associated Cryptosporidium was most strongly related to agricultural water pollution in the fall (conditions also associated with elevated odds ratios of other zoonotic pathogens occurring in water in relation to all sampling conditions), whereas wildlife/unknown sources of Cryptosporidium were geospatially associated with smaller watercourses where urban/rural development was relatively lower. Conditions that support wildlife may not necessarily increase overall human infection risks associated with Cryptosporidium since most Cryptosporidium genotypes classed as wildlife in this study (e.g., muskrat I and II genotype) do not pose significant infection risks to humans. Consequently, from a human health perspective, land use practices in agricultural watersheds that create opportunities for wildlife to flourish should not be rejected solely on the basis of their potential to increase relative proportions of wildlife fecal contamination in surface water. The present study suggests that mitigating livestock fecal pollution in surface water in this region would likely reduce human infection risks associated with Cryptosporidium and other zoonotic pathogens.
INTRODUCTION
Cryptosporidium spp. are a serious cause of diarrheal disease and a major concern for the production of safe drinking water. Sixty percent of 120 major worldwide waterborne parasitic protozoan outbreaks that occurred from 2004 to 2010 were due to Cryptosporidium spp. (1). Nonoutbreak cases have been increasing in the United States, from about 3,400 in 2004 to over 8,000 in 2007 (2). Cryptosporidium is particularly resistant to disinfection, and water treatment systems may not be able to fully eliminate this parasite from drinking water. In response, some jurisdictions have implemented extensive Cryptosporidium monitoring at water treatment facilities to help gauge and mitigate risks (3).
The Cryptosporidium genus is very diverse, with species and genotypes that vary in their host specificities and in their virulence potentials for humans (4). For instance, Cryptosporidium parvum (5) and Cryptosporidium hominis pose a significant threat to human health (6), whereas, Cryptosporidium andersoni, most commonly associated with adult cattle, is considered to pose a much lower risk (7). As a result, the assessment and/or reduction of Cryptosporidium species exposure risks in water requires species/genotype quantification and species/genotype identification, due to potential species/genotype virulence variation with respect to human infection (8–11).
Generalized host specificity of Cryptosporidium (12) has enabled the use of Cryptosporidium as a microbial source tracking (MST) agent. In watersheds, which are settings that can have multiple sources of fecal contamination (9, 13), identifying the host sources of Cryptosporidium species contamination in water could dramatically increase the capacity to identify how this parasite manifests itself in water resources as a result of the effects of different land use and environmental factors (14, 15). Moreover, since other zoonotic pathogens can be associated with the occurrence of this parasite in water (16–18), it follows that Cryptosporidium species/genotype information could serve as an MST tool for assessing the sources of other cooccurring pathogens.
The primary objective of the present study was to identify spatial and temporal relationships between land use, season, and environmental variables from 6 years of Cryptosporidium species and genotype data derived from surface water sampled from several mixed-activity watersheds in eastern Ontario, Canada. A secondary objective of the study was to explore relationships between specific Cryptosporidium species or genotypes and the cooccurrence of other zoonotic pathogens in contaminated water. Identifying when, where, and ultimately why certain Cryptosporidium species/genotypes are detected in surface waters, and the associated occurrence of other zoonotic pathogens, will help inform risk assessment and risk management practices that reduce public health risks.
MATERIALS AND METHODS
Study site description.
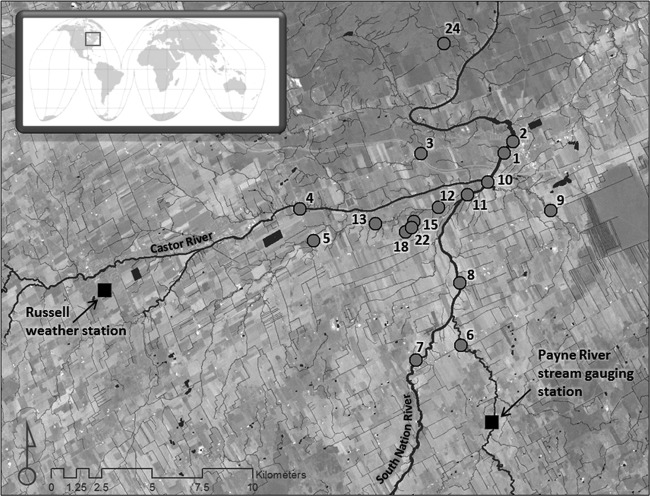
The South Nation River basin is located in eastern Ontario, Canada, and covers an area of approximately 3,900 km2 (Fig. 1). A variety of land uses prevail in this river basin that are relevant to this study, including dairy and beef cattle farming, cash and livestock crop production, livestock pasturing, and fall and spring manure applications as well as nonagricultural land uses related to urban and rural development (19–21). The water sampling sites are located on the South Nation River proper and river tributaries in an ∼200-km2 area of the South Nation River basin (Fig. 1) (21).
Fig 1.
Map of the study area and location of water sample sites. The top left inset map with square region indicates where the study area is situated globally.
Water sample collection and analysis.
Seventeen of a total of 24 long-term surface water sampling sites were visited on a biweekly basis between April and December of each year. Water sampling began in October 2004 and ceased in July 2009 (6 years) for Cryptosporidium. In any given year, 5 to 17 sites were visited and sampled over 2 to 3 seasons. One liter of water was collected in tandem with all other samples and analyzed for ammonia and ammonium, dissolved reactive phosphorus, nitrite and nitrate, total Kjeldahl nitrogen, total phosphorus, turbidity, water temperature, specific electrical conductivity, dissolved oxygen, and oxidation-reduction potential following Wilkes et al. (21). One liter of water was collected by submerging sterile polyethylene terephthalate (PET) bottles (Systems Plus, Woodstock, Ontario, Canada) to a depth of approximately 0.5 to 1 m below the water surface in a manner that did not disturb bottom sediment. Samples were placed on ice in coolers and shipped by overnight air courier to the Public Health Agency of Canada's Laboratory for Foodborne Zoonoses (Lethbridge, Alberta, Canada) for the isolation of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 according to the methods of Jokinen et al. (22).
Cryptosporidium oocyst and Giardia cyst quantification, genotyping Cryptosporidium, and Cryptosporidium species identification.
Twenty liters of water was collected in Nalgene containers (2240-0050 Nalgene Jerricans; Thermo Fisher Scientific, Rochester, NY) and refrigerated (4°C) overnight. The following day, the water was passed through Filta-Max filters (IDEXX, Westbrook, MN) and shipped by overnight delivery to the Alberta Provincial Laboratory for Public Health (Calgary, Alberta, Canada), where they were processed using U.S. Environmental Protection Agency method 1623 (23). Elution of parasites was carried out using a phosphate-buffered saline (0.01%) solution followed by immuno-magnetic separation (IMS) with a DynaBeads G-C Combo Kit (Invitrogen Canada Inc., Burlington, Ontario, Canada). A modification, including a secondary washing procedure, was included to achieve slides with less debris for easier and more accurate microscopy (24). Slides were dried and fixed with methanol and stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO), followed by fluorescently labeled monoclonal antibodies (EasyStain; Biotechnology Frontiers, North Ryde, New South Wales, Australia). Parasites identified by fluorescence microscopy were confirmed to be Cryptosporidium based on DAPI staining characteristics or on their size, shape, and morphology using differential interference contrast (DIC) microscopy.
Oocyst removal from slides, lysis, DNA extraction, and replicate nested PCR-restriction fragment length polymorphism (RFLP) and DNA sequencing were carried out as previously described (24). All raw sequence data were imported into Seqscape, version 2.6.1 (Applied Biosystems, Foster City, CA), with base-calling performed using the KB basecaller. Bidirectional sequence data were assembled, and a consensus sequence was produced and exported as FASTA-formatted files. Multiple sequence alignments and phylogenetic analysis were carried out using software packages (Clustal-W and PHYLIP) offered by the Sun Center of Excellence for Visual Genomics (COE) at the University of Calgary through a web interface accessible via the Secure Global Desktop. Phylogenetic analysis of the sequences from this study along with the species and genotype identification has been thoroughly described in Ruecker et al. (24).
The species or genotypes of the Cryptosporidium oocysts detected in each water sample were grouped into broad source classes (livestock, avian, human, wildlife/unknown [including muskrat genotypes], and just muskrat [genotypes]) and four human infection risk categories (high, medium, low, and no human infection risk) (Table 1) (24). Unknown genotypes were grouped with wildlife in the broad wildlife/unknown source class (8, 24) for data mining analysis. To assign the human infection risk categories, the available literature was reviewed. The evidence used to develop these risk classes comes primarily from published United Kingdom studies, where genotyping of human stools is routine and centralized for both sporadic and outbreak-related human cases of infection (25, 26). While the evidence is not complete, particularly in Canada, the available literature reports reflect our current state of knowledge on this infection. There is one Canadian study of human cryptosporidiosis cases and detected genotypes, and the results follow those of the United Kingdom studies, though the sample size was small (n = 11) (27). The literature suggests that C. hominis and C. parvum pose the largest risk to humans, and these were categorized as high risk in this study (28), while Cryptosporidium meleagridis and Cryptosporidium ubiquitum were categorized as medium risk (29). C. andersoni and the skunk genotype were categorized as low risk (7), and the remaining species and genotypes were considered nonpathogenic (i.e., no risk) to humans and were categorized as no risk (4, 30, 31). When a sample contained more than one source class (n = 53) or species/genotype (n = 78), the highest risk class was assigned to that sample. Classification of these genotypes into risk groupings is important to help elucidate fecal pollution drivers and prioritize intervention efforts on the basis of the sources of fecal pollution that represent the greatest public health significance.
Table 1.
Cryptosporidium species and genotypes detected in surface water samples, their known hosts and assigned generalized host, and human health risk classes
| Cryptosporidium species or genotype | Known host(s) (genus or species) | Generalized host class from known host(s) | Known human infection risk class |
|---|---|---|---|
| C. bailey | Bird (Aves) | Avian | No risk |
| C. meleagridis | Bird (Aves) | Avian | Medium risk |
| C. andersoni | Cattle (Bos primigenius) | Livestock | Low risk |
| C. parvum | Cattle, human (Bos primigenius, Homo sapiens) | Livestock | High risk |
| C. ubiquitum | Sheep, deer, rodent (Ovis aries, Cervidae, Rodentia) | Livestock | Medium risk |
| C. hominis | Human (Homo sapiens) | Human | High risk |
| Deer mouse III genotype | Deer mice (Peromyscus) | Wildlife/unknown | No risk |
| Fox genotype | Fox (Volpini) | Wildlife/unknown | No risk |
| Muskrat I genotype | Muskrat, vole (Ondatra zibethicus, Arvicolinae) | Wildlife/unknown | No risk |
| Muskrat II genotype | Muskrat, vole (Ondatra zibethicus, Arvicolinae) | Wildlife/unknown | No risk |
| Shrew genotype | Shrew (Soricidae) | Wildlife/unknown | No risk |
| Skunk genotype | Skunk (Mephitidae) | Wildlife/unknown | Low risk |
| Vole genotype | Vole (Arvicolinae) | Wildlife/unknown | No risk |
| W12 genotype | Unknown (?) | Wildlife/unknown | No risk |
| W18 genotype | Unknown (?) | Wildlife/unknown | No risk |
| W19 genotype | Unknown (?) | Wildlife/unknown | No risk |
| W25 genotype | Unknown (?) | Wildlife/unknown | No risk |
| W27 genotype | Unknown (?) | Wildlife/unknown | No risk |
| W28 genotype | Unknown (?) | Wildlife/unknown | No risk |
Environmental and land use variables.
Environmental, seasonality, water quality, and land use variables that were deemed to have a possible association with the occurrence and diversity of Cryptosporidium species/genotypes in the region (21) are given in Table S1 in the supplemental material. Over 50 distinct land use variables were produced within a geographic information system (GIS) from roadside survey data and land use classification data; the land use classification data were generated from remotely sensed satellite imagery.
Statistical analyses.
As per Wilkes et al. (21), land use and environmental variable (see Table S1) associations with target water quality endpoints were examined quantitatively using classification and regression tree (CART) analysis (CART Pro, version 6.0; Salford Systems, San Diego, CA) in classification tree mode. CART target, or dependent variables in this study, were the presence and absence of Cryptosporidium species/genotype generalized source classes (e.g., livestock presence and absence data) and species/genotype human infection risk classes defined on the basis of species/genotypes that are known human health risks. Briefly, CART is a nonparametric, automated test-and-learn, recursive partitioning procedure that splits dependent variables into homogeneous groups, referred to as nodes here, based on independent variable data-splitting criteria. Herein we use the word criteria to describe independent variable (land use or environmental) “greater-than” (>) and “less-than-or-equal to” (≤) conditions associated with designating how these nodes of dependent data are generated or split further into two subsequent groups (i.e., child nodes). CART tests for the best discriminating split criteria using all possible independent variable-splitting possibilities. A root node of a tree consists of all available data for analysis (the first node in a tree, designated node 1 in the tree figures), and CART can split these data into two child nodes. Each child node can be further split into subsequent child nodes repeatedly. Nodes that can no longer be split further due to user or CART-defined node-generating stopping criteria are designated terminal nodes (T nodes). CART classification trees were generated to define source and risk class groups (nodes) based on land use, weather, season, water physical-chemical, and stream order variable criteria (variables are given in Table S1 in the supplemental material). We employed the CART default Gini data-splitting criteria for this purpose. Gini is a data-splitting methodology that puts the largest variable class into one pure node and all others into the opposite node. Each data node split criterion has an improvement score associated with it, which is an index of how well the split generated group homogeneity or group purity in the child nodes as a result of a parent node split. The highest improvement score generated by data-splitting criteria represents the best relative performance in terms of dividing data into homogeneous groups, in the case of this study, the presence and absence of a source or risk class of Cryptosporidium. For purposes of tree model interpretation brevity, and given the exploratory nature of the work, we selected CART trees for interpretation between a minimum of 2 to a maximum of 4 tree levels (assuming the root node is 1 tree level). These would produce, respectively, a minimum of 2 and a maximum of 8 terminal nodes for a given tree model. For all CART analyses, a 10 V-fold cross-validation (10% test, 90% learning approach) was used for estimating the error rate of subtrees generated when a maximum CART tree is grown, and then pruned back, for a target Cryptosporidium source or risk class. Here, we selected the tree models with the lowest cross-validated error between 2 and 4 tree levels. For all CART analyses, the minimum number of cases in a terminal node was limited to 10 cases to limit data-sparse terminal nodes. The top competitor and surrogate variable split criteria associated with optimal data split criteria (the ones with the highest improvement score) are presented. The top competitor split criteria for each nodal split are considered the next best split criteria, relative to the optimal split criteria. Strong surrogate split criteria are those that mimic optimal split criteria on a case-by-case basis. Surrogate split criteria help reveal intercorrelations between predictor variables as well as robust predictors. Top competitor or surrogate split criteria are provided in the context of their improvement scores divided by the optimal split improvement score (multiplied by 100).
An additional analysis was conducted to evaluate associations among Cryptosporidium source and risk classes and the occurrence of other zoonotic pathogens. Odds ratios (ORs) and 95% confidence interval (CI) limits (Taylor series) were determined to assess the relative odds of detecting the presence of a zoonotic pathogen in a water sample taken when a particular Cryptosporidium source or risk class was present compared to when species/genotypes of that source or risk class were not present in a water sample. Further, odds ratios were also used to evaluate the relative odds of detecting the presence of a zoonotic pathogen in a water sample taken when specific environmental or land use criteria in the CART analysis described above (CART terminal node conditions) were met, relative to the global detection of these pathogens in all water samples collected (root node data which is designated node 1 in CART model figures).
RESULTS
Detection of Cryptosporidium species/genotypes.
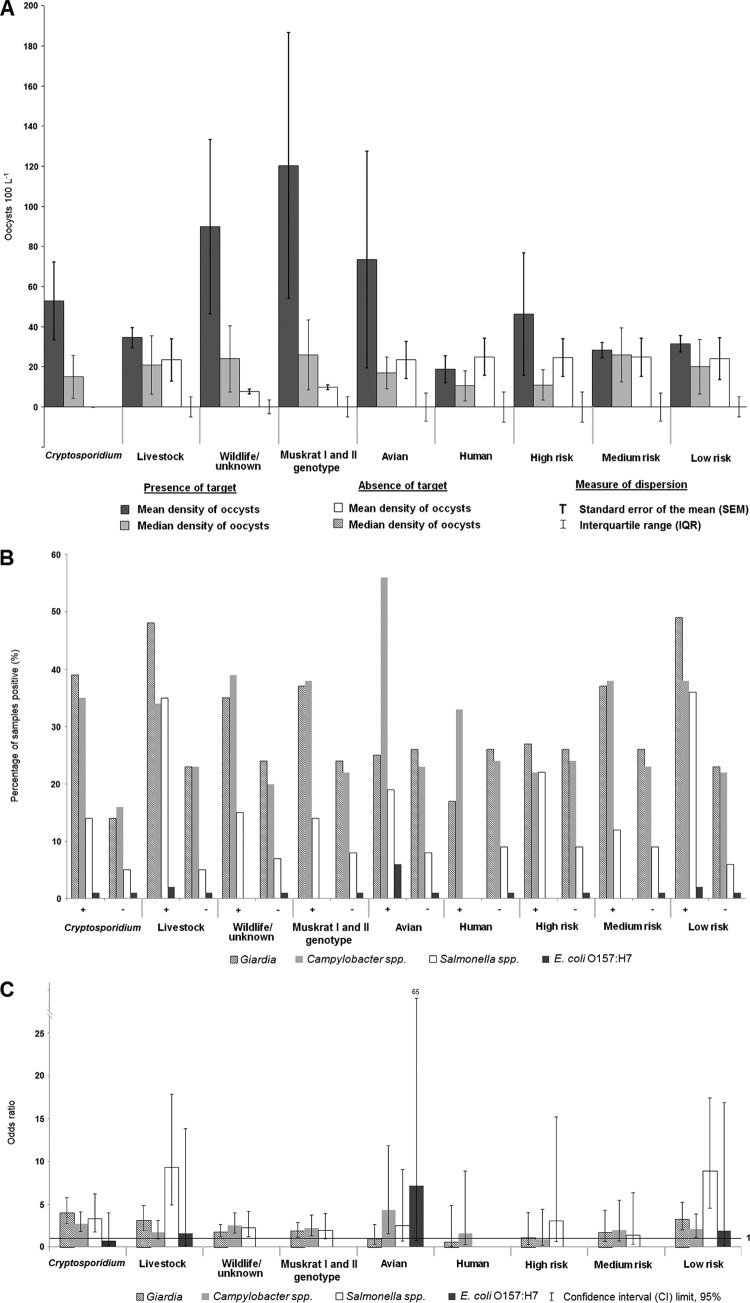
Species and genotypes that were evaluated, and their frequency of detection in 687 water samples, are summarized in Tables 1 and 2. Briefly, C. andersoni and C. parvum represent typical bovine-derived species, and they were detected in 75 and 5 water samples, respectively (11 and <1% of samples). C. parvum has both human and livestock known hosts, but for purposes of brevity in analysis, it is classed as a livestock host in this study. The avian species Cryptosporidium bailey and C. meleagridis were detected in 14 (2%) and 6 (<1%) of the samples, respectively. C. hominis, associated with human hosts, was detected in <1% of the samples. In all cases, C. hominis was detected downstream of rural homes located on or near a stream or river. C. ubiquitum was detected in ∼2% of the samples. Wildlife and livestock can both be hosts of C. ubiquitum (Table 1), but here and in Ruecker et al. (24) it has been given a livestock host designation for purposes of streamlining the data mining analysis. Rodent genotypes are represented by muskrat I, muskrat II, deer mouse III, and vole and were detected in 38 (6%), 66 (10%), 4 (<1%), and 8 (1%) surface water samples, respectively. Six unknown host genotypes were identified (W12, W18, W19, W25, W27, and W28). The various unknown genotypes were detected in less than 5% to less than 1% of samples. Cryptosporidium species presenting a high and medium risk to humans were observed in 2 and 3% of samples, respectively, with associated oocyst mean ± standard error densities of 46 ± 31 and 28 ± 4 oocysts 100 liters−1, respectively (Table 2 and Fig. 2A). The mean densities of Cryptosporidium oocysts when a livestock, wildlife/unknown, muskrat genotype, avian, and human source was detected in a sample were 35 ± 5, 90 ± 43, 120 ± 66, 74 ± 54, and 19 ± 7 oocysts 100 liters−1, respectively.
Table 2.
The number and percentage of water samples from which a Cryptosporidium oocyst, assigned host source, or human health risk class was detected
| Target in sample | Total no. of samples | No. of detections in samples | Percentage of detections in samples |
|---|---|---|---|
| Cryptosporidium (morphology or PCR)a | 687 | 330 | 48 |
| Livestock source | 687 | 90 | 13 |
| Wildlife/unknown source | 687 | 144 | 21 |
| Avian source | 687 | 20 | 3 |
| Human source | 687 | 6 | <1 |
| High human infection risk | 687 | 11 | 2 |
| Medium human infection risk | 687 | 17 | 3 |
| Low human infection risk | 687 | 71 | 10 |
The category Cryptosporidium (morphology or PCR) is a sum of the samples where a detection was made by morphology, PCR, or morphology and PCR.
Fig 2.
(A) Mean ± standard error and median plus interquartile range of Cryptosporidium oocysts per 100 liters for specific host source classes of Cryptosporidium and human infection risk classes. (B) The percentage of samples positive for a zoonotic pathogen (Giardia, Campylobacter spp., Salmonella spp., and E. coli O157:H7) out of the total number of samples tested for a zoonotic pathogen when a specific Cryptosporidium source or risk class occurs (+) or does not occur (−). (C) Odds ratio estimates (with 95% confidence intervals) calculated as the odds of the presence of a zoonotic pathogen occurring when a specific Cryptosporidium generalized source or risk class occurs, relative to the odds of the presence of a zoonotic pathogen occurring when the respective Cryptosporidium source or risk classes do not occur.
Relationships among environmental and land use variables and livestock-associated Cryptosporidium species.
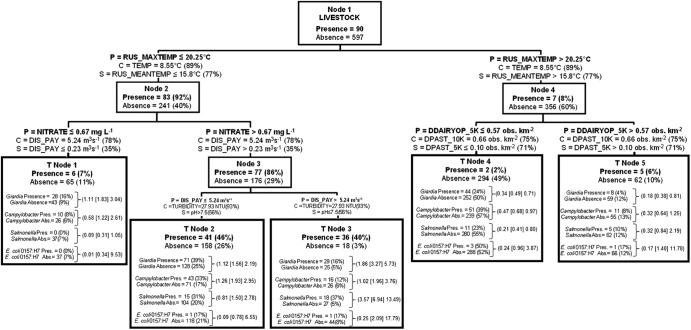
The legend of Fig. 3 provides guidance on how to interpret CART tree diagrams. Maximum daily air temperature (RUS_MAXTEMP; where RUS indicates Russell meteorological station) of 20.25°C (the 48th percentile for maximum temperature) was the optimal root node (node 1) split criterion in the CART analysis (Fig. 3). Ninety-two percent of all livestock-associated species were detected in water samples taken when the maximum daily air temperature was at or below 20.25°C; only 8% were detected when the temperature was above this threshold. Samples collected during the cooler maximum air temperatures were largely made in the fall (number of samples in the fall [nfall] = 274) and spring (nspring = 50). In contrast, samples taken when the temperature was above this threshold were primarily taken during the summer (nsummer = 242) but also spring and fall (nspring = 109; nfall = 12). Hence, season and temperature variables were important surrogate and competitor variables (Fig. 3) for this root node split of Cryptosporidium source classes.
Fig 3.
Classification tree delineating the presence and absence of livestock-classed Cryptosporidium species derived from independent variables given in Table S1 in the supplemental material. The tree begins with all data (node 1), which is then split into two child nodes by independent criteria, until node-splitting-stopping conditions are met. P, optimal data split criterion for node; C, competitor variable; S, surrogate variable. Percentages shown in parentheses for C and S were calculated as follows: top competitor and surrogate variable improvement score divided by the optimal node split criteria improvement score, multiplied by 100. The percentage in parentheses for node presence or absence of livestock-classed Cryptosporidium species was calculated as follows: the node total presence or absence divided by the respective node 1 presence and absence data multiplied by 100. CART tree results can be interpreted as follows using T node 1 as an example: total data in T node 1 is 71, with 6 livestock-classed Cryptosporidium species present and 65 absent. This group of 71 data was associated with the independent variable criteria: RUS_MAXTEMP of ≤20.25°C and nitrate of ≤0.67 mg liter−1. For each terminal node of data, the number of other zoonotic pathogens present or absent and the percent present or absent based on total presence or total absence for the entire data set (node 1) are given. Odds ratios (data in parentheses are arranged, respectively, as the lower 95% CI limit [odds ratio estimate] upper 95% CI limit) for zoonotic pathogens are given for each terminal node. They were calculated as the odds of a pathogen occurring in a given terminal node in relation to the odds of the same pathogen occurring in node 1.
For samples collected when maximum daily air temperatures were ≤20.25°C, water nitrate concentration was a node-splitting criterion at 0.67 mg liter−1, or the 45th percentile. The dominant competitor and surrogate variable split criterion for the nitrate node 2 split (Fig. 3) was river discharge (DIS_PAY, where PAY indicates Payne hydrometric station), which was positively associated with nitrate concentrations for this nodal split. Forty percent (presence/absence ratio of 2) of all livestock-associated species detections (T node 3) were found during maximum daily air temperatures of ≤20.25°C (node 1 split) and nitrate concentrations of >0.67 mg liter−1 (node 2 split) and river discharge of >5.24 m3 s−1 (node 3 split). The livestock species presence/absence ratio for T node 2 is 0.26. River discharges of 5.24 m3 s−1 represent the 92nd percentile of this variable.
At maximum daily air temperatures of >20.25°C, livestock-associated species in water were not detected as frequently. Only 7 samples were detected positive for livestock-associated species above this temperature threshold, representing 8% of all positive livestock source detections overall. Of the 7 detections for this water temperature condition, 5 (6% of all positive livestock detections) were found in water samples with maximum daily air temperatures of >20.25°C (48th percentile for maximum temperature) and where density of dairy operations in catchment areas upstream of a site within a maximum upstream flow length of 5 km (DDAIRYOP_5K) was >0.57 observations (obs) km−2 (82nd percentile for this dairy operation variable). Maximum air temperatures above 20.25°C represent all summer, many spring, and several fall samples (node 4); and the dairy operation density of 0.57 obs km−2 data split criterion partitions samples taken in areas where dairy operation density upstream of a sample site is relatively low (T node 4) and high (T node 5). “Pasturing type” surrogate and competitor variables related to the CART split given in Fig. 3, node 4, were related to dairy operation type-independent variables. This illustrates that the dairy operation and pasturing variables were correlated and behaved similarly when higher and lower occurrences of livestock Cryptosporidium sources in surface water at higher relative maximum air temperatures were partitioned.
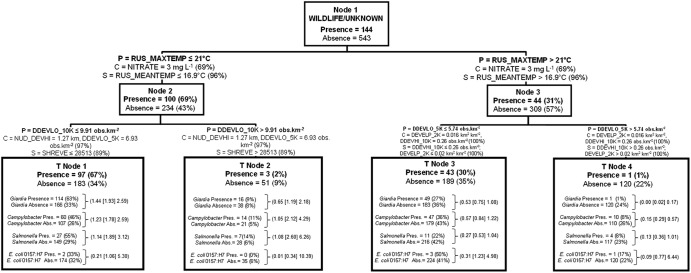
Relationships among environmental and land use variables and wildlife/unknown and muskrat I- and II-associated Cryptosporidium genotypes.
The majority of wildlife/unknown source detections (97 of 144 total source hits; 67%) occurred when maximum daily air temperatures were ≤21°C (50th percentile) and where the density of low development (e.g., rural residential or light urban development) within 10 km upstream of sampling sites (DDEVLO_10K) was ≤9.91 obs km−2 (85th percentile) (Fig. 4). Very few water samples taken downstream of areas that had higher densities of development were positive for wildlife/unknown sources. Nitrate concentrations in water and mean daily temperature were modest to strong surrogate and competitor variables for the optimal root node data split (node 1), with respective improvement scores of 69 and 96%. Shreve stream order served as strong surrogate variable split criteria for the split of node 2 data, possessing an improvement score of ∼90% of the optimal splitting criteria. Surrogate variable information indicated that sites with lower stream orders were associated with lower densities of land with low development (low-developed land) and, vice versa, that sites with higher stream orders were associated with higher densities of low-developed land for node 2 data. The surrogate stream order split criteria for node 2 partitioned strongly the sites on the South Nation River proper (Shreve of >28,513) from all other sites of lower order (small drains and streams, intermediate streams, and smaller rivers). When maximum daily temperatures were ≤21°C (node 2), the majority of samples were from the fall (nspring = 50, nsummer = 10, and nfall = 274). In contrast, when the maximum daily temperature was >21°C, the majority of samples were from the spring-summer (nspring = 109, nsummer = 232, and nfall = 12). In this predominantly spring-summer period (node 3), 43 of the wildlife/unknown source detections were observed when the density of low development in sample site catchment areas within a maximum upstream flow length of 5 km (DDEVLO_5K) was ≤5.74 obs km−2 (63rd percentile of this variable). The node 3 splitting criteria (DDEVLO_5K of ≤5.74 obs km−2 and DDEVLO_5K of >5.74 obs km−2) effectively split sites with Shreve stream orders ranging from 36 to 3,942 (T node 3), while T node 4, representing 1% of the wildlife/unknown genotype detections, was composed of sites with Shreve stream orders ranging from 48 to 50,855.
Fig 4.
Classification tree delineating the presence and absence of wildlife/unknown-classed Cryptosporidium genotypes derived from independent variables given in Table S1 in the supplemental material. The tree begins with all data (node 1), which is then split into two child nodes by independent criteria until node-splitting-stopping conditions are met. P, optimal split criterion for node; C, competitor variable; S, surrogate variable. The percentages in parentheses for C and S were calculated as follows: top competitor and surrogate variable improvement score divided by the optimal node split criteria improvement score, multiplied by 100. The percentage in parentheses for node presence or absence of wildlife/unknown-classed Cryptosporidium was calculated as the node total presence or absence divided by the respective node 1 presence and absence data, multiplied by 100. For each terminal node of data, the number of other zoonotic pathogens present or absent and the percent present or absent based on total presence or total absence for the entire data set (node 1) are given. Odds ratios (data in parentheses are arranged, respectively, as the lower 95% CI limit [odds ratio estimate] upper 95% CI limit) for zoonotic pathogens are given for each terminal node. They were calculated as the odds of a pathogen occurring in a given terminal node in relation to the odds of the same pathogen occurring in node 1. See the legend of Fig. 3 for instructions on how to read tree models.
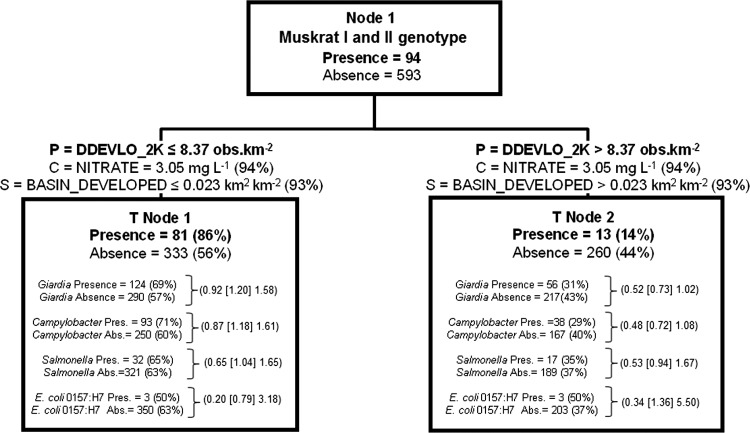
Given that about two-thirds of all the wildlife/unknown detections were muskrat I or muskrat II genotypes (number of muskrat genotype detections [nmuskrat genotype] = 94), consistent with their frequent observation in the experimental area, CART analysis was employed on just muskrat genotype data (Fig. 5). Eighty-six percent of all muskrat genotype detections were found in regions of the study area where the density of low development in sample site catchment areas within a maximum upstream flow length of the catchment of 2 km (DDEVLO_2K) was ≤8.37 obs km−2 (60th percentile of this variable), versus only 14% of muskrat genotype detections above this development density threshold. Strong surrogate variable split criteria were related to developed land and stream order, and nitrate concentration was a strong competitor split criterion.
Fig 5.
Classification tree delineating the presence and absence of muskrat I and II genotypes derived from independent variables given in Table S1 in the supplemental material. The tree begins with all data (node 1), which is then split into two child nodes by independent criteria until node-splitting-stopping conditions are met. P, optimal split criterion for node; C, competitor variable; S, surrogate variable The percentages in parentheses for C and S were calculated as follows: top competitor and surrogate variable improvement score divided by the optimal node split criteria improvement score, multiplied by 100. The percentage in parentheses for node presence or absence of muskrat I and II genotypes is calculated as the node total presence or absence divided by the respective node 1 presence and absence data, multiplied by 100. For each terminal node of data, the number of other zoonotic pathogens present or absent and the percent present or absent based on total presence or total absence for the entire data set (node 1) are given. Odds ratios (data in parentheses are arranged, respectively, as the lower 95% CI limit [odds ratio estimate] upper 95% CI limit) for zoonotic pathogens are given for each terminal node. They were calculated as the odds of a pathogen occurring in a given terminal node in relation to the odds of the same pathogen occurring in node 1. See the legend of Fig. 3 for instructions on how to read tree models.
Relationships among environmental and land use variables and human infection risk classes.
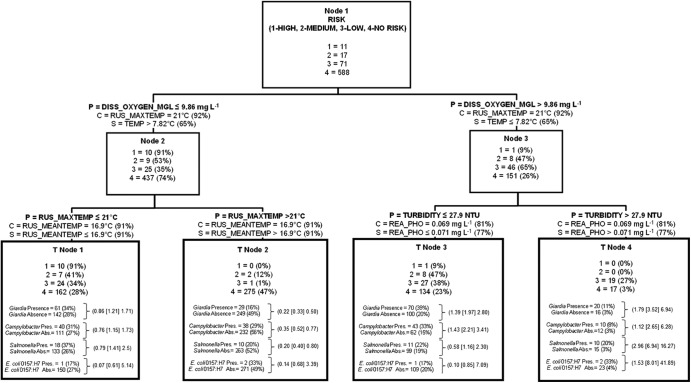
The oocysts analyzed were ascribed a human health risk class, and factors potentially affecting the spatiotemporal distribution of the various risk classes were explored (Fig. 6). The majority of samples with high risk (n = 10; 91% of high-risk samples) and many with medium risk (n = 7; 41% of medium-risk samples) for human infection were found when dissolved oxygen concentrations were ≤9.86 mg liter−1 and maximum daily temperatures were ≤21°C (the 66th and 50th percentiles of these variables, respectively) (Fig. 6). Air temperature variables were strong competitor split criteria of the root node (node 1) and child node 2. Samples with temperatures below this value and below this dissolved oxygen level represent a majority of fall samples (for T node 1, nspring = 49, nsummer = 44, and nfall = 110), whereas below this oxygen level and above this temperature value, the majority of samples were in the summer (for T node 2, nspring = 74, nsummer = 186, and nfall = 18). The majority of the remaining water samples posing high (n = 1; 9% of sample detections) and medium (n = 8; 47%) risks for human infection were observed when dissolved oxygen concentrations were >9.86 mg liter−1 and turbidity was ≤27.9 nephelometric turbidity units (NTU; the 86th percentile of this variable) (T node 3).
Fig 6.
Classification tree delineating human infection risk classes of Cryptosporidium derived from independent variables given in Table S1 in the supplemental material. The tree begins with all data (node 1), which is then split into two child nodes by independent criteria until node-splitting-stopping conditions are met. P, optimal split criterion for node; C, competitor variable; S, surrogate variable. The percentages in parentheses for C and S were calculated as follows: top competitor and surrogate variable improvement score divided by the optimal node split criteria improvement score, multiplied by 100. The percentages in parentheses for node presence or absence of human infection risk classes are the node total presence or absence of the risk class divided by the respective root node presence and absence data, multiplied by 100. For each terminal node of data, the number of other zoonotic pathogens present or absent and the percent present or absent based on total presence or total absence for the entire data set (node 1) are given. Odds ratios (data in parentheses are arranged, respectively, as the lower 95% CI limit [odds ratio estimate] upper 95% CI limit) for zoonotic pathogens are given for each terminal node. They were calculated as the odds of a pathogen occurring in a given terminal node in relation to the odds of the same pathogen occurring in node 1. See the legend of Fig. 3 for instructions on how to read tree models.
Maximum air temperature, surface water discharge, and density of developed land variable relationships with Cryptosporidium species/genotypes and human infection risk classes.
Taking into consideration the CART analyses, 3 key environmental or land use variables emerged as being particularly important with respect to the spatiotemporal distribution of Cryptosporidium species/genotypes within the study area. These were river discharge at sampling time (Fig. 7), maximum daily air temperature (Fig. 8), and density of developed land upstream of a sample location (Fig. 9). CART was not undertaken on the avian and human species as these were too infrequently detected. However, all oocysts are considered in the following analysis.
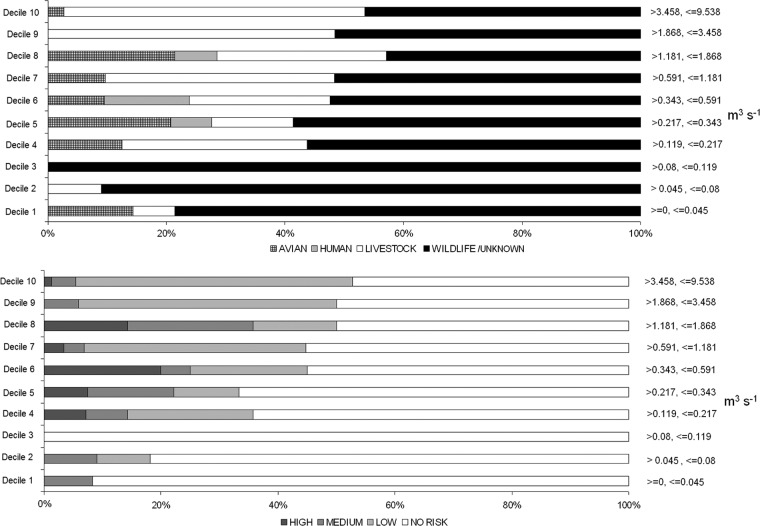
Fig 7.
Percentage of water samples with a Cryptosporidium host source or human infection risk class detection out of the total number of water samples with a source or risk class detection for various deciles of mean daily discharge (m3 s−1) measured at the Payne River hydrometric station.
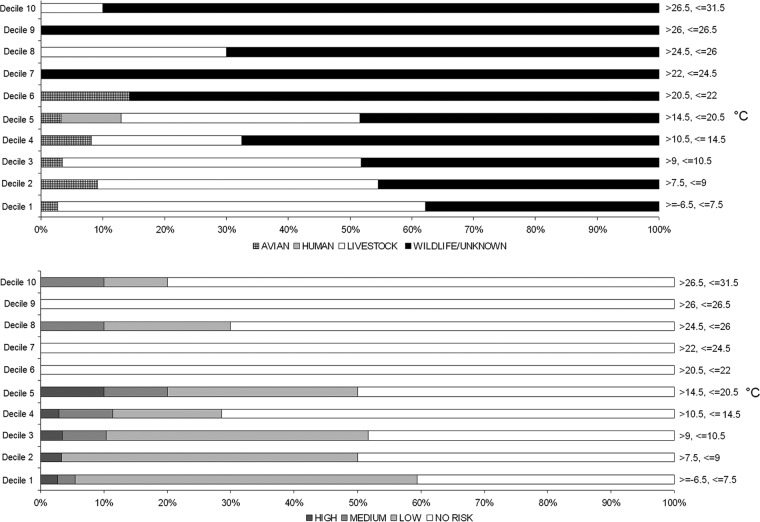
Fig 8.
Percentage of water samples with a Cryptosporidium host source or human infection risk class detection out of the total number of water samples with a source or risk class detection for various deciles of maximum daily temperature (°C) at the Russell meteorological station.
Fig 9.
Percentage of water samples with a Cryptosporidium host source or human infection risk class detection out of the total number of water samples with a source or risk class detection for various quartiles of density of low development in a catchment area with a maximum upstream flow length of 2 km upstream of sampling site (units are obs km−2).
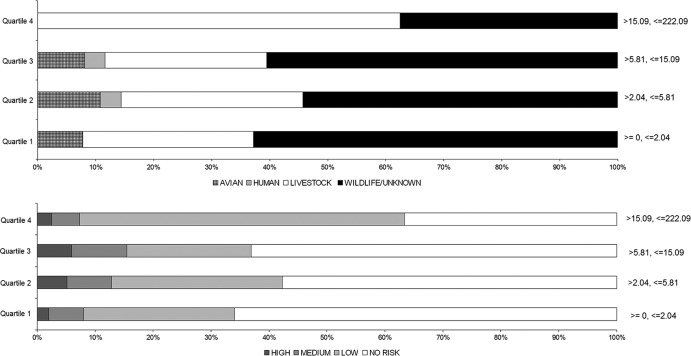
The generalized source classes were parsed according to river discharge (average daily) at time of sampling (Fig. 7). Avian sources of Cryptosporidium comprised ≤21% of the source detections in all deciles of mean daily discharge. Mean daily discharge decile associations with C. hominis did not have notable proportional trends, but there were very few observations of C. hominis during the study. Livestock-associated Cryptosporidium oocysts were present in percentages of ≤9% of positive samples in the first 3 deciles (lower discharge) of mean daily discharge. But livestock Cryptosporidium detection ranged from 14 to 39% in deciles 4 to 8 and from 48 to 51% for deciles 9 and 10 (Fig. 7). Wildlife/unknown Cryptosporidium genotypes were detected in all deciles of mean daily discharge, with the highest percentage occurrence in deciles 1 to 3 at 79 to 100%. Cryptosporidium samples assigned as high risk for human infection occurred at discharge deciles >4 (Fig. 7). No more than 9% of the sources posing a medium risk for human infection occurred below the 4th decile of mean daily discharge.
Avian-classed Cryptosporidium species were observed consistently in deciles 1 to 6 of maximum daily temperature at percentages of ≤14% of collected samples (Fig. 8), and avian species were not observed in maximum daily temperature deciles >6 (>22°C). There were too few C. hominis observations to detect any temperature-dependent trends. Livestock-associated Cryptosporidium species had the highest proportion of occurrence at maximum daily temperature deciles 1 to 5, ranging between 24 and 59%, with very few detections at higher temperatures. In contrast, wildlife/unknown Cryptosporidium genotypes typically had the highest proportion of occurrence at maximum daily temperature deciles 6 to 10, with proportions of 70 to 100%. Species which pose the highest risk to humans occurred primarily at lower maximum daily temperatures, represented by deciles 1 to 5 (Fig. 8).
Proportions of wildlife/unknown genotype occurrence tended to decrease with an increase in density of developed land upstream of a sample location (Fig. 9); the converse modestly occurred for livestock. These trends were reflected in the human health risk class, given that a majority of the low-risk detections were of livestock origin, and the majority of no-risk detections were of wildlife origin. High- and medium-risk classes occurred in all quartiles of developed land density, with no apparent trends.
Pathogenic bacteria and Giardia associations with Cryptosporidium source and human infection risk classes.
With the exception of the infrequently occurring C. hominis, Fig. 2B predominantly illustrates that the percentage of Giardia, Campylobacter spp., and Salmonella spp. is higher when Cryptosporidium (overall detection), and livestock, wildlife/unknown, and muskrat I and II species/genotypes of Cryptosporidium are present relative to when they are not. Of particular note is the especially large percentage of Campylobacter spp. in samples where avian Cryptosporidium was detected. Figure 2C shows that the odds ratios for all pathogens, except the infrequently occurring E. coli O157:H7, are between 2.7 to 4 with 95% CI limits of ≥1 when Cryptosporidium is present in water in relation to when Cryptosporidium is not present in water. In other words, the odds of these pathogens occurring were 2.7 to 4 times greater when Cryptosporidium occurred in water than when Cryptosporidium did not occur in water. Odds ratios and associated 95% CI limits for all pathogens, except Campylobacter spp., were ≥1 when livestock-associated Cryptosporidium species were present compared to when livestock-associated species were absent. The odds ratio for Salmonella spp. in this livestock grouping was 9.3, the highest odds ratio for all pathogens for all Cryptosporidium class associations. Odds ratios for pathogens with CI limits of ≥1 for when wildlife/unknown source classes and muskrat genotypes were detected in water, in relation to when they were not detected in water, were associated only with Giardia and Campylobacter spp. For avian-sourced Cryptosporidium, the odds ratio for Campylobacter spp. detection in water, versus when it was not in water, was 4.3, with CI limits of >1. When low-risk Cryptosporidium species were detected in water, in relation to when they were not detected in water, odds ratios, with CI limits of ≥1, were ≥2 for Giardia, Campylobacter spp., and Salmonella spp. There was no significant increase in the odds of pathogen presence for the high-risk grouping in relation to the grouping without high risk.
Figure 3 illustrates that T node 3, which had the largest livestock Cryptosporidium species presence/absence ratio of all terminal nodes (ratio of 2), was the terminal node that was also associated exclusively with odds ratios of >1 for zoonotic pathogens. Thus, the odds of detecting Giardia, Salmonella spp., Campylobacter spp., and E. coli O157:H7 (confidence ranges for this pathogen were very large) for environmental conditions (independent variable split criteria) associated with T node 3 were on average around 3.5 times more likely than for all sampling conditions (node 1 conditions).
Figure 4 shows that T node 1 was the terminal node that had the highest wildlife/unknown Cryptosporidium genotype presence/absence ratio (0.53) of all the terminal nodes in the model. Moreover, the odds of detecting Giardia, Salmonella spp., and Campylobacter spp. (E. coli O157:H7 occurrence was generally dispersed among all terminal node data groupings) in water for the environmental/land use CART split criteria for T node 1 were 1.9 times more likely than for all sampling conditions (node 1, or the root node). Odds ratio CI limits were >1 for all three of these pathogens. Figure 5 presents the muskrat genotype I and II CART model, with only two terminal nodes. In T node 1, with the highest muskrat genotype presence/absence ratio (0.24) in the tree, Giardia, Salmonella spp., and Campylobacter spp. odds ratios were all >1, with the exception of E. coli O157:H7; however, all lower 95% CI values were below a value of 1. Thus, the odds of detecting these pathogens for this terminal node land use criterion were not significantly higher than the odds of detecting those pathogens during all spatiotemporal conditions observed (node 1 data).
For the CART model regarding the human infection risk classes (Fig. 6), T node 4, where there were no high- or medium-risk Cryptosporidium species present, was the terminal node of data where all pathogens had odds ratios that were >1 (and with >1 lower 95% CI limit values). Hence, the odds of detecting Giardia, Campylobacter spp., Salmonella spp., and E. coli O157:H7 were significantly greater for T node 4 data with associated independent split criteria than for all other conditions, including when the highest risk of Cryptosporidium species/genotypes was almost exclusively present in surface water (T node 1).
DISCUSSION
The outcome of nearly 690 Cryptosporidium water samplings for a 6-year period (2004 to 2009) and identification of Cryptosporidium species and genotypes in these samples via molecular and phylogenetic analysis methods (24) indicate that livestock and wildlife are the dominant sources of Cryptosporidium contamination in these mixed-use watersheds (Table 1). With a rate of detection of over 20% in total number of samples collected, wildlife/unknown-associated genotypes appear to prevail as the most significant contributor of Cryptosporidium contamination of surface water in the South Nation River basin study area. Muskrat I and II genotypes (muskrat and vole) were detected, respectively, in over 5 and 9% (total, 14%) of water samples collected. In total, these two muskrat genotypes are more prevalent than all other genotypes/species, including C. andersoni (13% of water samples) (24).
Globally, wildlife/unknown-classed genotypes were proportionally more abundant during lower discharge conditions and higher maximum daily temperatures than livestock-specific species (Fig. 7 and 8). In these low-gradient surface water systems, low-flow or even no-flow conditions are not uncommon, especially in the summer, and lower discharge would reduce significant hydrologic “flushing,” dilution, and transport of wildlife-derived Cryptosporidium that accumulated from direct wildlife fecal release into the stream or near stream areas. This reasoning follows since wildlife in the study area are more of a direct fecal source to streams than livestock, with the latter having more limited and usually geographically constrained access to watercourses. Moreover, indirect inputs of livestock fecal contamination will be reduced in summer due to limited manure inputs on land and more limited subsurface drainage and runoff from adjacent fields than in the fall and spring (19).
The vole genotype was detected in only 1% of water samples, yet voles can be significant hosts of the muskrat I and II genotypes (12). Nevertheless, direct observation of muskrat activity in watercourses supports at least a potential for muskrats to contribute Cryptosporidium to raw water collected in this study. Muskrats often travel along watercourses and stream banks and swim in surface water. They live around the riparian zone and often defecate on stream banks, on logs, and directly into water (32). Muskrats prefer low water velocities, shallower water depths, and streams bordered by agricultural crops, and they breed prolifically (33, 34). They prefer cattail-type riparian environments over nonvegetated or forested areas (35). Voles can be found and proliferate in similar mesic, wet, and marsh-like habitats, where some species are known to swim (36). These environmental conditions and actual muskrat and vole habitation were observed upstream of sample locations, especially in smaller watercourses (e.g., Shreve order of ≤38) (37). The CART analyses (Fig. 3 to 6) indicated that most wildlife/unknown sources of Cryptosporidium, for which muskrat and/or vole was apparently a significant component, occurred under both higher- and lower-temperature conditions at sites where densities of developed land were, in a relative sense, low. Visual inspection of the sites meeting these geographic criteria indicated that they were generally associated with habitat and environmental conditions conducive to rodents and hydrophilic fauna (19). These areas dominate a large part of the surface water network in the region. Cryptosporidium wildlife genotypes and specifically muskrat genotypes had a much stronger association with spatial features in the landscape (land use density and location and stream order) than Cryptosporidium livestock-associated species, perhaps reflecting wildlife habitat constraints in these agriculturally dominated watersheds.
In a 10-week fall season pilot study in this same watershed study area, mature cattle were identified as the dominant source of Cryptosporidium in surface waters (38). The presence of numerous dairy and several beef cattle farming operations in the study area is consistent with the widespread detection of C. andersoni in sampled water. Ruecker et al. (38) had previously detected other host sources, notably muskrat I and II genotypes. Both studies indicated higher frequency of C. andersoni detection in the fall. But the CART analysis indicated that a vast majority of the Cryptosporidium livestock-associated species during the seasonally cooler temperature conditions occurred when river discharge was relatively high and when nitrate concentrations in the water were also relatively high. Relative to wildlife genotype spatiotemporal hot spots, it appears that Cryptosporidium from livestock manifested more strongly in surface water during elevated runoff/drainage conditions and when nitrate concentrations were relatively high in the fall, which are cumulative conditions indicative of agriculturally based (or initially derived) water contamination in the region (e.g., fall manure applications). Livestock manure is frequently applied in the spring and/or fall, and field amendments can be introduced into surface water via runoff and artificial subsurface drainage systems, which are prolific in the region and tend to drain more frequently in fall and spring (in summer manure is not readily applied to fields in the region) (39).
Different genotypes and species of Cryptosporidium present various risks to human infection. C. hominis and C. parvum pose a higher risk of human infection than other species or genotypes of Cryptosporidium, including C. andersoni and muskrat I and II genotypes. Morgan-Ryan et al. (40) detail C. hominis and C. parvum host associations. Both C. hominis and C. parvum were detected in a small number of water samples (number of C. hominis-positive samples [nC. hominis] = 6; number of C. parvum-positive samples [nC. parvum] = 5). However, C. hominis in this study was detected downstream of homes along streams. Septic system problems at some of these homes were observed during the course of the study period, but there was no clear weather or hydrological affinities with C. hominis occurrence.
We had previously speculated that enhancement and protection of riparian zones as a means to reduce surface water pollution by pasturing livestock could increase the abundance of Cryptosporidium oocysts shed by wildlife that take advantage of newly protected riparian habitat (19). However, an increase in oocyst densities, resulting from rodents and other wildlife proliferating in that riparian habitat, would not necessarily heighten human infection risks related to Cryptosporidium. The fact that muskrat I and II genotype detections were prevalent in raw water in the agriculturally dominated mixed-use watersheds in this study may have also been due to general reductions in habitat of natural predators as a result of agricultural and urban land use activities (41). Additional source tracking data would be required in concert with Cryptosporidium genotype/species data to help elucidate such effects. Even so, land use practices that protect or generate wildlife habitat such as wetlands, riparian buffers, and forested areas may increase fecal pollution pressure from wildlife, without necessarily increasing human infection risks related to Cryptosporidium. The fact that C. ubiquitum (livestock and wildlife serve as known hosts) (Table 1) was detected in only about 2% of the samples further supports this contention.
Identifying the sources of pathogens in water is one cornerstone of water quality risk assessment. Multiple MST tools can be employed to draw relationships among pathogens and dominant fecal sources (38, 42), and identification of the fecal source can be used to help inform other fecal pollution-driving mechanisms in a landscape. Relying on generalized specificity of known hosts, Cryptosporidium genotype and species information is one source-tracking tool. In the present study, associations between source-tracking information and the occurrence of a suite of zoonotic bacterial pathogens and Giardia were explored. This approach is supported by the fact that multiple pathogens can be shed during a specific fecal release by a host or be released by pathogen reservoirs in particular environment settings (43, 44).
In the present study, Cryptosporidium genotype/species information was useful in helping to identify spatiotemporal trends in other zoonotic pathogens in surface water. Overall, when Cryptosporidium occurs in water, the odds of detecting Giardia, Campylobacter spp., and Salmonella spp. are greater than when the parasite is absent in the water (Fig. 2). Moreover, the highest significant (CI limits did not bracket unity) odds ratios for Giardia and Salmonella spp. occurred exclusively when livestock Cryptosporidium was present in water versus when livestock-associated Cryptosporidium was absent in water. The highest odds ratios for Campylobacter spp. were associated with avian source presence versus avian source absence of Cryptosporidium, suggesting that birds could be a significant source of Campylobacter spp. For wildlife/unknown genotypes of Cryptosporidium, primarily fall conditions and watershed settings where rural and urban development was relatively low (Fig. 4, T node 1) were environmental/land use conditions associated with the greatest wildlife genotype presence/absence ratios (0.53) and were conditions where the odds of detecting zoonotic pathogens (less the infrequently occurring E. coli O157:H7) were around two times greater than all wildlife genotype presence and absence data (Fig. 4, node 1). Thus, wildlife fecal inputs in these spatiotemporal hot spots could be formative regarding increasing marginally the odds of occurrence of Giardia, Campylobacter spp., and Salmonella spp. in water. Yet muskrats and/or voles are a clear dominant culprit in the shedding of wildlife-derived Cryptosporidium, and, by extension, it could be assumed that these rodents shed other zoonotic pathogens that may be more important, in terms of human infection, than the Cryptosporidium they shed. When muskrat I and II genotypes were present, compared to muskrat I and II genotype absence, there were significantly elevated relative odds of Giardia and Campylobacter spp. occurring in water (similar to overall wildlife trends) (Fig. 2). However, zoonotic pathogens detected for the land use conditions identified in the CART analysis where the muskrat I and II genotype presence/absence ratio was highest (0.24) (Fig. 5, T node 1) indicate that there is generally no greater odds (odds ratio 95% CI limit bracket unity) of detecting other examined zoonotic pathogens in these Cryptosporidium genotype spatiotemporal hot spots than in the full pool of muskrat I and II genotype detection sites in the study region.
Analyses revealed that there was coherence between the environmental conditions where the highest livestock-derived Cryptosporidium species presence/absence ratios (ratio of 2) occurred and greater odds (odds ratios ranging from 2 to 7 times greater) of occurrence of zoonotic bacteria pathogens and Giardia (Fig. 3, T node 3). These relationships are in agreement with correlations among Cryptosporidium oocyst occurrence in water and the occurrence of other pathogens in that same water (20). Cumulative fecal inputs in surface water systems mobilized by wetter fall conditions, times of increased manure application to land in fall, and water pollution-driving mechanisms accompanied with higher discharge and higher relative nitrate concentrations are factors linked to livestock fecal inputs in the region.
Weighing the results from the two most dominant generalized host classes of Cryptosporidium in the study area, namely, wildlife/unknown (21% occurrence in water samples) and livestock (13% occurrence in water samples), best management practices (BMPs) that alleviate livestock inputs to surface water and that create opportunities for wildlife to survive or perhaps proliferate should not be rejected solely on the basis of a potential for heightened proportional inputs of wildlife fecal matter. This contention is supported by the following findings from the present study: (i) livestock source-classed Cryptosporidium can have low-, medium-, and high-risk species, whereas wildlife/unknown sources provide only known no- to low-risk genotypes (Tables 1 and 2); (ii) livestock host class Cryptosporidium is associated with, on average, higher ORs associated with zoonotic pathogens occurring in water (less Campylobacter spp.) than the wildlife/unknown host class (ORs here are the odds of pathogen presence when host class is present/odds of pathogen presence when host class is absent) (Fig. 2C); (iii) there was no E. coli O157:H7 that occurred when wildlife/unknown source class Cryptosporidium occurred in water (Fig. 2B); (iv) E. coli O157:H7 was detected when livestock-associated Cryptosporidium was observed in water (Fig. 2B); (v) spatiotemporal hot spots where livestock-associated Cryptosporidium dominated were associated exclusively with higher significant odds ratios of other zoonotic pathogens occurring in water (Fig. 3, T node 3) than wildlife/unknown spatiotemporal hot spots (Fig. 4, T node 1), where such odds were not exclusively as high as their respective host class absence categories; and (vi) there was a uniformly low presence of C. ubiquitum, a medium-risk Cryptosporidium commonly associated with livestock.
The highest human infection risk class specifically related to Cryptosporidium, and as provided by CART analysis, was most frequently detected (91% of total highest risk class observations) when water oxygen was relatively low (≤9.9 mg liter−1) and when maximum air temperatures were relatively cool (<21°C). The highest risk class of Cryptosporidium includes both C. parvum and C. hominis. C. parvum is hosted primarily by cattle or humans, while C. hominis is hosted more exclusively by humans. Thus, it is possible that high-risk contamination originated from both livestock and human sources (knowing that different pollution driving processes likely influenced inputs of these different species if derived from different sources). Alternatively, high-risk classes may have originated from human sources exclusively since all C. parvum and C. hominis oocysts could have originated from human sources. The 9.9 mg liter−1 oxygen level represents the 65th percentile, suggesting that a majority of the high and medium human infection risk Cryptosporidium species occurred in more oxygen-limited water. The significance of the lower dissolved oxygen condition is likely multifactorial as Spearman rank correlations between dissolved oxygen concentrations and water/air temperature variables were approximately −0.58 to −0.65 (P ≤ 0.05). Perhaps this reflects a seasonal effect where Cryptosporidium inputs to water occur at some of the lower-stream-order sites at higher water temperatures (e.g., surrogate node 1 split criteria) (Fig. 6). Moreover, dissolved oxygen was modestly, but significantly, positively correlated with stream order (Spearman ρ, 0.18; P ≤ 0.10), indicating roughly that the lower stream orders, where biological oxygen demand may have been greater, were spatial/temporal hot spots for species/genotypes of human health risk potential. Yet such conditions can occur in larger watercourses during the summer months as well. The mean Shreve stream order of water samples collected meeting the CART “greater-than” dissolved oxygen split criterion defined above was 13,211, whereas the mean Shreve stream order for the CART “less-than-or-equal to” dissolved oxygen split criterion was 10,669. The correlation between river discharge and dissolved oxygen was a Spearman ρ of 0.62 (P < 0.05). Overall, lower river discharge primarily in the fall, warmer water/lower oxygen concentrations, and generally smaller stream orders could be conditions where there is greater potential for multiple source inputs (e.g., human and/or bovine) to surface water of higher-risk Cryptosporidium oocysts.
The present study represents a comprehensive analysis of the geospatial and temporal prevalence of Cryptosporidium species and genotypes in surface water in mixed-use, but agriculturally dominated, watersheds. Very few of the Cryptosporidium species and genotypes detected posed a high risk for human infection. Given the geographic disposition of C. hominis occurrence, practices that mitigate septic system leakages, a documented concern in the region, may be a means to reduce some exposure risks associated with this pathogen even when oocyst densities associated with C. hominis were relatively low. Muskrat I and II genotypes were the dominant sources of wildlife-classed Cryptosporidium species, and their presence was coherent with habitat. Muskrat genotypes, and the majority of the genotypes in the generalized wildlife/unknown Cryptosporidium host class (Table 1) are not known to be human infective. Therefore, land use practices that facilitate protection and proliferation of wildlife should not necessarily augment human infection risks for Cryptosporidium in the region. Avian types of Cryptosporidium species were not as prevalent as expected given that the watershed is located in the Atlantic fly zone for migrating waterfowl (45). Considering that C. meleagridis (medium risk for human infection) occurred in only <1% of all samples in the present study, C. meleagridis was not deemed problematic in terms of exposure risk potential. Livestock-related species of Cryptosporidium were observed less often than wildlife-related sources of Cryptosporidium, and their occurrence was most strongly related to agricultural non-point source pollution conditions in the fall. The odds of detecting Giardia, Campylobacter spp., and Salmonella spp. in water were roughly 2.7 to 4 times greater when Cryptosporidium oocysts were detected in water than when they were not. The greatest significant odds ratios (odds of pathogen presence when host class is present/odds of pathogen presence when host class is absent) for Giardia spp., Campylobacter spp., and Salmonella spp. in water were associated, respectively, with livestock (odds ratio of 3.1), avian (4.3), and livestock (9.3) host classes.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by Agriculture and Agri-Food Canada through the Sustainable Agriculture Environmental Systems (SAGES) program, the Watershed Evaluation of Beneficial Management Practice (WEBs) program, the National Water Quality Surveillance Research Initiative through an agreement with Health Canada, the National Agri-Environmental Standards Initiative (NAESI) program of Environment Canada, and the Alberta Water Research Institute.
We thank Diane Medeiros (Health Canada) and the South Nation Conservation Authority for helping to facilitate this research. We also thank Public Health Agency of Canada personnel in Lethbridge, Alberta, for their excellent assistance with laboratory analyses, and the OIE Salmonella Reference Laboratory of the Public Health Agency of Canada in Guelph, Ontario, for providing serotyping results.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at 10.1128/AEM.01924-12.
REFERENCES
- 1. Baldursson S, Karanis P. 2011. Waterborne transmission of protozoan parasites: review of worldwide outbreaks: an update 2004–2010. Water Res. 45:6603–6614 [DOI] [PubMed] [Google Scholar]
- 2. Collinet-Adler S, Ward HD. 2010. Cryptosporidiosis: environmental, therapeutic, and preventive challenges. Eur. J. Clin. Microbiol. Infect. Dis. 29:927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smeets PWMH, van Dijk JC, Stanfield G, Rietveld LC, Medema GJ. 2007. How can the UK statutory Cryptosporidium monitoring be used for quantitative risk assessment of Cryptosporidium in drinking water? J. Water Health 5:107–118 [DOI] [PubMed] [Google Scholar]
- 4. Chalmers RM, Davies AP. 2010. Minireview: clinical cryptosporidiosis. Exp. Parasitol. 124:138–146 [DOI] [PubMed] [Google Scholar]
- 5. Graczyk TK, Fayer R, Trout JM, Jenkins MC, Higgins J, Lewis EJ, Farley CA. 2000. Susceptibility of the Chesapeake Bay to environmental contamination with Cryptosporidium parvum. Environ. Res. 82:106–112 [DOI] [PubMed] [Google Scholar]
- 6. Xiao L, Ryan UM. 2008. Molecular epidemiology, p 119–163 In Fayer R, Xiao L. (ed), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 7. Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin 2006. J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J. Med. Microbiol. 55:703–707 [DOI] [PubMed] [Google Scholar]
- 8. Jiang J, Alderisio KA, Xiao L. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jellison KL, Hemond HF, Schauer DB. 2002. Sources and species of Cryptosporidium oocysts in the Wachusett reservoir watershed. Appl. Environ. Microbiol. 68:569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ong CSL, Eisler DL, Alikhani A, Fung VWK, Tomblin J, Bowie WR, Isaac-Renton JL. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pintar KDM, Fazil A, Pollari F, Charron DF, Waltner-Toews D, McEwan SA. 2010. A risk assessment model to evaluate the role of fecal contamination in recreational water on the incidence of cryptosporidiosis at the community level in Ontario. Risk Anal. 30:49–64 [DOI] [PubMed] [Google Scholar]
- 12. Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L. 2007. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73:6475–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ong C, Moorehead W, Ross A, Isaac-Renton J. 1996. Studies of Giardia spp. and Cryptosporidium spp. in two adjacent waters. Appl. Environ. Microbiol. 62:2798–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graczyk TK, Evans BM, Shiff CJ, Karreman HJ, Patz JA. 2000. Environmental and geographical factors contributing to watershed contamination with Cryptosporidium parvum oocysts. Environ. Res. 82:263–271 [DOI] [PubMed] [Google Scholar]
- 15. Ziegler PE, Wade SE, Schaaf SL, Stern DA, Nadareski CA, Mohammed HO. 2007. Prevalence of Cryptosporidium species in wildlife populations within a watershed landscape in southeastern New York State. Vet. Parasitol. 147:176–184 [DOI] [PubMed] [Google Scholar]
- 16. Edge T, El-Shaarawi A, Gannon V, Jokinen C, Kent R, Khan UH, Koning W, Lapen D, Miller J, Neumann N, Phillips R, Robertson W, Schreir H, Scott A, Shtepani I, Topp E, Wilkes G, van Bochove E. 2012. Investigation of an Escherichia coli environmental benchmark for waterborne pathogens in agricultural watersheds in Canada. J. Environ. Qual. 41:21–30 [DOI] [PubMed] [Google Scholar]
- 17. LeChevallier MW, Norton WD, Lee RG. 1991. Occurrence of Giardia and Cryptosporidium spp. in surface water supplies. Appl. Environ. Microbiol. 57:2610–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ryu H, Abbaszadegan M. 2008. Long-term study of Cryptosporidium and Giardia occurrence and quantitative microbial risk assessment in surface waters of Arizona in the U. S. A. J. Water Health 6:263–273 [DOI] [PubMed] [Google Scholar]
- 19. Sunohara M, Topp E, Wilkes G, Gottschall N, Neumann N, Ruecker N, Jones TH, Edge T, Marti R, Lapen DR. 2012. Impacts of riparian zone protection from cattle on nutrient, bacteria, f-coliphage, Cryptosporidium, and Giardia water contamination in small agricultural stream. J. Environ. Qual. 41:1301–1314 [DOI] [PubMed] [Google Scholar]
- 20. Wilkes G, Edge T, Gannon V, Jokinen C, Lyautey E, Medeiros D, Neumann N, Ruecker N, Topp E, Lapen DR. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res. 43:2209–2223 [DOI] [PubMed] [Google Scholar]
- 21. Wilkes G, Edge T, Gannon V, Jokinen C, Lyautey E, Neumann N, Ruecker N, Scott A, Sunohara M, Topp E, Lapen DR. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. 45:5807–5825 [DOI] [PubMed] [Google Scholar]
- 22. Jokinen C, Edge T, Ho S, Koning W, Laing C, Mauro W, Medeiros D, Miller J, Robertson W, Taboada E, Thomas JE, Topp E, Ziebell K, Gannon V. 2011. Molecular subtypes of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res. 45:1247–1257 [DOI] [PubMed] [Google Scholar]
- 23. U. S. Environmental Protection Agency (USEPA) 2003. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 815-R-03-XXX. Office of Ground Water and Drinking Water Technical Support Center, U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 24. Ruecker NJ, Matsune JC, Wilkes G, Lapen DR, Topp E, Edge T, Sensen CW, Xiao L, Neumann NF. 2012. Molecular and phylogenetic approaches for tracking host sources of Cryptosporidium contamination in raw water. Water Res. 46:5135–5150 [DOI] [PubMed] [Google Scholar]
- 25. Chalmers RM, Smith R, Elwin K, Clifton-Hadley FA, Giles M. 2011. Epidemiology of anthroponotic and zoonotic human cryptosporidiosis in England and Wales, 2004–2006. Epidemiol. Infect. 139:700–712 [DOI] [PubMed] [Google Scholar]
- 26. Lake IR, Nichols G, Bentham G, Harrison FC, Hunter PR, Kovats SR. 2007. Cryptosporidiosis decline after regulation, England and Wales, 1989–2005. Emerg. Infect. Dis. 13:623–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine S, Martin SW, Nydam DV, Jamieson F, Xiao L. 2006. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol. Res. 99:346–352 [DOI] [PubMed] [Google Scholar]
- 28. Hunter PR, Hadfield SJ, Wilkinson D, Lake IR, Harrison FCD, Chalmers RM. 2007. Correlation between subtypes of Cryptosporidium parvum in humans and risk. Emerg. Infect. Dis. 13:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedraza-Díaz S, Amar C, McLauchlin J. 2000. The identification of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189–194 [DOI] [PubMed] [Google Scholar]
- 30. Elwin K, Hadfield SJ, Robinson G, Chalmers RM. 2012. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol. Infect. 140:673–683 [DOI] [PubMed] [Google Scholar]
- 31. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]
- 32. Halfpenny JC. 1986. A field guide to mammal tracking in western America. Big Earth Publishing, Boulder, CO [Google Scholar]
- 33. Allen AW, Hoffmann RD. 1984. Habitat suitability index models: muskrat. Publication FWS/OBS-82/10.46. U.S. Fish and Wildlife Service, Washington, DC [Google Scholar]
- 34. Errington PL. 1951. Concerning fluctuations in populations of the prolific and widely distributed muskrat. Am. Nat. 85:273–292 [Google Scholar]
- 35. Kadlec RH, Pries J, Mustard H. 2007. Muskrats (Ondatra zibethicus) in treatment wetlands. Ecol. Eng. 29:143–153 [Google Scholar]
- 36. Tamarin RH. 1985. Biology of new world microtus. American Society of Mammalogists, Shippensburg, PA [Google Scholar]
- 37. Shreve RL. 1966. Statistical law of stream numbers. J. Geol. 74:17–37 [Google Scholar]
- 38. Ruecker NJ, Braithwaite SL, Topp E, Edge T, Lapen DR, Wilkes G, Robertson W, Medeiros D, Sensen CW, Neumann NF. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl. Environ. Microbiol. 73:3945–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cicek H, Sunohara M, Wilkes G, McNairn H, Pick F, Topp E, Lapen DR. 2010. Using vegetation indices from satellite remote sensing to assess corn and soybean response to controlled tile drainage. Agric. Water Manag. 98:261–270 [Google Scholar]
- 40. Morgan-Ryan UM, Fall A, Ward LA, Hijjawi N, Sulaiman I, Fayer R, Thompson RCA, Olson M, Lal A, Xiao L. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433–440 [DOI] [PubMed] [Google Scholar]
- 41. McLaughlin A, Mineau P. 1995. The impact of agricultural practices on biodiversity. Agric. Ecosyst. Environ. 55:201–212 [Google Scholar]
- 42. Marti R, Zhang Y, Lapen DR, Topp E. 2011. Development and validation of a microbial source tracking marker for the detection of fecal pollution by muskrats. J. Microbiol. Methods 87:82–88 [DOI] [PubMed] [Google Scholar]
- 43. Hoar R, Atwill ER, Elmi C, Farver TB. 2001. An examination of risk factors associated with beef cattle shedding pathogens of potential zoonotic concern. Epidemiol. Infect. 127:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lemarchand K, Lebaron P. 2003. Occurrence of Salmonella spp. and Cryptosporidium spp. in a French coastal watershed: relationship with fecal indicators. FEMS Microbiol. Lett. 218:203–209 [DOI] [PubMed] [Google Scholar]
- 45. Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 4:177–189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.