Abstract
Cholix toxin (ChxA) is a recently discovered exotoxin in Vibrio cholerae which has been characterized as a third member of the eukaryotic elongation factor 2-specific ADP-ribosyltransferase toxins, in addition to exotoxin A of Pseudomonas aeruginosa and diphtheria toxin of Corynebacterium diphtheriae. These toxins consist of three characteristic domains for receptor binding, translocation, and catalysis. However, there is little information about the prevalence of chxA and its genetic variations and pathogenic mechanisms. In this study, we screened the chxA gene in a large number (n = 765) of V. cholerae strains and observed its presence exclusively in non-O1/non-O139 strains (27.0%; 53 of 196) and not in O1 (n = 485) or O139 (n = 84). Sequencing of these 53 chxA genes generated 29 subtypes which were grouped into three clusters designated chxA I, chxA II, and chxA III. chxA I belongs to the prototype, while chxA II and chxA III are newly discovered variants. ChxA II and ChxA III had unique receptor binding and catalytic domains, respectively, in comparison to ChxA I. Recombinant ChxA I (rChxA I) and rChxA II but not rChxA III showed variable cytotoxic effects on different eukaryotic cells. Although rChxA II was more lethal to mice than rChxA I when injected intravenously, no enterotoxicity of any rChxA was observed in a rabbit ileal loop test. Hepatocytes showed coagulation necrosis in rChxA I- or rChxA II-treated mice, seemingly the major target for ChxA. The present study illustrates the potential of ChxA as an important virulence factor in non-O1/non-O139 V. cholerae, which may be associated with extraintestinal infections rather than enterotoxicity.
INTRODUCTION
Vibrio cholerae is a Gram-negative, curved-rod-shaped, motile bacterium with a monotrichous flagellum, and it is transmitted through contaminated water. To date, >200 serogroups of this species have been reported based on the surface O antigen. V. cholerae strains which express O1 or O139 surface antigens and possess two major virulent genes, the cholera toxin (CT, encoded by the ctx gene) and the toxin-coregulated pilus (TCP, encoded by the tcpA gene), are mainly responsible for acute diarrheal disease affecting millions of people every year (1). Strains other than O1 or O139 are commonly referred to as non-O1/non-O139. The non-O1/non-O139 V. cholerae generally does not carry ctx and tcpA genes, but some strains of this group can be potentially virulent in humans, causing sporadic cases of diarrhea and extraintestinal infections (2–4). The toxigenic potential of some of these strains is attributed to the type III secretion system (T3SS), type VI secretion system (T6SS), heat-stable enterotoxin (NAG-ST), hemagglutinin protease (HAP), and zonula occludens toxin (ZOT) (5–10). However, many non-O1/non-O139 strains isolated from patients do not contain any of these potential virulence factors. Some additional genes encoding hemolysin (HLY), repeats in toxin (MARTX), etc., are ubiquitously present in V. cholerae strains and are not considered to contribute to diarrhea directly, but they have redundant roles in promoting persistent gut colonization. The accessory toxins, especially HLY and MARTX, act not as direct mediators of adherence but by altering the local host environment and, in part, by enabling V. cholerae to survive innate immune clearance (11, 12).
Several pathogenic bacteria (Corynebacterium diphtheriae, Pseudomonas aeruginosa, Clostridium botulinum, Staphylococcus aureus, V. cholerae, Escherichia coli, etc.) produce protein toxins, which possess ADP-ribosyltransferase (ADPRT) activity as a key mechanism to modify the properties of host cell proteins to induce disease. Bacterial ADP-ribosylating exotoxins can be differentiated into various families on the basis of their target molecules (eukaryotic elongation factor, GTP binding proteins, actin, etc.) (13). Recently, Jørgensen et al. (14) reported the presence of a eukaryotic elongation factor 2 (eEF2)-specific ADP-ribosylating factor termed cholix toxin (protein designation, ChxA) in non-O1/non-O139 V. cholerae strains. The evaluation of the ability of the chxA gene to produce an exotoxin was pioneered by the discovery of an open reading frame (ORF) in an environmental non-O1/non-O139 V. cholerae strain which showed close sequence identity with the toxA gene of P. aeruginosa encoding the exotoxin A (ExoA) (15). The mode of action of ChxA in V. cholerae, i.e., causing cytotoxicity by targeting eEF2, is similar to that of ExoA from P. aeruginosa and diphtheria toxin (DT) from C. diphtheriae (16). However, ChxA and ExoA are more closely related and their domain orientation, receptor recognition, and translocation mechanisms are different from those of DT (17).
The chxA gene encodes a 666-amino-acid (aa)-residue protein (70.7 kDa; 634-aa mature protein) including a 32-aa residue leader sequence. ChxA consists of three structural domains with structures similar to that of ExoA: a receptor-binding domain (domain Ia, aa 1 to 264; domain Ib, aa 387 to 423 [unknown function]), the parts of which together form a 13-stranded anti-parallel β-jellyroll, a translocation domain (domain II, aa 265 to 386) consisting of a bundle of six α-helices, and a catalytic domain (domain III, aa 424 to 634) with an α/β-fold topology (14). Studies of the toxigenic properties of ChxA are in an earlier stage. There is also little information about the abundance and genetic diversity of the chxA gene among V. cholerae strains isolated from diarrheal patients and about their pathogenic mechanisms.
The present study has extensively investigated the occurrence and genetic diversity of the chxA gene in clinical and environmental V. cholerae strains belonging to O1 and O139 as well as non-O1/non-O139 serogroups. In addition, the virulence properties of ChxAs were evaluated by cytotoxicity, rabbit ileal loop (RIL), and mouse lethality assays. Our findings illustrate the potential of ChxA as an important virulence factor among non-O1/non-O139 V. cholerae strains. We have observed the high genetic diversity of the chxA gene and unveiled the presence of at least three toxinotypes with varied biological activities. To the best of our knowledge, this is the first comprehensive study of ChxA at the genetic and protein levels to understand its role in the pathogenesis of V. cholerae infection.
(This study was performed in partial fulfillment of the requirements of a Ph.D. thesis for Sharda Prasad Awasthi from the Graduate School of Life and Environmental Sciences, Osaka Prefecture University, Osaka, Japan.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Vibrio cholerae strains used in this study were randomly selected from our laboratory collection. A total of 765 V. cholerae strains of clinical and environmental origins, including O1 (n = 485), O139 (n = 84), and non-O1/non-O139 (n = 196), were screened for the presence of the chxA gene (Table 1). Some of them were originally isolated from diarrheal patients in India, Bangladesh, Brazil, and Peru, and the others were of environmental origin. The identities of the strains were confirmed by molecular methods (18). The strains stored in glycerol stock at −80°C were grown in alkaline peptone water (APW) and subsequently on thiosulfate-citrate-bile salts-sucrose (TCBS) agar at 37°C when needed.
Table 1.
Abundance of chxA (cholix toxin) gene among Vibrio cholerae strains used in this study
| Serogroup | Country | Source | No. of strains | Presence or absencea of chxA |
|---|---|---|---|---|
| O1 (n = 485) | India | Clinical | 260 | − |
| India | Environmental | 208 | − | |
| Brazil | Clinical | 10 | − | |
| Bangladesh | Clinical | 4 | − | |
| Peru | Clinical | 3 | − | |
| O139 (n = 84) | India | Clinical | 61 | − |
| Bangladesh | Clinical | 23 | − | |
| Non-O1/non-O139 (n = 196) | India | Clinical | 64 | − |
| India | Clinical | 18 | + | |
| India | Environmental | 79 | − | |
| India | Environmental | 35 | + | |
| Total | 765 | 53 |
−, absence; +, presence.
Chemicals and enzymes.
Chemicals were purchased from Nacalai Tesque (Kyoto, Japan), Wako Pure Chemical Industries (Tokyo, Japan), or Sigma Chemical Co. (St. Louis, MO). Restriction enzymes and TaKaRa Ex Taq were purchased from TaKaRa Bio Inc. (Shiga, Japan). The QIAquick PCR product purification kit was from Qiagen (Hilden, Germany). The pET vector system was purchased from Novagen (New Canaan, CT) and ProTEV protease from Promega (Madison, WI). Luria-Bertani (LB) broth was purchased from Difco Laboratories (Detroit, MI). Tryptic soya agar (TSA) and APW were purchased from Nissui Pharmaceutical Co. Ltd. (Tokyo, Japan), and TCBS agar was from Eiken Chemical Co. Ltd. (Tokyo, Japan). SeaKem Gold agarose was from FMC Bioproducts (Rockland, ME), and SeaKem LE agarose was from Lonza (Rockland, ME). Pulsed-field certified agarose and low-melting agarose were from Bio-Rad Laboratories Inc. (Hercules, CA). Molecular weight markers were purchased from Bio-Rad Laboratories Inc. or Nippon Genetics Co. Ltd. (Tokyo, Japan). Sequencing reagents such as BigDye Terminator, buffers, etc., were purchased from Applied Biosystems Inc. (Foster City, CA), and the Clean Seq kit was from Agencourt Bioscience Corporation, Beckman Coulter (Beverly, MA). Nickel Sepharose was purchased from GE Healthcare Life Sciences (Little Chalfont, Buckinghamshire, United Kingdom). Minimal essential medium (MEM) was purchased from Nissui Pharmaceutical Co. Ltd. (Tokyo, Japan). MEM-α, Glutamax-I supplement, nonessential amino acids (NEAA), and fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA).
Serotyping.
Preparation of O antisera and determination of serogroups by slide agglutination were performed as previously described (19). In brief, the serogroup reference strains were cultured overnight in infusion broth, heated at 100°C for 2 h, and washed twice with physiological saline. After centrifugation, pellets were resuspended in 20 ml physiological saline and intravenously administered to rabbits at 4-day intervals. Seven days after the last immunization, the rabbits were anesthetized by intramuscular injection of 45 mg kg−1 ketamine (Ketalar; Daiichi Sankyo Co., Ltd., Tokyo, Japan) and 5 mg kg−1 xylazine (Selactar; Bayer Healthcare, Leverkusen, Germany), and whole blood was collected from the rabbits. For O antigen determination of target bacterial strains, tube and slide agglutination tests were carried out with a cell suspension heated at 100°C for 1 h.
Colony hybridization.
Distribution of the virulence genes (Tables 1 and 2) was examined by colony hybridization assay as described previously (20). In brief, strains were grown on nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany) overlaid on LB agar at 37°C for 4 to 6 h. Then, colonies were lysed, and DNA was denatured in situ by the alkaline lysis method followed by UV cross-linking. Specific DNA probes for target genes were prepared by PCR using respective primer sets (see Table S2 in the supplemental material). These probes were labeled with [α-32P]dCTP (PerkinElmer, Wellesley, MA) by a random priming method using the Multiprime DNA labeling system (GE Healthcare Life Sciences). The processed nitrocellulose membranes were hybridized with the target probes under suitable buffer conditions, and radioactivity was visualized by the BAS FLA-3000 system (Fuji Film, Tokyo, Japan). For detection of T3SS genes, three T3SS-specific genes, vcsN2, vcsC2, and vopF, were used as a mixed probe for hybridization. Later on, the presence of each gene was confirmed by PCR.
Table 2.
Virulence gene profiles of Vibrio cholerae strains harboring the chxA gene
| No. of strainsa | Presence or absenceb |
||||||
|---|---|---|---|---|---|---|---|
| ctx | tcpA | vcsN2-vcsC2-vopF | stn | rtx | zot | hly | |
| 43 | − | − | − | − | + | − | + |
| 4 | − | − | + | − | + | − | + |
| 3 | − | − | + | + | + | − | + |
| 1 | − | − | − | + | + | − | + |
| 1 | + | + | + | − | + | + | + |
| 1 | − | + | − | − | + | − | + |
| 53c | 1 | 2 | 8 | 4 | 53 | 1 | 53 |
All strains were of the non-O1/non-O139 serogroup.
−, absence of gene(s); +, presence of gene(s). ctx, cholera toxin; tcpA, toxin-coregulated pilus; vcsN2-vcsC2-vopF, type III secretion system; stn, heat-stable enterotoxin; rtx, repeats in toxin; zot, zonula occludens toxin; hly, hemolysin.
Totals of columns are in last row.
DNA template preparation.
A DNA template of V. cholerae strains for PCR was prepared by boiling the overnight cultures (18). In brief, a single yellow colony from the TCBS plate was inoculated into 3 ml LB broth and incubated at 37°C overnight with shaking (180 rpm). The culture was diluted 10 times with TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and boiled for 10 min followed by snap cooling on ice. After centrifugation at 8,900 × g for 3 min, the supernatant was used as a DNA template and stored at −30°C for future use.
PCR amplification of the chxA gene for DNA probe preparation and sequencing.
In a 50-μl PCR mixture, 1 μl of boiled DNA template was added to 1.25 U of TaKaRa Ex Taq DNA polymerase, its buffer system (TaKaRa Bio Inc.), and the following PCR primers. Based on the analysis of a published chxA gene sequence (GenBank accession no. AY876053), a forward primer, chxAU (5′-TGTGTGATGATGCTTCTGG-3′), and a reverse primer, chxAR1 (5′-TTATTTCAGTTCATCTTTTCGC-3′), were designed and used for PCR. The PCR was carried out in a TaKaRa PCR Thermal Cycler Dice (TaKaRa Bio Inc.) with an initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 1 min and a final extension step at 72°C for 7 min. The PCR products were subjected to 1.0% LE agarose gel electrophoresis in TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]). The gels were stained in an ethidium bromide solution (2 μg ml−1) followed by destaining in distilled water, each for 5 to 10 min. Gel images were captured with a Gel-Doc 2000 (Bio-Rad Laboratories Inc.).
A PCR-amplified chxA gene (∼2.0 kb) was purified using the QIAquick PCR purification kit and then used as a gene probe for hybridization as well as sequencing. Purified DNA (10 ng) was subjected to cycle sequencing using 12 sequencing primers (see Table S2 in the supplemental material) and the BigDye Terminator v 1.1 cycle sequencing kit. The product was then purified using a Clean Seq kit and subjected to sequencing in an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems Inc.). The nucleotide sequences were aligned and analyzed using a Lasergene DNASTAR (Madison, WI) software package.
Genome walking.
Genome walking was performed as described by Asakura et al. (21) to sequence the chxA gene from strains that were positive by colony hybridization but did not produce amplicons by PCR (chxA III) using a chxAU/chxAR1 primer set. First, an ∼1.5-kb chxA gene was amplified by a chxAU/chxAR2 primer set and sequenced (see Table S2 in the supplemental material). The remaining ∼0.5-kb sequence was amplified and sequenced by genome walking. In brief, 50 ng genomic DNA was randomly extended for only 1 cycle of 5 min at 94°C, 30 s at 30°C, and 30 s at 72°C, using a 10 μM chx random1 primer (see Table S2). Further PCR amplification was performed using a 10 μM chx target1 primer (see Table S2) for 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C. Ex Taq DNA polymerase was used. The amplified fragments were sequenced with 5 μM chx seq1 primer (see Table S2) and analyzed by the DNA Lasergene software package.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed according to the Pulse Net USA protocol (www.cdc.gov/pulsenet/protocols.htm) with slight modifications. Briefly, freshly cultured V. cholerae cells were embedded into 0.5% SeaKem Gold agarose followed by in situ lysis with 0.5 mg ml−1 proteinase K (P8044-5G; Sigma) and 1.0% sarcosine (Sigma) at 54°C for 1 h. Agarose blocks containing genomic DNA were digested with 30 U of NotI restriction endonuclease at 37°C for 3 h. DNA fragments were separated on a CHEF Mapper (Bio-Rad Laboratories Inc.) as described by Yamasaki et al. (22). Gels were stained for 30 min and destained twice for 15 min, and pictures were captured as described before. Lambda ladder (Bio-Rad Laboratories Inc.) was used as a molecular mass standard. PFGE fingerprints were analyzed by Fingerprinting II software (Bio-Rad Laboratories Inc.). The unweighted-pair group method with arithmetic means (UPGMA) was applied during dendrogram analysis following the band-based (Dice coefficient) option.
Cloning of chxA gene and expression of rChxA.
chxA genes representing the three toxinotypes, chxA I (strain C9), chxA II (strain Vc106), and chxA III (strain Vc36), were amplified by PCR. The amplified DNA fragments were cloned downstream of the His6 tag sequence of the pET28a+ vector (Novagen). Just upstream of the chxA DNA fragment, a TEV protease recognition sequence was incorporated. This recombinant plasmid was transformed into E. coli BL21(DE3). After induction with isopropyl β-d-1-thiogalactopyranoside (IPTG), each construct yielded a considerable amount of fusion protein (His6-TEV-cholix). These fusion proteins were purified with a Ni-Sepharose column. The purified fusion proteins were digested with ProTEV protease to remove His6-TEV from recombinant cholix toxin (rChxA). The digested product was checked by SDS-PAGE and again passed through the Ni-Sepharose column to obtain the purified rChxA without a His6 tag.
Antiserum against ChxA.
Antiserum against purified rChxA was basically prepared as described previously (23). In brief, 100 μg of purified rChxA I in phosphate-buffered saline (PBS) (pH 7.0) was emulsified with an equal volume of Freund complete adjuvant (Difco Laboratories, Detroit, MI). The emulsion was administered intramuscularly to an adult New Zealand White rabbit (2 kg). Subsequently, two booster doses of 100 μg toxin were given in similar fashion at 30 and 42 days after first immunization. The serum antibody titer was regularly monitored by a Ouchterlony gel diffusion assay as described previously (23). Up to a dilution of 1:16 of the antisera, a precipitin line against 1 μg of homologous toxin could be observed. At the end, the rabbits were anesthetized by intramuscular injection of 45 mg kg−1 ketamine (Ketalar; Daiichi Sankyo Co., Ltd.) and 5 mg kg−1 xylazine (Selactar; Bayer Healthcare), whole blood was collected from the rabbits, and the serum was separated and stored at −80°C for further use.
Cytotoxicity assay.
Eukaryotic cells were routinely cultured in their respective media: HeLa and Vero in MEM with 5% FBS, Int-407 and Hep-2 in MEM with 10% FBS, Caco-2 in MEM with 10% FBS and NEAA, CHO in MEMα with 10% FBS, Y-1 in Ham's F-12 with 5% FBS, and NIH-3T3 in Dulbecco MEM (DMEM) with 10% FBS. The cells were incubated with 0.25% trypsin in 1 mM EDTA at 37°C for 5 min, and the concentration of cells was determined using a hemocytometer. The cells (100 μl of 105 cells ml−1) were cocultured with various amounts (0.05 to 1 μg) of purified rChxA in the presence or absence of antiserum against rChxA I in a 96-well culture plate (Orange Scientific, Braine-l'Alleud, Belgium) at 37°C in a water-jacketed incubator with 5.0% carbon dioxide in air for 24 to 48 h, and cytotoxic effects were observed under a Leica DMI6000 B microscope (Leica Microsystems, Mannheim, Germany).
Competition assay.
To examine the binding competition between ChxA toxinotypes, a competition assay using HeLa cells was performed. In brief, HeLa cells were cultured, harvested, and seeded to 96-well culture plates as described previously and incubated to form a monolayer. Then, cells were washed with PBS (pH 7.0), and rChxA I (200 ng or 1 μg well−1) was added in the presence or absence of excess (up to 20 μg well−1) rChxA II or rChxA III. Cytotoxicity was observed under a microscope after 48 h of incubation.
Rabbit ileal loop assay.
A rabbit ileal loop (RIL) assay was carried out as described earlier (24). Adult 2-kg New Zealand White rabbits were used in this assay. The rabbits were anesthetized by intramuscular injection of 45 mg kg−1 ketamine (Ketalar; Daiichi Sankyo Co., Ltd.) and 5 mg kg−1 xylazine (Selactar; Bayer Healthcare). Laparotomy was performed on anesthetized animals from the lower liver margin and 8 loops (8 cm long) with a 3-cm inter loop were ligated. The first loop was injected with CT (3 μg) and the last loop with PBS (pH 7.0) as positive and negative controls, respectively. The intermediate loops were inoculated with rChxA I, rChxA II, or rChxA III at varied concentrations (10, 50, 100, or 500 μg loop−1). Loops were placed back in the peritoneal cavity. The animals were euthanized by injection of 200 mg kg−1 pentobarbital (Nembutal; Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan) 16 h after inoculation, and the loops were examined for fluid accumulation.
Mouse lethality assay.
Lethality assays were performed as described by Yutsudo et al. (25) with some modifications. rChxA I and rChxA II were used for analysis of mouse lethality. rChxA III was not analyzed for mouse lethality, as it failed to cause cytotoxicity or enterotoxicity. For each assay, 7 male ICR mice with an average body weight of approximately 20 g (Nihon Clea Co., Japan) were intravenously injected with 1, 5, 10, or 25 μg mouse−1 (0.05, 0.25, 0.5, or 1.25 mg kg−1) of either rChxA I or rChxA II. Mice were observed for the specific time interval, and the number of mice that died within this time interval was recorded. The major organs (heart, kidney, lung, liver, and spleen) were removed, and histopathological analysis was performed. PBS (pH 7.0) was used as a carrier control. All animal experiments were performed according to the Guidelines for Animal Experimentation of Osaka Prefecture University and approved by the Animal Experiment Committee of Osaka Prefecture University.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the chxA genes have been deposited in the DNA Data Bank of Japan (DDBJ) with accession numbers AB754424 to AB754476 (53 entries).
RESULTS
Prevalence of chxA among Vibrio cholerae strains.
A total of 765 V. cholerae strains, including O1 (n = 485), O139 (n = 84), and non-O1/non-O139 (n = 196) were screened by colony hybridization for the presence of the chxA gene. Among them, 53 (27%) non-O1/non-O139 strains from diverse serogroups harbored the chxA gene. All O1 and O139 strains tested were negative for this gene (Table 1; see Table S1 in the supplemental material). The 53 chxA gene sequences were grouped into three clusters designated chxA I (n = 33), chxA II (n = 16), and chxA III (n = 4) (Fig. 1). The 33 isolates from chxA I belonged to 14 different serogroups, whereas the 16 isolates from chxA II were from 11 distinct serogroups. All 4 isolates from chxA III belonged to the O64 serogroup (see Table S1). The V. cholerae isolates positive for the chxA gene were from both clinical (22%; 18 of 82) and environmental (30%; 35 of 114) origins. Among 18 clinical strains, 16 chxA genes belonged to chxA I, 2 to chxA II, and none to chxA III. In strains of environmental origin (n = 35) 17, 14, and 4 chxA genes belonged to chxA I, chxA II, and chxA III, respectively.
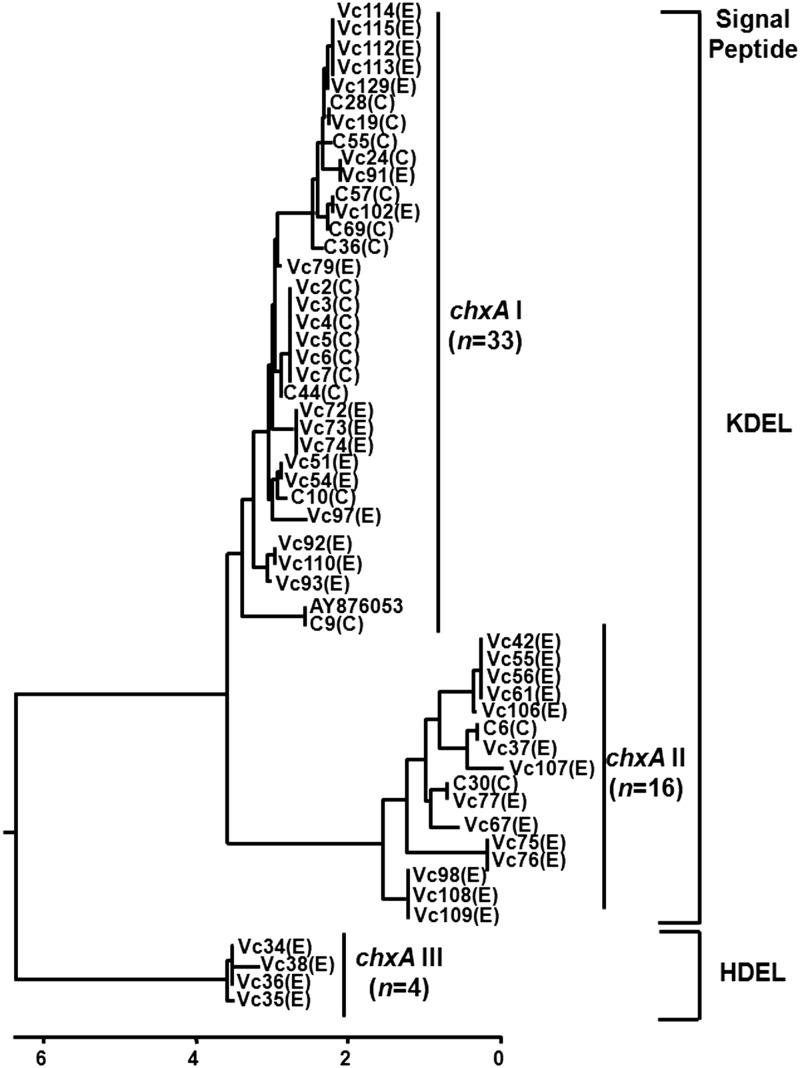
Fig 1.
Dendrogram depicting the diversity of the chxA gene from V. cholerae non-O1/non-O139 strains. Overall, 29 different chxA subtypes were observed among 53 sequences. These strains were grouped into three sequence clusters: chxA I (n = 33), chxA II (n = 16), and chxA III (n = 4). The chxA I sequences are close to the published prototype sequence (accession no. AY876053). Letters in parentheses show the origin of the isolate: C and E, clinical and environmental, respectively. At protein level, the signal peptide in ChxA I and II is KDEL whereas ChxA III has HDEL.
Furthermore, the chxA gene-positive strains (n = 53) were screened for the presence of other reported virulence genes (ctx [CT], tcpA [TCP], vcsN2, vcsC2, and vopF [T3SS], stn [NAG-ST], zot [ZOT], rtx [MARTX], hly [HLY]) in V. cholerae. All the strains were positive for rtx and hly genes. The majority of chxA gene-positive strains (43 of 53 non-O1/non-O139) did not have other tested virulence genes. Of the remaining 10 chxA gene-positive non-O1/non-O139 strains, one strain from the O141 serogroup was positive for ctx, tcpA, vcsN2/vcsC2/vopF (T3SS), and zot, three were positive for vcsN2/vcsC2/vopF (T3SS) and stn, four carried only vcsN2/vcsC2/vopF (T3SS), one had only stn, and one possessed tcpA (Table 2).
Diversity in the cholix toxin gene sequence.
The chxA genes from all the positive strains (n = 53) were sequenced, aligned, and compared with a published prototype sequence of the chxA gene (GenBank accession no. AY876053). The length of the chxA gene (without leader sequence) in the test strains varied from 1,887 bp (628 aa) to 1,905 bp (634 aa). Deletion, insertion, and/or substitution of nucleotides affected amino acid substitutions. Phylogenetic analysis based on the chxA gene sequence differentiated 53 chxA genes into 29 subtypes and further grouped them into three major clusters designated chxA I, chxA II, and chxA III (Fig. 1). Sequence types belonging to chxA I prevailed in the majority of chxA gene-positive V. cholerae strains (33 of 53; ∼63%). The prototype chxA gene sequence belonged to this cluster. About one-third (16 of 53; ∼30%) of the chxA sequences belonged to the chxA II cluster. The chxA sequences belonging to the chxA III cluster were less abundant (4 of 53; ∼7.5%) among the tested strains. Despite the presence of differences in sequences, the GC content of the chxA genes (43% to 44%) was almost conserved (Fig. 1 and Table 1).
The chxA gene sequences obtained in this study as well as the prototype sequence were translated using the Editseq program from DNASTAR, and deduced amino acid (aa) sequences were analyzed to understand the diversity of ChxA at the amino acid sequence level. ChxA I sequences had up to 97.6% similarity among themselves, and ChxA sequence from one strain (C9; see Table S1 in the supplemental material) showed 100% similarity to the prototype sequence. The amino acid sequences of ChxA II had 91.8% to 93.7% similarity whereas the ChxA III sequences had 91.2% to 92.0% similarity to the prototype sequence (Fig. 1). ChxA III possessed a different signal peptide, i.e., an HDEL in place of KDEL, compared to all other sequences of ChxA I and ChxA II (Fig. 1 and 2).
Fig 2.
Alignment of the amino acid sequences representing the three clusters of ChxA (ChxA I, II and III) from V. cholerae strains. Only one ChxA sequence from each cluster, along with the published prototype sequence (AY876053), is shown here for simplicity. The ChxA I sequences are identical or similar to prototype ChxA. The ChxA II sequences clearly define the amino acid changes in the receptor binding domain (RBD) and ChxA III in the catalytic domain (CD). All the sequences of ChxA II possess an identical deletion of 6 amino acids (GLALCW) in the RBD, and 6 sequences have a 5-amino-acid (AIAKE) insertion in CD. ChxA I and II possess KDEL whereas ChxA III possesses an HDEL signal peptide at the C-terminal end. Serine-to-glycine substitution can be observed at the P5 position of the furin binding site in ChxA III. *, conserved amino acid residue critical for catalytic activity. Matching amino acids are indicated by dots and deleted ones by dashes.
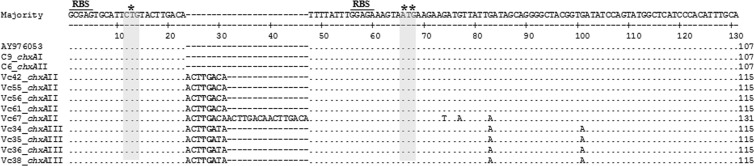
The leader sequence of ChxA (32-aa polypeptide) is encoded by a 96-nucleotide region just upstream of the chxA sequence in the prototype sequence. Analysis of the leader sequence revealed that it was conserved in all the chxA genes from chxA I (n = 33) and 11 of 16 genes from chxA II. The leader gene sequence from 5 of 16 chxA II variants and all sequences of chxA III (n = 4) had insertions of nucleotide repeats between nucleotide positions 12 and 13 compared to the prototype sequence (Fig. 3). The 4 and 1 chxA II leader gene sequences had insertions of single or 3 copies of the tandem octamer (ACTTGACA) repeats, respectively, between positions 12 and 13. All chxA III leader sequences also had insertions of single copies of nearly identical octamers (ACTTGATA) between the same positions. This insertion of nucleotides resulted in incorporation of a termination codon (TGA) at 16 to 18 nucleotide positions (Fig. 3). However, the bead enzyme-linked immunosorbent assay (ELISA) developed in our laboratory for detection of ChxA indicated that all these strains express ChxA extracellularly (unpublished data), indicating that the putative initiation codon might be different.
Fig 3.
chxA leader gene sequence alignment for the strains with nucleotide repeat insertion. One chxA leader gene sequence from chxA I and chxA II, along with the published prototype sequence (AY876053), is shown here for comparison. All the chxA I leader gene sequences are conserved. The chxA II leader sequences from 5 strains have insertions of single or 3 copies of octamer repeats, and all 4 chxA III also have insertions of single copies of nearly identical octamers. The bold characters show the termination codon; *, initiation codon in the prototype sequence; **, proposed initiation codon with probable ribosome binding site (RBS). Matching amino acids are indicated by dots and deleted ones by dashes.
Diversity at the RBD.
The receptor-binding domain (RBD; 264 aa residues in the prototype sequence) of ChxA I was conserved in length (264 aa), and there were 3 to 4 amino acid substitutions at several positions (Fig. 2; see Table S1 in the supplemental material) in comparison to the prototype sequence. In the case of the RBD of ChxA II, there was a common deletion of 6 amino acids (204GLALCW209) in comparison to that of ChxA I or ChxA III. Therefore, the length of the RBD in ChxA II was shortened to 258 amino acids. Moreover, there were also 28 to 32 amino acid substitutions at several positions (Fig. 2; see Table S1 in the supplemental material). The RBD of ChxA III was of a length identical to that of ChxA I (264 aa) but substitutions of 12 amino acids at varied positions were observed compared to the prototype sequence. Overall, the RBD of ChxA II was highly divergent in comparison to that of ChxA I or ChxA III. In some strains, the total number of amino acid replacements was the same (see Table S1) but the positions of substituted amino acids were either the same or different (details not shown).
Diversity at the UFD.
The length of the unknown function domain (UFD; 37 aa residues in the prototype sequence) of ChxA was conserved in all three toxinotypes, and the sequence diversity was also less. For example, the amino acid sequence of the UFD was conserved in 11 of 16 ChxA II variants and there was only one amino acid substitution in each of the remaining 5 compared to the prototype sequence (see Table S1 in the supplemental material). Among the 33 sequences belonging to ChxA I, 4 did not have any change, and the rest had 1 or 2 amino acid substitutions in comparison to the prototype sequence. In ChxA III, the UFD had 2 amino acid substitutions with respect to the prototype sequence (see Table S1).
Diversity at the TD.
The translocation domain (TD; 122 aa residues in the prototype sequence) in all sequences of ChxA comprised 122 amino acids, and there were 5 to 8 amino acid substitutions at different positions compared to the prototype sequence (see Table S1 in the supplemental material). Among 33 ChxA I sequences, 21, 11, and 1 had 5, 6, and 7 amino acid substitutions, respectively, in the TD. All ChxA II (n = 16) sequences differed by 5 amino acid substitutions in the TD except for 2 ChxA sequences which had 7 amino acid substitutions. In the case of ChxA III, a total of 8 amino acid substitutions were observed in the TD (see Table S1). In comparison to ChxA I and ChxA II, all the ChxA III (n = 4) sequences had serine (polar amino acid) to glycine (nonpolar amino acid) substitutions at the P5 position of the furin binding site, which is located inside the furin binding pocket (P1 to P6 and P1′ to P2′) (Fig. 2).
Diversity at the CD.
Among the observed V. cholerae strains possessing the chxA gene, the catalytic domain (CD; 211 aa residues in the prototype sequence) of ChxA I was conserved in length (211 aa) while ChxA II and ChxA III had insertions and deletions of some amino acids in this domain. Out of 33 ChxA I sequences, 13, 11, and 1 carried 3, 1, and 2 amino acid substitutions in the CD, respectively, at various positions with respect to the prototype sequence (see Table S1 in the supplemental material). ChxA II (n = 16) could be divided into two categories based on the diversity at CD: the first category had 211 amino acids (n = 10), like ChxA I, while the second category had 216 amino acids (n = 6). Among the first category of ChxA II there were up to 3 amino acid replacements at various positions of the CD. On the other hand, the second category of ChxA II carried an identical insertion of 5 amino acids (AIAKE) between 617E and 618Q and additionally 8 amino acid replacements at other positions with respect to the CD of the prototype sequence (Fig. 2; see Table S1). ChxA III (210 aa) had substantial modifications in the CD, with 29 to 32 amino acid changes along with an identical deletion of a lysine (629K) compared to the prototype sequence (Fig. 2; see Table S1). This deletion of lysine and a prior replacement of a histidine altered the signal peptide of ChxA III into HDEL, while ChxA I and ChxA II had KDEL as a signal peptide (Fig. 1 and 2). The five amino acids (H460, Y493, Y504, E574, and E581) which have been reported to play a vital role in the catalysis in all ADP-ribosyltransferases (14) were conserved among all the ChxA sequences, irrespective of their diversity (Fig. 2).
PFGE analysis.
The clonality of the V. cholerae strains harboring the chxA gene was examined by PFGE after digestion with NotI (see Fig. S1 in the supplemental material). The PFGE patterns exhibited the distribution of 14 to 23 bands over a size range of 20 to 500 kbp. A high level of genetic diversity was observed among the strains, although some of the strains showed nearly identical PFGE patterns (see Fig. S1). Dendrogram analysis revealed that at an 85% cutoff value, the 53 chxA-positive strains could be classified into 41 pulsotypes. No clear-cut correlation was observed between chxA sequence diversity, serogroup, and clonality.
Cytotoxicity assay.
After assessing the diversity of ChxA, three strains were selected as representatives of ChxA I (C9), ChxA II (Vc106), and ChxA III (Vc36). The ChxA sequence of strain C9 was 100% identical to the prototype sequence, whereas strain Vc106, representing ChxA II, possessed a high level of amino acid changes (37 aa changes) in the RBD of ChxA in comparison to that of C9 (Fig. 4; see Table S1 in the supplemental material). The Vc36 strain representing ChxA III had a highly diverse CD of ChxA with an altered furin-binding site (Fig. 2). Each chxA gene was PCR amplified, cloned, and expressed in E. coli using the pET vector system as described in Materials and Methods. The rChxA proteins were purified and used for a cytotoxicity assay against multiple eukaryotic cells. Among the 8 cell lines listed in Table 3, rChxA I (C9) showed strong cytotoxicity against HeLa and Y1 cells (death within 24 to 48 h), but rChxA II (Vc106) failed to show any such effect on the same cell lines (Table 3). In contrast, when CHO or Caco-2 cells were cocultured with these toxins there was no apparent cell death at the early stage (24 to 48 h). However, microscopic observation revealed that both rChxA I and rChxA II inhibited (at similar sensitivity levels) the cell growth (Table 3). When NIH-3T3, Vero, Int-407, and HEp-2 cells were cultured with rChxA I and rChxA II, both toxinotypes showed strong cytotoxicity. This cytotoxicity by rChxA I and rChxA II could be neutralized by ChxA I antisera (data not shown). In contrast to the cytotoxicity patterns of rChxA I and rChxA II, rChxA III (Vc36) failed to cause cytotoxicity in any of the tested cell lines. A competition assay using HeLa cells between rChxA I and rChxA II/rChxA III revealed that rChxA III could partially suppress rChxA I-induced cytotoxicity, whereas no suppression of the rChxA I cytotoxicity was observed when cells were cocultured with rChxA I in the presence of rChxA II (data not shown).
Fig 4.
Number of amino acid changes at each domain of the three ChxA toxinotypes in comparison to the prototype sequence (AY876053). The length of ChxA peptide is 634 amino acids for ChxA I and 633 for ChxA III. The peptide length of ChxA II varies from 628 to 633 amino acids. The receptor binding domain (RBD) of ChxA II with 34 to 38 amino acid changes and the catalytic domain (CD) of ChxA III with 29 to 32 amino acid changes are the most diverse domains. TD, translocation domain; UFD, unknown function domain. †, common deletion of 6 amino acids in all strains; ‡, insertion of 5 amino acids in 6 strains; ¶, deletion of 1 amino acid in all strains.
Table 3.
Summary of cytotoxicity assays of various cell lines
| Cell line | Origin | Source | Resulta |
||
|---|---|---|---|---|---|
| ChxA I | ChxA II | ChxA III | |||
| HeLa | Human | Cervical cancer | O | X | X |
| Y-1 | Mouse | Adrenocortical tumor | O | X | X |
| Int-407 | Human | Embryonic intestine | O | O | X |
| Hep-2 | Human | Epidermoid cancer | O | O | X |
| NIH-3T3 | Mouse | Embryonic fibroblast | O | O | X |
| Vero | Monkey | Kidney epithelium | O | O | X |
| Caco-2 | Human | Colorectal adenocarcinoma | Δ | Δ | X |
| CHO | Hamster | Ovary | Δ | Δ | X |
O, cytotoxic; X, non-cytotoxic; Δ, cell growth retardation.
Rabbit ileal loop assay.
Inoculation of 10 and 100 μg loop−1 of rChxA I (C9), rChxA II (Vc106), or rChxA III (Vc36) into the rabbit ileum did not cause any fluid accumulation irrespective of the ChxA type (data not shown). In a subsequent experiment, a higher dose (500 μg loop−1) of rChxA I, rChxA II, and rChxA III was used, which also could not stimulate any fluid accumulation (data not shown).
Mouse lethality assay.
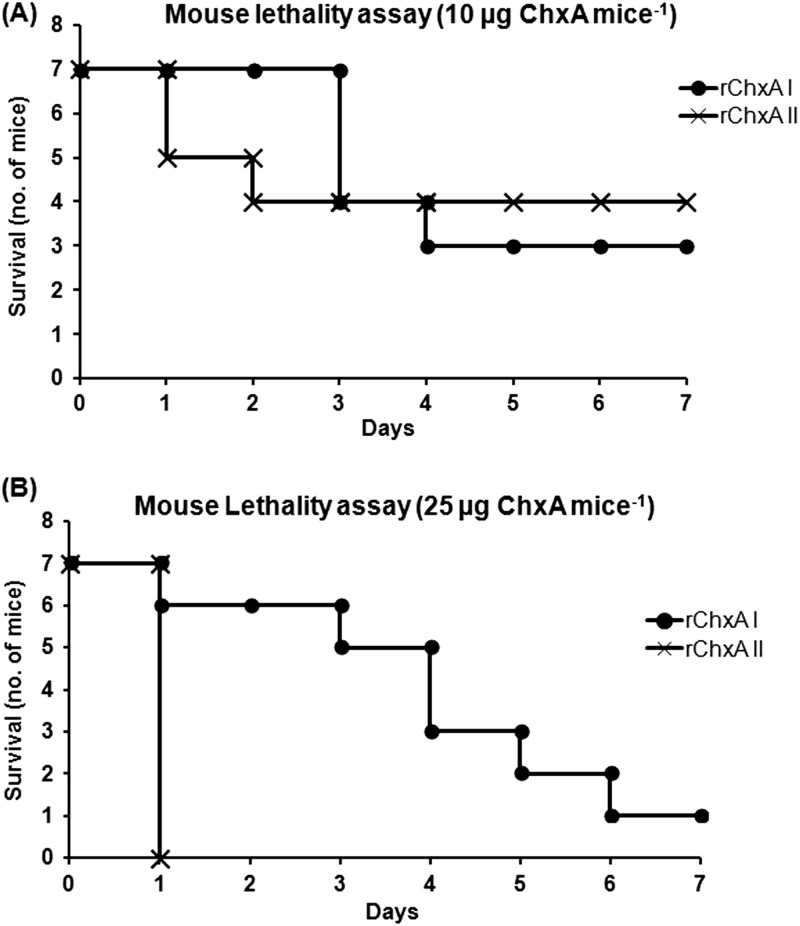
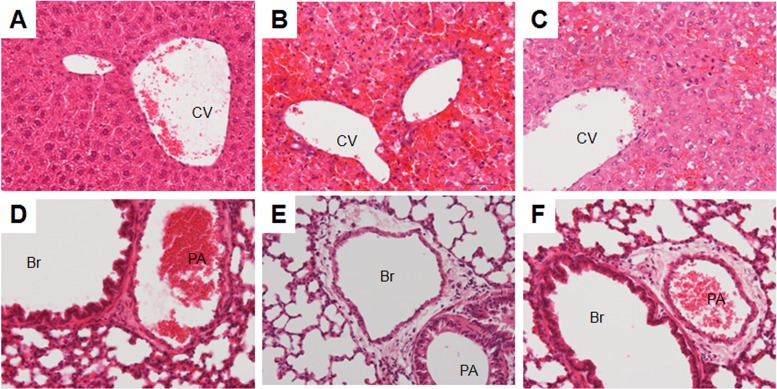
Intravenous injection of rChxA I (C9) and rChxA II (Vc106) administered to mice at a dose of 1 or 5 μg mouse−1 (50 or 250 μg kg−1) had no lethal effect, and all mice (n = 7 for each toxinotype) survived during 10 days of observation. Injection of a dose of 10 μg mouse−1 (500 μg kg−1) of rChxA I and rChxA II led to the deaths of 4 and 3 mice, respectively, of 7 during the 1-week observation period (Fig. 5). rChxA I at a dose of 25 μg mouse−1 (1.25 mg kg−1) caused the deaths of 6 mice at regular time intervals from days 1 to 7, whereas rChxA II at a similar dose killed all mice (n = 7; mortality rate, 100%) within 24 h of the injection (Fig. 5). The five major organs, viz., heart, kidney, lung, liver, and spleen, were examined for histopathology. Visual inspection of these organs clearly showed severe damage to the liver but no other organs when injected with either rChxA I or rChxA II (data not shown). Histopathology also revealed severe coagulation necrosis of the hepatocytes in mice injected with either toxinotype, suggesting that the liver could be the major target of ChxA-mediated cytotoxicity (Fig. 6). Interstitial edema with mild inflammation was also observed around the arterioles in the lungs (Fig. 6) of mice injected with either rChxA I or rChxA II. However, none of the other organs, including heart, kidney, or spleen, were damaged. The mouse lethality for rChxA III was not evaluated, as the representative rChxA III (Vc36) failed to cause any cytotoxicity or enterotoxicity.
Fig 5.
Mouse lethality assay with rChxA I and rChxA II. Specific-pathogen-free male ICR mice (n = 7, each group) with an average body weight of 20 g were intravenously injected with rChxA I and rChxA II. The survival was observed for 1 week. Survival curves with 10 μg mouse−1 (500 μg kg−1) rChxA (A) and 25 μg mouse−1 (1.25 mg kg−1) rChxA (B) are shown here.
Fig 6.
Hematoxylin-and-eosin-stained sections of mouse liver (A, B, and C) and lung (D, E, and F). (A and D) Control group. (B and E) rChxA I-treated group. (C and F) rChxA II-treated group. Severe coagulation necrosis of the hepatocytes and interstitial edema with mild inflammation around the arteriole in lungs can be observed in both the groups treated with either rChxA I or rChxA II. CV, central vein; Br, bronchiole; PA, pulmonary arteriole. Magnification, ×200.
DISCUSSION
Although V. cholerae serogroups O1 and O139, which produce CT, are clinically the most important, non-O1/non-O139 V. cholerae strains, which normally do not produce CT, are also isolated from hospitalized patients with severe diarrhea (2). There are also reports of the occurrence of toxigenic non-O1/non-O139 V. cholerae in the environment. Several virulence factors have been identified in non-O1/non-O139 V. cholerae, such as T3SS, NAG-ST, HLY, etc. (5, 7, 11). However, pathogenic mechanisms of non-O1/non-O139 V. cholerae in diarrheal diseases as well as extraintestinal infection are still unclear. Recent identification of a chxA gene, encoding ChxA, has added a new member to the group of potentially virulent genes among V. cholerae strains (14).
The present study has revealed the presence of the chxA gene among a large number (53 of 196, 27%) of non-O1/non-O139 V. cholerae strains of clinical and environmental origin (Table 1). These 53 isolates belonged to 27 diverse serogroups (see Table S1 in the supplemental material). By sequencing the entire chxA genes of 53 isolates, we further identified 2 variants of the chxA gene, designated chxA II and chxA III, besides the prototype chxA gene sequence, representing the chxA I toxinotype, reported by Jørgensen et al. (14). We have also observed high sequence diversity in chxA sequences (29 subtypes among 53 sequences). The deduced amino acid sequences revealed that the RBD of ChxA II and the CD of ChxA III (Fig. 1 and 2) were significantly diverse compared to that of the ChxA I or prototype sequence. This diversity in the RBD and CD may facilitate the targeting of various hosts by chxA-positive V. cholerae strains. The identification of at least three different toxinotypes (chxA I, chxA II, and chxA III) of this exotoxin with extensive genetic diversity gives us new insights into ChxA-mediated V. cholerae pathogenicity.
As mentioned in Results, 5 of 16 chxA II and all 4 chxA III leader gene sequences had insertions of nucleotide repeats which resulted in the incorporation of the termination codon in the chxA gene sequence (Fig. 3). This insertion should lead to nonexpression of this toxin by these strains, but the bead ELISA for ChxA revealed that all these isolates express ChxA (unpublished data). On analyzing the DNA sequence carefully, we found that a possible initiation codon (ATG) and probable ribosome binding site (RBS) were present in the leader sequences of all the chxA genes (Fig. 3). Thus, we propose that, at least for these 9 chxA genes, the ATG could be used as an initiation codon rather than CTG as documented by previous authors for the prototype sequence. This change of the initiation codon resulted in a decrease in the leader sequence polypeptide length from 32 to 22 amino acids. We also assume that the proposed ATG may act as an initiation codon for all the chxA gene sequences.
The occurrence of the chxA gene among V. cholerae strains is mostly independent of the presence of other major virulence genes, e.g., those related to CT, TCP, T3SS, NAG-ST, etc. (Table 2; see Table S1 in the supplemental material). The presence of the chxA gene among a large number of genetically and serotypically divergent non-O1/non-O139 strains isolated from patients as well as the environment and the absence of any other important virulence genes (Tables 1 and 2; see Table S1) illustrate the potential of ChxA as an important virulence factor in non-O1/non-O139 V. cholerae. Among non-O1/non-O139 V. cholerae strains, the O141 serogroup is predominantly reported as a causative agent in sporadic cases of diarrhea. The majority of O141 isolates are reported to have ctx and tcpA genes (26, 27). In this study, a V. cholerae isolate possessing multiple virulence factors, viz., ctx, vcsN2/vcsC2/vopF (T3SS), and chxA, was found to be from the O141 serogroup (see Table S1). Although the virulence profile analysis of the chxA gene-positive V. cholerae strains revealed that the existence of chxA is independent of other virulence factors, it also coexists with factors like ctx, vcsN2/vcsC2/vopF (T3SS), stn, etc. (Table 2), which are associated with enterotoxicity. The present study clearly demonstrates a strong cytotoxic effect of rChxA I on HeLa and Y1 cells but no effect of rChxA II (Table 3). Interestingly, both of the toxinotypes have similar growth retardation effects on CHO and Caco-2 cells (Table 3). On the other hand, both of the toxinotypes showed cytotoxicity on Vero, Int-407, Hep-2, and NIH-3T3 cells. rChxA III failed to cause cytotoxicity to any cell lines used in this study (Table 3). The hallmark catalytic residues (H460, Y493, Y504, E574, and E581) for ADPRT activity (14) are conserved among all the ChxA sequences observed in our study. Therefore, the variations in cytotoxicity on HeLa and Y1 cells by rChxA I and rChxA II may be due to the large amino acid differences in the RBD that reflect on their receptor recognition. To confirm this, we performed a competition assay by using HeLa cells between different ChxA toxinotypes. rChxA I-induced cytotoxicity was suppressed by rChxA III but not by rChxA II, indicating that ChxA I and ChxA III possibly share a receptor on HeLa cells whereas ChxA II may not bind to the same receptor (data not shown). The high-resolution crystal structures of ChxA I at 2.1 Å have been previously described (Protein Data Bank [PDB]: 2Q5T) (14). In silico structural analyses of ChxA II and ChxA III with ChxA I suggest that the CDs of all three ChxA toxinotypes are nearly identical, with no significant differences. Similarly, the RBDs of three ChxA toxinotypes are nearly identical; however, ChxA II has an altered β-jellyroll chain in the particular region (corresponding to amino acid residues H200 to T238 and N200 to T238 in ChxA I and ChxA II, respectively [Fig. 2]), where the primary structure is also diverse between these toxinotypes, which resulted in altered conformation of this domain. Thus, the diversity of the RBD sequence, competition assay data, and structural analysis of ChxA toxinotypes further support our assumption that varied cytotoxicity of ChxA II may be due to diversity of the RBD. However, the growth retardation effect of both rChxA I and rChxA II on CHO and Caco-2 cells and cytotoxicity on other cell lines illustrates the probable presence of an alternative mechanism to target host cells or the possession of varied receptors by these cell lines. These variable effects suggest that there may be more than one biological activity of ChxA, a possibility which demands further detailed investigations. The in silico analysis also revealed that although the CD of ChxA III is highly diverse, structurally ChxA I and ChxA III are nearly identical, with no significant differences. Moreover, the failure of ChxA III to cause cytotoxicity may not be related to its binding ability or catalytic activity because rChxA III could inhibit rChxA I-induced cytotoxicity (data not shown).
Furins are expressed in all tissues/cell lines and process latent precursor proteins into their biologically active products (28). The ExoA and DT are activated within host cells by furin cleavage (29, 30). The ChxA proteins also possess the furin cleavage site for cellular activation (14). The furin cleavage site is a 20-aa motif running from positions P14 to P6′ with a core region of 8 amino acids (P6 to P2′) which form the furin binding pocket and determine the binding strength (31). ChxA III has an altered furin binding site with a serine-to-glycine substitution at the P5 position, which is inside the furin binding pocket (P1 to P6 and P1′ to P2′). This mutation at the furin binding site hampers the proteolysis of rChxA III by furin in vitro (data not shown) and thus may repress cellular activation and subsequently translocation. This could explain the reason for the failure of ChxA III to induce cytotoxicity in the tested cell lines (Table 3).
The RIL assay is one of the most extensively used assays for the analysis of the enterotoxic potency of a particular toxin or pathogen (32–34). It was, however, observed in this study that rChxA factors failed to cause any fluid accumulation in this assay. Nevertheless, we cannot rule out the possibility that ChxA could mediate enterotoxicity in humans, as some of the chxA-positive strains used in this study are of clinical origin. ExoA is reported as an important virulence factor in P. aeruginosa infections like septicemia, lung, renal, and liver infections (35–38). The contribution of ChxA as a virulence factor to organ dysfunction has not yet been studied. The present study revealed that intravenous injection of rChxA into mice could result in their deaths (Fig. 5), probably via damage to the critical organs. Indeed, the occurrence of severe coagulation necrosis of the hepatocytes after injection of rChxA into mice (Fig. 6) suggests that the liver could be the primary target of ChxA, although interstitial edema and mild inflammation were also observed around the arterioles in the lungs of mice injected with either rChxA I or rChxA II (Fig. 6). The data obtained through the mouse lethality assay suggest that ChxA II could be more lethal than ChxA I (Fig. 5). Thus, taking together the results of the RIL and mouse lethality assays, it may be concluded that ChxA could be an important virulence factor of non-O1/non-O139 V. cholerae, which may be associated with extraintestinal infections rather than enterotoxicity. This hypothesis may be also supported by our recent isolation of a chxA-positive non-O1/non-O139 V. cholerae strain in a patient reported to have septicemia, disseminated intravascular coagulation, and multiple organ failure (unpublished data). Interestingly, this V. cholerae strain possesses only chxA as a virulence factor, and its involvement in septicemia demands detailed investigation. Thus, we can say that the infections caused by V. cholerae strains possessing chxA may be linked to systemic infections.
The present study shows the presence of a C-terminal KDEL endoplasmic reticulum (ER) retention sequence in all sequences of ChxA I and ChxA II (Fig. 1 and 2). The presence of the tetrapeptide KDEL may facilitate its comparatively low Km (3-fold lower than that of ExoA, with REDL as a C-terminal retention sequence) and higher affinity for the substrate (14). In the case of ExoA, replacement of REDL with KDEL can increase its cytotoxicity (39, 40). In that sense, ChxA can be an alternative candidate as a therapeutic immunotoxin for tumor suppression, which is expected to be more effective than ExoA. Sarnovsky et al. (41) have already constructed immunotoxin from ChxA as a possible therapy for tumor suppression, which may be of potential utility after a patient has developed neutralizing antibodies against ExoA-based immunotoxin. Interestingly, we have observed the presence of HDEL instead of KDEL among V. cholerae strains possessing ChxA III (Fig. 1 and 2). The receptor for KDEL is present mainly in animal cells, whereas the HDEL receptor is found in Saccharomyces cerevisiae and plants (42–44). Moreover, a receptor present in human cells has been shown to bind both the KDEL and HDEL sequences in vitro (45, 46). Therefore, ChxA may have functional capabilities to target diverse host systems. Like ExoA, all the ChxA sequences carry the C-terminal lysine residue (Fig. 2), and its removal from the C-terminal (K/H)DELK pentapeptide may be a prerequisite for the binding of ChxA to the receptor in the ER of host cells that ChxA invades (47).
Despite the existence of high genetic diversity among the subtypes of the chxA gene (Fig. 2; see Table S1 in the supplemental material), the GC content is almost conserved (43% to 44%) and is very close to that of the V. cholerae N16961 genome (47%) but far from that of the toxA gene (∼70%; GenBank accession no. AE004091) of P. aeruginosa. The GC content of the DT gene (42.5%; GenBank accession no. NC_002935) is close to that of the chxA gene, but a very low sequence identity and the reverse orientation of functional domains in comparison to ChxA rule out the possibility of horizontal transfer of the chxA gene from C. diphtheriae. The flanking sequence analyses of the chxA gene do not suggest the presence of any transposon- or phage-related genes and thus do not provide any known clue to its horizontal transmission (15, data not shown). Flanking sequence analysis also revealed that the insertion site of the chxA gene is the same for all the chxA-positive V. cholerae strains analyzed in this study (data not shown). Therefore, the presence of the chxA gene among a wide variety of genotypes (or serogroups) of V. cholerae (see Fig. S1 in the supplemental material) may be through vertical transmission or genetic exchange by homologous recombination from a close neighbor of the Vibrionaceae or just through capture as naked DNA in the environment by a chitin-induced DNA uptake process (48–50).
There are still a lot of undefined research queries about ChxA, including but not limited to the following. (i) What is the significance of ChxA in V. cholerae infection, whether systemic or intestinal? (ii) What are the other biological activities the toxin possesses in addition to cytotoxicity? (iii) What is the actual mode(s) of transmission of the chxA gene? (iv) What are the expression levels of ChxA in different chxA-positive V. cholerae strains?
In conclusion, chxA is prevalent among a large proportion of non-O1/non-O139 V. cholerae strains from diverse serogroups and possesses significant genetic diversity. This study reports for the first time the existence of at least three chxA toxinotypes (chxA I, chxA II, and chxA III). Among them, ChxA II and ChxA III possess a highly diverse RBD and CD, respectively, which may be attributable to their varied virulence patterns. Although none of the ChxA toxinotypes can cause enterotoxicity in rabbits, ChxA I and ChxA II can cause extensive damage to the internal organs of mice, especially the liver; this liver damage is lethal. This study suggests that ChxA I and ChxA II may be associated with extraintestinal infections rather than enterotoxicity at least in the animal model. Screening of a large number of V. cholerae strains isolated from patients with systemic infections and diarrhea for the presence of chxA would provide a better understanding of the importance of ChxA in its pathogenicity.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported, in part, by Health and Labor Sciences Research Grants for Research on global health issues from the Ministry of Health, Labor and Welfare, Japan. S.P.A., N.C., S.B.N., and H.M.G. were recipients of the Monbusho Scholarship for a Ph.D. program from the Ministry of Science, Culture and Sports of Japan.
Footnotes
Published ahead of print 10 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00982-12.
REFERENCES
- 1. WHO July 2012. Cholera. Media centre fact sheet no. 107. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs107/en/. [Google Scholar]
- 2. Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay AK, Basu A, Mitra R, Basu I, Bhattacharya SK, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair GB. 1998. Molecular analysis of Vibrio cholerae non-O1, non-O139 associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Faruque SM, Chowdhury N, Kamruzzaman M, Dziejman M, Rahman MH, Sack DA, Nair GB, Mekalanos JJ. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. U. S. A. 101:2123–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarkar B, Bhattacharya T, Ramamurthy T, Shimada T, Takeda Y, Nair GB. 2002. Preferential association of the heat-stable enterotoxin gene (stn) with environmental strains of Vibrio cholerae belonging to the O14 serogroup. Epidemiol. Infect. 129:245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh A, Saha DR, Hoque KM, Asakura M, Yamasaki S, Koley H, Das SS, Chakrabarti MK, Pal A. 2006. Enterotoxigenicity of mature 45-kilodalton and processed 35-kilodalton forms of hemagglutinin protease purified from a cholera toxin gene-negative Vibrio cholerae non-O1, non-O139 strain. Infect. Immun. 74:2937–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fasano A, Baudry B, Pumplin DW, Wasserman SS, Tall BD, Ketley JM, Kaper JB. 1991. Vibrio cholerae produces a second enterotoxin which affects intestinal tight junctions. Proc. Natl. Acad. Sci. U. S. A. 88:5242–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fasano A, Fiorentini C, Donelli G, Uzzau S, Kaper JB, Margaretten K, Ding X, Guandalini S, Comstock L, Goldblum SE. 1995. Zonula occludens toxin modulates tight junctions through protein kinase C-dependent actin reorganization, in vitro. J. Clin. Invest. 96:710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rui H, Ritchie JM, Bronson RT, Mekalanos JJ, Zhang Y, Waldor MK. 2010. Reactogenicity of live-attenuated Vibrio cholerae vaccines is dependent on flagellins. Proc. Natl. Acad. Sci. U. S. A. 107:4359–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Queen J, Satchell KJ. 2012. Neutrophils are essential for containment of Vibrio cholerae to the intestine during the proinflammatory phase of infection. Infect. Immun. 80:2905–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krueger KM, Barbieri JT. 1995. The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev. 8:34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jørgensen R, Purdy AE, Fieldhouse RJ, Kimber MS, Bartlett DH, Merrill AR. 2008. Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J. Biol. Chem. 283:10671–10678 [DOI] [PubMed] [Google Scholar]
- 15. Purdy A, Rohwer F, Edwards R, Azam F, Bartlett DH. 2005. A glimpse into the expanded genome content of Vibrio cholerae through identification of genes present in environmental strains. J. Bacteriol. 187:2992–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson BA, Collier RJ. 1992. Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: active site structure and enzymatic mechanism. Curr. Top. Microbiol. Immunol. 175:27–41 [DOI] [PubMed] [Google Scholar]
- 17. Collier RJ. 1975. Diphtheria toxin: mode of action and structure. Bacteriol. Rev. 39:54–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Nair GB, Shimada T, Takeda Y. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20:201–207 [DOI] [PubMed] [Google Scholar]
- 19. Shimada T, Arakawa T, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair GB, Albert MJ, Takeda Y. 1994. Extended serotyping scheme for Vibrio cholerae. Curr. Microbiol. 28:175–178 [Google Scholar]
- 20. Lin Z, Yamasaki S, Kurazono H, Ohmura M, Karasawa T, Inoue T, Sakamoto S, Suganami T, Takeoka T, Taniguchi Y. 1993. Cloning and sequencing of two new verotoxin 2 variant genes of Escherichia coli isolated from cases of human and bovine diarrhea. Microbiol. Immunol. 37:451–459 [DOI] [PubMed] [Google Scholar]
- 21. Asakura M, Samosornsuk W, Taguchi M, Kobayashi K, Misawa N, Kusumoto M, Nishimura K, Matsuhisa A, Yamasaki S. 2007. Comparative analysis of cytolethal distending toxin (cdt) genes among Campylobacter jejuni, C. coli and C. fetus strains. Microb. Pathog. 42:174–183 [DOI] [PubMed] [Google Scholar]
- 22. Yamasaki S, Nair GB, Bhatacharya SK, Yamamoto S, Kurazono H, Takeda Y. 1997. Cryptic appearance of a new clone of Vibrio cholerae serogroup O1 biotype El Tor in Calcutta, India. Microbiol. Immunol. 41:1–6 [DOI] [PubMed] [Google Scholar]
- 23. Yutsudo T, Nakabayashi N, Hirayama T, Takeda Y. 1987. Purification and some properties of a Vero toxin from Escherichia coli O157:H7 that is immunologically unrelated to Shiga toxin. Microb. Pathog. 3:21–30 [DOI] [PubMed] [Google Scholar]
- 24. De SN, Chatterjee DN. 1953. An experimental study of the mechanisms of action of Vibrio cholerae on the intestinal mucous membrane. J. Pathol. Bacteriol. 46:559–562 [DOI] [PubMed] [Google Scholar]
- 25. Yutsudo T, Honda T, Miwatani T, Takeda Y. 1986. Characterization of purified Shiga toxin from Shigella dysenteriae 1. Microbiol. Immunol. 30:1115–1127 [DOI] [PubMed] [Google Scholar]
- 26. Dalsgaard A, Serichantalergs O, Forslund A, Lin W, Mekalanos J, Mintz E, Shimada T, Wells JG. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39:4086–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crump JA, Bopp CA, Greene KD, Kubota KA, Middendorf RL, Wells JG, Mintz ED. 2003. Toxigenic Vibrio cholerae serogroup O141-associated cholera-like diarrhea and bloodstream infection in the United States. J. Infect. Dis. 187:866–868 [DOI] [PubMed] [Google Scholar]
- 28. Nakayama K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogata M, Chaudhary VK, Pastan I, FitzGerald DJ. 1990. Processing of Pseudomonas exotoxin by a cellular protease results in the generation of a 37,000-Da toxin fragment that is translocated to the cytosol. J. Biol. Chem. 265:20678–20685 [PubMed] [Google Scholar]
- 30. Lory S, Collier RJ. 1980. Expression of enzymic activity by exotoxin A from Pseudomonas aeruginosa. Infect. Immun. 28:494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tian S, Huajun W, Wu J. 2012. Computational prediction of furin cleavage sites by a hybrid method and understanding mechanism underlying diseases. Sci. Rep. 2:261 doi:10.1038/srep.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sedlock DM, Deibel RH. 1978. Detection of Salmonella enterotoxin using rabbit ileal loops. Can. J. Microbiol. 24:268–273 [DOI] [PubMed] [Google Scholar]
- 33. Duncan CL, Sugiyama H, Strong DH. 1968. Rabbit ileal loop response to strains of Clostridium perfringens. J. Bacteriol. 95:1560–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leitch GJ, Iwert ME, Burrows W. 1966. Experimental cholera in the rabbit ligated ileal loop: toxin-induced water and ion movement. J. Infect. Dis. 116:303–312 [DOI] [PubMed] [Google Scholar]
- 35. Hirakata Y, Furuya N, Tateda K, Kaku M, Yamaguchi K. 1993. In vivo production of exotoxin A and its role in endogenous Pseudomonas aeruginosa septicemia in mice. Infect. Immun. 61:2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doring G, Goldstein W, Roll A, Schiotz PO, Hoiby N, Botzenhart K. 1985. Role of Pseudomonas aeruginosa exoenzymes in lung infections of patients with cystic fibrosis. Infect. Immun. 49:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma S, Kaur R, Yadav V, Harjai K, Joshi K. 2004. Contribution of exotoxin A of Pseudomonas aeruginosa in acute and chronic experimental renal infection. Jpn. J. Infect. Dis. 57:119–120 [PubMed] [Google Scholar]
- 38. Forristal JJ, Thompson MR, Morris RE, Saelinger CB. 1991. Mouse liver contains a Pseudomonas aeruginosa exotoxin A-binding protein. Infect. Immun. 59:2880–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kreitman RJ, Batra JK, Seetharam S, Chaudhary VK, FitzGerald DJ, Pastan I. 1993. Single-chain immunotoxin fusions between anti-Tac and Pseudomonas exotoxin: relative importance of the two toxin disulfide bonds. Bioconjug. Chem. 4:112–120 [DOI] [PubMed] [Google Scholar]
- 40. Seetharam S, Chaudhary VK, FitzGerald D, Pastan I. 1991. Increased cytotoxic activity of Pseudomonas exotoxin and two chimeric toxins ending in KDEL. J. Biol. Chem. 266:17376–17381 [PubMed] [Google Scholar]
- 41. Sarnovsky R, Tendler T, Makowski M, Kiley M, Antignani A, Traini R, Zhang J, Hassan R, FitzGerald DJ. 2010. Initial characterization of an immunotoxin constructed from domains II and III of cholera exotoxin. Cancer Immunol. Immunother. 59:737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lewis MJ, Sweet DJ, Pelham HR. 1990. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell 61:1359–1363 [DOI] [PubMed] [Google Scholar]
- 43. Semenza JC, Hardwick KG, Dean N, Pelham HR. 1990. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61:1349–1357 [DOI] [PubMed] [Google Scholar]
- 44. Vitale A, Ceriotti A, Denecke J. 1993. The role of the endoplasmic reticulum in protein synthesis, modification and intracellular transport. J. Exp. Bot. 44:1417–1444 [Google Scholar]
- 45. Lewis MJ, Pelham HR. 1990. A human homologue of the yeast HDEL receptor. Nature 348:162–163 [DOI] [PubMed] [Google Scholar]
- 46. Wilson DW, Lewis MJ, Pelham HR. 1993. pH-dependent binding of KDEL to its receptor in vitro. J. Biol. Chem. 268:7465–7468 [PubMed] [Google Scholar]
- 47. Hessler JL, Kreitman RJ. 1997. An early step in Pseudomonas exotoxin action is removal of the terminal lysine residue, which allows binding to the KDEL receptor. Biochemistry 36:14577–14582 [DOI] [PubMed] [Google Scholar]
- 48. Bartlett DH, Azam F. 2005. Microbiology: chitin, cholera, and competence. Science 310:1775–1777 [DOI] [PubMed] [Google Scholar]
- 49. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 50. Udden SM, Zahid MS, Biswas K, Ahmad QS, Cravioto A, Nair GB, Mekalanos JJ, Faruque SM. 2008. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc. Natl. Acad. Sci. U. S. A. 105:11951–11956 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.