Abstract
Campylobacter jejuni is a natural commensal of the avian intestinal tract. However, the bacterium is also the leading cause of acute bacterial diarrhea worldwide and is implicated in development of Guillain-Barré syndrome. Like many bacterial pathogens, C. jejuni assembles complex surface structures that interface with the surrounding environment and are involved in pathogenesis. Recent work in C. jejuni identified a gene encoding a novel phosphoethanolamine (pEtN) transferase, EptC (Cj0256), that plays a promiscuous role in modifying the flagellar rod protein, FlgG; the lipid A domain of lipooligosaccharide (LOS); and several N-linked glycans. In this work, we report that EptC catalyzes the addition of pEtN to the first heptose sugar of the inner core oligosaccharide of LOS, a fourth enzymatic target. We also examine the role pEtN modification plays in circumventing detection and/or killing by host defenses. Specifically, we show that modification of C. jejuni lipid A with pEtN results in increased recognition by the human Toll-like receptor 4–myeloid differentiation factor 2 (hTLR4-MD2) complex, along with providing resistance to relevant mammalian and avian antimicrobial peptides (i.e., defensins). We also confirm the inability of aberrant forms of LOS to activate Toll-like receptor 2 (TLR2). Most exciting, we demonstrate that strains lacking eptC show decreased commensal colonization of chick ceca and reduced colonization of BALB/cByJ mice compared to wild-type strains. Our results indicate that modification of surface structures with pEtN by EptC is key to its ability to promote commensalism in an avian host and to survive in the mammalian gastrointestinal environment.
INTRODUCTION
Campylobacter jejuni is a major cause of bacterial diarrhea worldwide (1). Infection with the pathogen results in significant acute illness, as well as serious life-threatening consequences, such as Guillain-Barré syndrome (2). Like many bacterial pathogens, C. jejuni assembles complex surface structures critical for long-term commensal colonization of the avian host, as well as pathogenesis in humans. Two major surface structures, the glycolipid lipooligosaccharide (LOS) and flagella, both important for pathogenesis, are often targets for modification and/or phase variation, providing the bacteria with antigenic diversity and adaptability in inhospitable environments. For example, variation of surface-exposed LOS results in a form of molecular mimicry between bacterial surface and host peripheral nerve gangliosides, implicating them in postinfectious neuropathies (2, 3). Another example is posttranslational modification of membrane- and surface-exposed proteins targeted by the C. jejuni Pse and Pgl family of proteins responsible for O- and N-linked glycosylation, respectively. O-linked glycosylation of the flagellar filament proteins, FlaA/B, is thought to provide antigenic diversity but the role N-linked glycosylation plays in cellular processes is undetermined (4–6). Considering the importance of LOS in host-pathogen interactions, the requirement for flagellar locomotion in many pathogens, and the unknown cellular processes influenced by N-linked glycosylation, the role played by C. jejuni surface modification systems in its pathogenesis warrants further investigation.
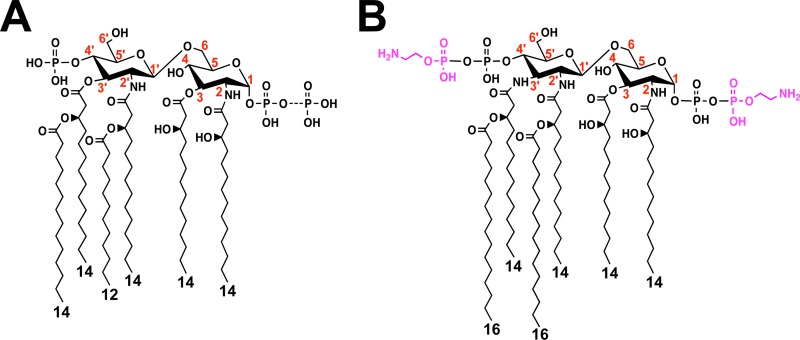
In most Gram-negative bacteria, LOS or lipopolysaccharide (LPS) is anchored to the bacterial surface by a unique lipid anchor, lipid A, whose structure is typified by the bis-phosphorylated hexa-acylated lipid A found in strains of Escherichia coli K-12 (Fig. 1A) (7). However, during transport to the surface, de novo LOS/LPS is often a target for modification, providing resistance to cationic antimicrobial peptides (CAMPs) secreted by the host and modulating detection by the host Toll-like receptor 4–myeloid differentiation factor 2 (TLR4-MD2) innate immune receptor, which is found on leukocytes and epithelial surfaces (7, 8). Recently, EptC (Cj0256) was found to catalyze the addition of a phosphoethanolamine (pEtN) residue to a variety of periplasmic targets, lipid A, the flagellar rod protein FlgG at residue Thr75, and the terminal sugar of N-linked heptasaccharides attached to several periplasmic proteins (9–11). Decoration of C. jejuni lipid A phosphate groups with pEtN residues (Fig. 1B) provided resistance to polymyxin B (PMB), a polypeptide whose mechanism of killing is thought to mimic that of CAMPs (i.e., defensins) (12). Modification of FlgG with pEtN was shown to be required for efficient flagellar production and motility (9). However, no phenotype has been attributed to pEtN modification on N-linked heptasaccharides.
Fig 1.
Chemical structures of the lipid A domains of E. coli K-12 (A) and C. jejuni (B). The dashed bonds indicate variable modifications of lipid A, and the lengths of acyl chains are shown. In wild-type E. coli, an additional phosphate group can be attached at the 1 position (49). In C. jejuni, the lipid A disaccharide backbone is modified at the 1 and 4′ positions with phosphoethanolamine (magenta), and the glucosamine disaccharide backbone can be replaced with the analogue 2,3-diamino-2,3-dideoxy-d-glucopyranose, resulting in two or three additional amide-linked acyl chains compared to E. coli (9). The numbers in red indicate the positions on the disaccharide backbone of lipid A.
LOS purified from C. jejuni shows reduced human TLR4-MD2 (hTLR4-MD2) activation compared to LPS purified from E. coli (13). These differences have been attributed to an increased number of amide-linked acyl chains at the 3 and 3′ positions of C. jejuni lipid A (Fig. 1B) (14). However, the role pEtN modification of C. jejuni lipid A plays in modulation of hTLR4-MD2 activation has not been examined. Furthermore, a variety of Gram-negative bacterial pathogens modify lipid A with pEtN, but to date, no studies have directly shown that pEtN modification of lipid A alters recognition by TLR4-MD2. Structural and experimental studies indicate the 4′-phosphate group is important for human and murine TLR4-MD2 (mTLR4-MD2) recognition of lipid A (15–17). C. jejuni eptC-deficient mutants produce a bis-phosphorylated hexa-acylated lipid A structure lacking pEtN modification at the 1 and 4′ positions (Fig. 1B), affording us the opportunity to examine the contribution of pEtN-modified lipid A to TLR4-MD2 activation (9). Considering the role of EptC in the modification of several bacterial surface structures important for virulence, the impact of pEtN modification on avian and mammalian host colonization is of interest and warranted further investigation.
In this work, we further characterize strains lacking pEtN-modified surface structures (i.e., LOS, FlgG, and N-linked glycans). Surprisingly, we identify a fourth enzymatic target for pEtN modification by EptC, the first heptose (Hep I) of the inner core region of LOS, adding to a growing list and variety of substrates. With the use of relevant avian- and human-derived CAMPs, we confirm our previous hypothesis that pEtN-modified LOS or other surface structures confer resistance to these host peptides. We demonstrate that pEtN-modified LOS contributes to increased recognition by hTLR4-MD2. Also, we show that pEtN modification of surface structures in C. jejuni is required for efficient colonization of a mammalian or avian host.
MATERIALS AND METHODS
Bacterial strains and growth.
A complete list and description of bacterial strains employed in this study can be found in Table 1. E. coli strains were grown routinely at 37°C in Luria-Bertani (LB) broth or on LB agar. C. jejuni strains were grown routinely at 37°C in Mueller-Hinton (MH) broth, on MH agar, or on tryptic soy agar (TSA) supplemented with 5% blood under microaerobic conditions.
Table 1.
Bacterial strains used in this study
| Strain | Genotype or descriptiona | Reference or source |
|---|---|---|
| E. coli | ||
| XL1 Blue | General cloning strain; recA1 endA1 gyrA96thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZM15::Tn10] Tetr | Stratagene |
| C. jejuni | ||
| 81-176 | Serotype HS: 23, 26 | S. A. Thompson |
| 81-176 eptC | 81-176 cj0256 Camr | 7 |
| 81-176 eptC eptC+ | 81-176 cj0256 astA::cj0256+ Camr Kanr | 7 |
| 81-176 waaF | 81-176 waaF Kanr | This study |
| 81-176 eptC waaF | 81-176 cj0256 waaF Camr Kanr | This study |
Antibiotics were used at the following concentrations: for E. coli cultures, 100 μg/ml ampicillin (Amp), 30 μg/ml kanamycin (Kan), 10 μg/ml tetracycline (Tet); for C. jejuni cultures, 25 μg/ml chloramphenicol (Cam) and 50 μg/ml kanamycin.
Construction of select mutants in C. jejuni.
Strains 81-176 waaF and 81-176 eptC waaF were created by transferring a previously published waaF mutation into the background strains wild-type 81-176 and 81-176 eptC, respectively (18). Genomic DNA harboring the deleted waaF allele was a generous gift from Erin Gaynor. During mutant construction, allelic replacement was achieved by natural transformation and selection on blood agar plates containing 50 μg/ml kanamycin. Confirmation was achieved by PCR of genomic DNA from isolated colonies and DNA sequencing.
LOS preparation.
LOS from C. jejuni was isolated from a 500-ml liquid culture grown under standard conditions to an A600 of ∼1.0 using the hot water-phenol method (19). Isolated LOS was further purified to remove contaminating immunostimulatory proteins (e.g., lipoproteins) using the previously described Hirschfeld method (20). Purified LOS was quantified using a Mettler Toledo XS105 Dual Range analytical balance (sensitivity ≥ 0.1 ng) and resuspended in HyPure Cell Culture Grade Endotoxin Free Water (HyClone) to a concentration of 5 mg/ml.
Capillary LC-ESI–MS-MS of LOS.
LOS samples for liquid chromatography-electrospray ionization–mass spectrometry (LC-ESI–MS) runs were prepared using LOS stocks (5 mg/ml) and diluted to a final concentration of 15 μM using a 50:50 mixture of methanol and water. All MS analysis was performed on a Bruker HCTultra ETDII 3-D ion trap mass spectrometer (Billerica, MA) with sample introduction and separation via a Dionex (Sunnyvale, CA) 3000 capillary LC system. An Agilent (Santa Clara, CA) Zorbax 300 Extend-C18 column (150 by 0.3 mm; 3.5-μm3 particle size) was used for all LC separations. Mobile phase A (MPA) consisted of a mixture of 62:36:2 methanol-water-chloroform plus 0.05% NH4OH, and mobile phase B (MPB) consisted of 80:20:2 methanol-chloroform-water plus 0.05% NH4OH. The flow rate was set to 2 μl/min, and the column was held at 40°C for the entire run. The column was initially held at 10% MPB for 4 min before applying a 50-min linear gradient ending at 30% MPB. The system was held at 30% MPB for 10 min before equilibrating the column at 10% MPB for 10 min. Injections of approximately 75 pmol were used for each sample. For all LC-MS runs, the first event was the full mass survey scan (m/z range, 600 to 2,800), followed by acquisition of multistage mass spectrometry (MS-MS) spectra for the three most abundant ions from the full mass scan. Another set of MS-MS spectra was collected for the three most abundant ions detected in the MS-MS scans. All collision-induced dissociation-MS events were acquired using a collisional energy setting of 1.25 V and an activation time of 30 ms. All solvents were high-performance liquid chromatography (HPLC) grade and were purchased from Fisher Scientific (Pittsburgh, PA), with the exception of HPLC grade 10% NH4OH in H2O, which was purchased from Sigma-Aldrich (St. Louis, MO).
Determination of MICs.
For MIC determination in liquid medium, the Hancock laboratory microtiter broth dilution method was used and modified as needed (21). C. jejuni strains were grown overnight in MH broth under standard growth conditions. The overnight cultures were inoculated into 96-well microtiter plates at a starting A600 of 0.05 in a total volume of 100 μl standard C. jejuni growth medium supplemented with a select concentration of CAMPs. The 96-well plates were incubated at 37°C in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2) with constant shaking, and the A600 of each well was determined using a Synergy-Mx monochromator-based multimode microplate reader at 24 h. Each experiment was repeated in triplicate. A positive growth control containing no CAMPs and a negative control containing no bacteria was performed with every replicate. The MIC was taken as the lowest concentration of CAMP that reduced growth (A600) by more than 50% compared to the positive growth control. The following CAMPs were purchased and used in the MIC experiment: polymyxin B (Sigma), human β defensin 2 (Phoenix Pharm), human cathelicidin LL-37 (Anaspec), histatin 5 P-113 (Sigma), and chicken gallinacin 6 (Gal-6) (Biomatik). All peptides were stored according to the manufacturers' instructions.
TLR signaling assay.
For TLR4 and TLR2, human epithelial kidney HEK-293 cells stably cotransfected with mTLR4 or hTLR4, mMD2 or hMD2, and mCD14 or hCD14 (designated HEK-m/hTLR4) or hTLR2 and hCD14 (designated HEK-hTLR2) were purchased from InvivoGen. All cell lines stably express secreted embryonic alkaline phosphatase (SEAP) under the control of a promoter inducible by NF-κB and activator protein 1 (AP-1). Thus, stimulation of m/hTLR4-MD2 or hTLR2 results in an amount of extracellular SEAP in the supernatant that is proportional to the level of NF-κB induction. All cell lines are commercially available from InvivoGen and were put through rigorous quality control and validation to ensure that extracellular SEAP did not influence TLR4-MD2 activation or the LOS structure. The cell lines were maintained in standard Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) (Gibco) supplemented with 4.5 g/liter glucose, 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1× HEK-Blue selection (InvivoGen) in a 5% saturated CO2 atmosphere at 37°C.
The induction of TLR signaling in HEK-m/hTLR4 and HEK-hTLR2 cell lines was assessed by measuring SEAP activity using a Quanti-Blue colorimetric assay (InvivoGen). The assay was performed according to the manufacturer's protocols. Briefly, cells were seeded in a 96-well plate in triplicate (2.5 × 104 cells/well for HEK-m/hTLR4 and 5 × 104 cells/well for HEK-hTLR2) in the presence or absence of 10-fold dilutions of purified LPS or bacterial cells grown under standard conditions. Additionally, for whole-cell experiments, cells were pelleted, washed in phosphate-buffered saline (PBS), and inoculated into each well based on the A600. CFU were confirmed by plating serial dilutions on MH agar. The antibiotic erythromycin (50 μg/ml) was included to prevent bacterial growth during incubation. Controls included Rhodobacter sphaeroides LPS (TLR4 antagonist; InvivoGen), E. coli W3110 LPS (TLR4 agonist), and the synthetic triacylated lipoprotein Pam3CSK4 (TLR2 agonist; InvivoGen). After 20 to 24 h of incubation, the supernatants (20 μl) were transferred to a 96-well plate and incubated at 37°C with Quanti-Blue (180 μl) for 1 to 2 h. SEAP activity was measured by reading the optical density at 655 nm with a Synergy-Mx multimode microplate reader (BioTek).
Chick colonization experiments.
The abilities of select C. jejuni 81-176 strains to colonize the ceca of chicks after oral inoculation were determined as previously described (22). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 days at 37.8°C with appropriate humidity and rotation in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12 to 36 h after hatching, the chicks were orally infected with 100 μl of PBS containing approximately 1 × 102 CFU of a single wild-type or mutant C. jejuni strain. To prepare them for infection, C. jejuni strains were suspended from plates after growth at 37°C under microaerobic conditions and diluted in PBS to obtain the appropriate inoculum for oral gavage of the chicks. Dilutions of the inocula were spread on MH agar to determine the number of bacteria in each inoculum. Seven days postinfection, the chicks were sacrificed, and the cecal contents were recovered and suspended in PBS. Serial dilutions were spread on MH agar containing trimethoprim (10 μg/ml) and cefoperazone (30 μg/ml). Bacteria were grown for 72 h at 37°C under microaerobic conditions and then counted to determine the number of CFU per gram of cecal contents. The mean of the colonization densities for each experimental group was calculated, and the statistical significance was determined by the Mann-Whitney statistical test using Prism software (GraphPad Software, San Diego, CA).
Mouse colonization experiments.
The abilities of select C. jejuni 81-176 strains to colonize mice after oral inoculation were determined as previously described (23, 24). Briefly, wild-type, eptC, and eptC eptC+ C. jejuni (81-176) strains were administered by oral gavage to BALB/cByJ mice (Jackson Laboratory) at a dose of 1 × 109 CFU. After the indicated number of days, C. jejuni bacteria shed in fecal pellets were enumerated on MH agar plates containing cephaperazone (20 μg/ml), vancomycin (10 μg/ml), and amphotericin B (2 μg/ml) to determine the total numbers of bacteria and reported as CFU/g of fecal material. Variation in the animal numbers within each cohort occurred, because mice became ill or died during the course of an experiment and therefore were excluded from the data set. The number of mice used in each cohort (5 to 7) was sufficient to obtain solid statistical comparisons. The mean of colonization densities for each experimental group was calculated, and statistical significance was determined by the Mann-Whitney statistical test using Prism software (GraphPad Software, San Diego, CA).
RESULTS
EptC catalyzes the addition of phosphoethanolamine to the first heptose sugar (Hep I) of the inner core oligosaccharide of C. jejuni LOS.
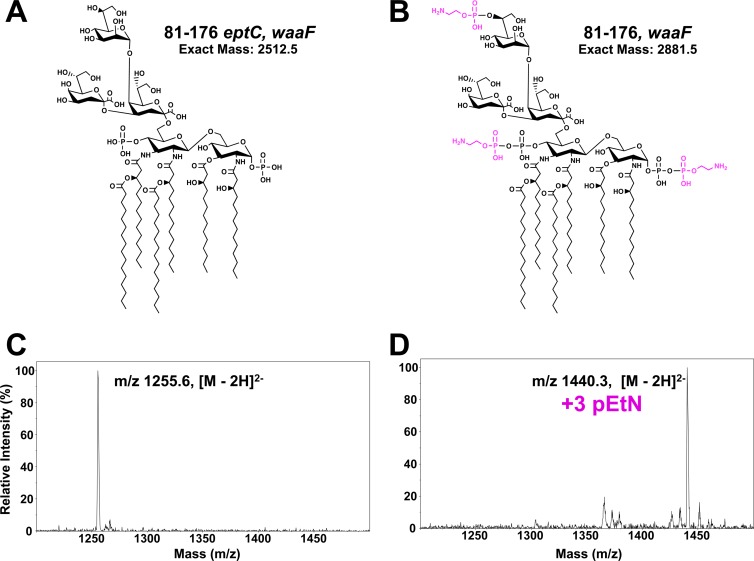
Several organisms, including Salmonella enterica, E. coli, and Neisseria meningitidis, modify their LPSs with pEtN moieties (25–28). The pEtN transferases of these organisms appear to be selective for a single target modifying a sugar in the core oligosaccharide of LPS. Structural studies of the C. jejuni LOS structure revealed a pEtN attached to heptose I of LOS (18). Considering the promiscuous nature of EptC in modifying several surface structures and the fact that the C. jejuni genome contains no other pEtN transferase homologs, we hypothesized that EptC may also modify the inner core of Campylobacter LOS. To test this hypothesis, we transferred a previously published waaF mutation (18) into the background strains wild-type 81-176 and 81-176 eptC, creating 81-176 waaF and 81-176 eptC waaF, respectively. WaaF is the ADP-heptose-LPS heptosyltransferase II responsible for the addition of the second heptose (Hep II) of the C. jejuni inner core. Deletion of waaF results in a truncated form of LOS consisting of lipid A, two 3-deoxy-d-manno-octulosonic acid sugars (Kdo2), and a single heptose (Hep I) (Fig. 2B) (18). Since structural analysis of C. jejuni LOS can be difficult due to the heterogeneity of the core sugar composition, use of waaF mutants greatly simplifies our analysis.
Fig 2.
LC-ESI mass spectra of LOS of C. jejuni strains. (A and B) Proposed structures of the LOS from C. jejuni 81-176 eptC waaF (A) and C. jejuni 81-176 waaF (B). (C and D) LCI-ESI mass spectra of LOS purified from C. jejuni 81-176 eptC waaF (C) and C. jejuni 81-176 waaF (D). The LOS was characterized by LC-ESI–MS in negative mode using collision-induced dissociation as the ion activation method to generate fragmentation ions. Analysis of LOS from strain 81-176 eptC waaF and 81-176 waaF revealed dominant doubly charged molecular ions of 1,255.6 and 1,440.3 [M-2H]2−, respectively. The observed masses are consistent with the proposed structures in panels A and B, and the difference between the dominant molecular ions (184.7 [M-2H]2−) is consistent with the addition of three pEtNs to the LOS in strain 81-176 waaF. The results agree between at least two experimental replicates of a single biological sample.
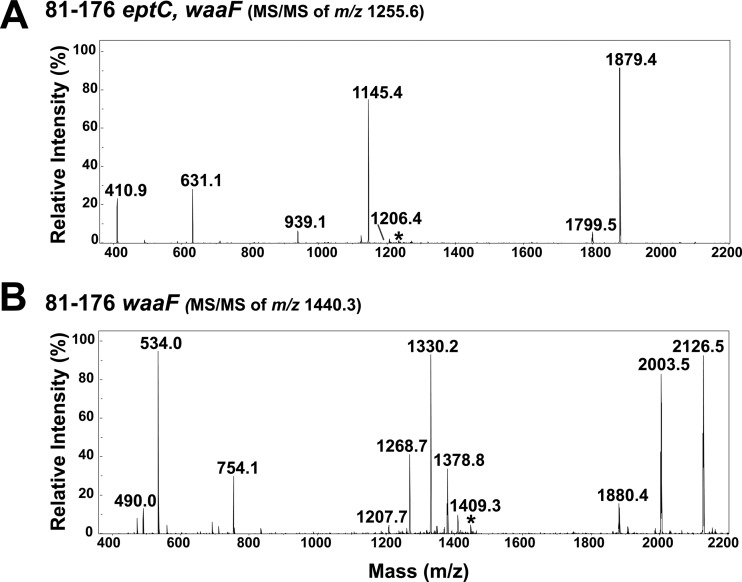
LOS purified from strains 81-176 waaF and 81-176 eptC waaF was characterized by capillary LC-ESI–MS-MS in the negative mode using collision-induced dissociation. The results of LC-ESI–MS of C. jejuni strains 81-176 waaF and 81-176 eptC waaF are shown in Fig. 2. Analysis of LOS purified from 81-176 eptC waaF (Fig. 2C) and waaF (Fig. 2D) revealed prominent unmodified and triply pEtN-modified species, respectively, consistent with the expected molecular weights of the proposed structures (Fig. 2A and B). The mass shift difference (369 Da) between the dominant LOS species in Fig. 2C and D is consistent with the addition of three pEtNs to the LOS of C. jejuni. MS-MS and multistage MS-MS-MS (MS3) experiments confirmed the proposed C. jejuni lipid A core structures; the spectra are shown in Fig. 3 and Fig. S1 in the supplemental material for unmodified 81-176 eptC waaF LOS (Fig. 3A) and pEtN-modified 81-176 waaF (Fig. 3B) LOS, respectively. Figures S2 and S3 in the supplemental material show each LOS with the proposed cleavage points that lead to the diagnostic fragment ions. Upon collisional activation, the unmodified 81-176 eptC waaF LOS yields several characteristic fragment ions with two of the most prominent (observed at m/z 631.1 and 1,879.4 in Fig. 3A and Fig. S2 in the supplemental material) arising from cleavage of the Kdo-lipid A glycosidic bond, releasing the truncated inner core domain [m/z 631.1 (1−) singly deprotonated] and the lipid A moiety (m/z 1,879.4 if singly charged or 939.1 if doubly charged). The other dominant ion (m/z 1,145.4) observed in Fig. 3A and Fig. S2 in the supplemental material is consistent with the loss of one Kdo sugar. Subsequent collisional activation of this key ion of m/z 1,145.4 results in the MS3 spectrum shown in Fig. S1A in the supplemental material, which also exhibits the same Kdo-lipid A glycosidic bond cleavage noted for the intact LOS, generating fragment ions of m/z 410.8 and 1,879.4. The fragment ion of m/z 410.8 corresponds to the remaining two sugars of the LOS (i.e., Hep I plus Kdo).
Fig 3.
LC-ESI–MS-MS of precursor ion m/z 1,255.6 (2−) from 81-176 eptC waaF (A) and precursor ion m/z 1,440.3 (2−) from 81-176 waaF (B) LOS. (A) Upon collisional activation, unmodified precursor ion m/z 1,255.6 (81-176 eptC waaF) yields several characteristic fragment ions, with the two most prominent, m/z 631.1 and 1,879.4, arising from cleavage of the Kdo-lipid A glycosidic bond, releasing the truncated inner core domain and the lipid A moiety. The other dominant ion observed, m/z 1,145.4, is consistent with the loss of one Kdo sugar. All ion fragments lack pEtN modification. (B) Upon collisional activation, pEtN-modified precursor ion m/z 1,440.3 (81-176 waaF) yields several characteristic fragment ions, m/z 1,378.8, 1,330.2, and 1,268.7, arising from the loss of one pEtN moiety, the loss of one Kdo group, or the loss of one pEtN and one Kdo group, respectively. The ions of m/z 2,126.5 and 754.1 are consistent with a lipid A-Kdo glycosidic cleavage, similar to that seen in unmodified LOS but modified with pEtN. The ion of m/z 533.9 confirms that one of the pEtN modifications remains on the heptose-plus-Kdo sugar substructure of the inner core domain, according to the proposed fragmentation pathway (see Fig. S3 in the supplemental material). The selected precursor ions are labeled with asterisks. The results agree between at least two experimental replicates of a single biological sample.
The fragmentation patterns for the triply pEtN-modified 81-176 waaF LOS are shown in Fig. 3B and Fig. S1B in the supplemental material, and the companion fragmentation map is shown in Fig. S3 in the supplemental material. The series of prominent fragment ions of m/z 1,378.8, 1,330.2, and 1,268.7 in Fig. 3B arise via loss of one pEtN moiety, loss of one Kdo group (analogous to the same loss observed for the unmodified 81-176 eptC waaF LOS), or loss of one pEtN and one Kdo group. The ions of m/z 754.1 and 2,126.5 are complementary, meaning that a single glycosidic cleavage yields both ions, analogous to the m/z 631.1 and 1,879.4 pair observed for the unmodified LPS (see Fig. S3 in the supplemental material). The ion of m/z 1,330.2, which is postulated to develop upon the loss of one Kdo sugar (a pathway illustrated in Fig. S3 in the supplemental material), was isolated and subjected to a second stage of collisional activation, yielding the MS3 spectrum shown in Fig. S1B in the supplemental material. Two of the key fragment ions, m/z 533.9 and 2,126.3, are directly analogous to the complementary pair observed in Fig. S1A in the supplemental material (m/z 410.8 and 1,879.4) and correspond to the same glycosidic-bond cleavage. Most importantly, the ion of m/z 533.9 confirms that one of the pEtN modifications remains on the (Hep I plus Kdo) substructure of the inner core domain, whereas the other two pEtN groups remain on the lipid A structure. Together, the combination of MSn and comparison of the fragmentation patterns of the unmodified 81-176 eptC waaF and triply pEtN-modified 81-176 waaF LOS allow confident determination that the inner core domain is modified by one pEtN, whereas two pEtN groups are attached to the lipid A portion. These data confirm an additional enzymatic target for pEtN modification by EptC.
Phosphoethanolamine addition to C. jejuni surface structures by EptC results in resistance to relevant avian and mammalian CAMPs.
Resistance to CAMPs is of great importance to all commensal and pathogenic bacteria for survival in a host. In order to persist within the digestive tract of chickens and survive the transition to a human gastrointestinal (GI) environment, C. jejuni must be resistant to CAMPs found in both environments. Previous work from our laboratory demonstrated that C. jejuni was highly resistant to PMB, a polypeptide whose mechanism of killing is thought to mimic that of CAMPs (i.e., defensins), with a MIC for wild-type strains of ∼18.0 μg/ml (9). Furthermore, we demonstrated that addition of pEtN to the lipid A component of LOS by EptC was a mechanism used by C. jejuni to resist the antimicrobial activity of PMB (9). However, it was not determined if modification of LOS with pEtN by EptC actually provided resistance to relevant CAMPs found in the human and avian GI tracts.
For this study, we chose a number of peptides found throughout the human and avian GI tracts, including (i) human cathelicidin LL-37, produced by both leukocytes and epithelial cells; (ii) human β-defensin 2 (HBD-2), found throughout the GI tract; (iii) P-113, a fragment of human histatin 5 found within the oral cavity; and (iv) the chicken Gal-6, a β6-defensin produced in the digestive tract of chickens (29, 30). To determine the MICs, a standard microtiter broth dilution method was utilized. As expected, C. jejuni 81-176 wild type was highly resistant to all CAMPs tested (Table 2). In contrast, the MICs of all human and avian CAMPs examined for the eptC mutant showed significant decreases (2.4- to 23-fold) (Table 2). CAMP resistance was restored to wild-type levels in the complemented eptC eptC+ strain (Table 2). Previous MICs were determined using Polymyxin B Etest strips (bioMérieux) on agar plates. The PMB MIC using the microtiter broth dilution method was lower than previous determinates using the Etest strips, but the fold changes when comparing the wild type and the eptC mutants were similar (∼20-fold decrease), validating our results. These findings confirm that modification of surface structures with pEtN by EptC is essential for resistance to CAMPs found in the human and avian hosts.
Table 2.
MICs of cationic antimicrobial peptides against C. jejuni strains
| Background (81-176) | MIC (μg/ml)a |
||||
|---|---|---|---|---|---|
| PMB | HBD-2 (hβ-Def) | LL-37 | P-113 | GAL-6 (cβ-Def) | |
| Wild type | 11.7 ± 1.2 | 48.3 ± 2.9 | 18.0 ± 1.2 | 46.7 ± 2.9 | 56.7 ± 2.9 |
| eptC | 0.5 ± 0.1 | 20.0 ± 5.0 | 2.0 ± 0.2 | 11.0 ± 2.0 | 12.1 ± 3.1 |
| eptC eptC+ | 11.7 ± 1.2 | 46.7 ± 2.9 | 18.0 ± 1.3 | 45.0 ± 0.0 | 58.3 ± 2.9 |
MICs are the averages of three independent experiments using the broth dilution method for determining MICs. hβ-Def, human β-defensin; cβ-Def, chicken β-defensin.
Phosphoethanolamine addition to C. jejuni LOS by EptC results in attenuated human TLR4-MD2 activation and differential recognition by murine TLR4-MD2.
The ability of LPS/LOS from a variety of organisms to activate hTLR4-MD2 has been extensively studied. It is well documented that Gram-negative bacterial pathogens can alter the basic hexa-acylated bis-phosphorylated lipid A structure, like that found in E. coli K-12 (Fig. 1A), in an attempt to circumvent detection and clearance from the host (8). For example, Helicobacter pylori, a related epsilonproteobacterium implicated in the development of peptic ulcer disease, modified de novo lipid A from a hexa-acylated bis-phosphorylated form to a tetra-acylated form bearing a single pEtN residue (31–34), rendering its LPS immunologically silent (17). LOS purified from C. jejuni also shows reduced hTLR4-MD2 activation compared to LPS purified from E. coli (13). The role that pEtN modification of C. jejuni LOS plays in modulation of hTLR4-MD2 activation has not been examined. Furthermore, a variety of Gram-negative bacterial pathogens modify LOS/LPS with pEtN, but to date, no studies have directly shown that pEtN modification of LOS/LPS alters recognition by TLR4-MD2. The previously characterized C. jejuni eptC mutant lacking pEtN-modified LOS afforded us this opportunity.
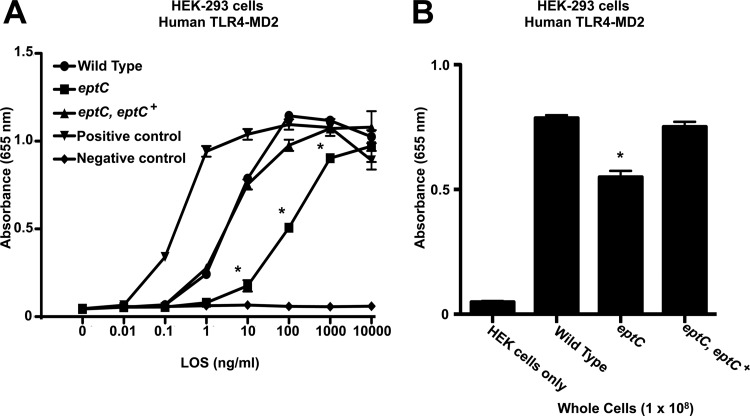
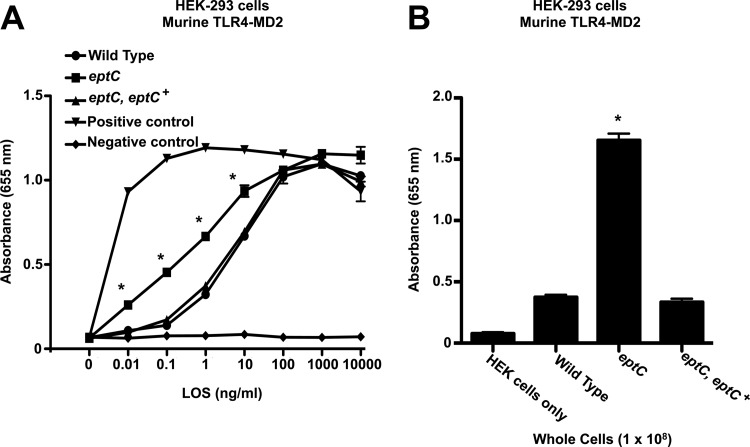
The Toll activation profiles of intact LOS were examined using samples purified using the Hirschfeld method, which allows removal of potential contaminating lipoproteins (20). Activation of TLRs was monitored using HEK-Blue 293 cells stably transfected with hTLR4 machinery and a SEAP reporter gene placed under the control of an NF-κB-inducible promoter, allowing easy detection of hTLR activation using a colorimetric assay. Compared to LOS from wild-type C. jejuni, LOS from the eptC mutant showed a significant decrease in hTLR4-MD2 activation, requiring 100-fold more LOS to reach maximal activation (Fig. 4A). hTLR4-MD2 activation by LOS prepared from the eptC eptC+ complemented strain was identical to that by wild-type 81-176 (Fig. 4A). These findings were confirmed using whole cells. Compared to wild-type C. jejuni or the eptC eptC+ complemented strain, the eptC mutant cells showed a significant decrease in hTLR4-MD2 activation (Fig. 4B). Titration curves for whole-cell experiments were performed and revealed results similar to those for purified LOS (data not shown). For all experiments, LPS from R. sphaeroides, a known hTLR4-MD2 antagonist (35), and E. coli were used as negative and positive controls for activation of hTLR4-MD2, respectively (Fig. 4A).
Fig 4.
Activation of human TLR4-MD2 by C. jejuni LOS and whole cells. Activation of TLR4 was monitored using HEK-293 cells stably transfected with human TLR4-MD2-CD14. TLR activation was monitored colorimetrically using a SEAP reporter gene placed under the control of an NF-κB-inducible promoter. (A) HEK-293 cells were stimulated for 24 h with the indicated ligands. Highly purified LOS from the indicated strains of C. jejuni was added to each well in triplicate at the indicated concentrations. The values are the means of results from triplicate wells ± standard deviations. Compared to the wild type, LOS from the eptC mutant showed a significant decrease in hTLR4-MD2 activation, requiring 100-fold more LOS to reach maximal activation, 100 ng/ml for the wild type versus 10,000 ng/ml for eptC. hTLR4-MD2 activation by LOS prepared from the eptC eptC+ complemented strain was identical to that of the wild type. (B) These results were confirmed using whole cells, where purified LOS was replaced with C. jejuni cells (1 × 108). Compared to the C. jejuni wild-type or eptC eptC+ strain, the eptC mutant cells showed a significant decrease in hTLR4-MD2 activation. Titration curves for whole-cell experiments were performed and revealed results similar to those for purified LOS (data not shown). (A) For all experiments, LPSs from R. sphaeroides and E. coli were used as negative and positive controls for activation of hTLR4-MD2, respectively. *, P ≤ 0.001. Statistical comparisons of mean activations were made between the wild type and select mutants (LOS/whole cells). The data were generated from three experimental replicates of a single biological replicate. However, all results agreed between at least two biological replicates.
The obvious progression of this research was to determine if pEtN modification of substrates in C. jejuni is required for colonization of various hosts. For C. jejuni, a murine model of infection has been established to investigate colonization of a mammalian host. However, it is well documented in structural and experimental studies that mTLR4-MD2 displays differential recognition of LPS compared to the activation profiles of hTLR4-MD2 (15, 16). This presents a problem for attempting to determine the role TLR4-MD2 activation plays in virulence using a murine host. For this reason, we used purified LOS from select C. jejuni strains to determine the mTLR4-MD2 recognition profile of pEtN-modified LOS. Briefly, mTLR4-MD2 activity was monitored using HEK-293 cells stably transfected with mTLR4-MD2-CD14 and quantified using an identical SEAP reporter method described above. Surprisingly, compared to wild-type 81-176, LOS from the eptC mutant showed a significant increase in mTLR4-MD2 activation (Fig. 5A), requiring 100-fold less LOS for significant activation (1 ng/ml for the wild type versus 0.01 ng/ml for eptC mutants). mTLR4-MD2 activation by LOS prepared from the eptC eptC+ complemented strain was identical to that of the wild type (Fig. 5A). Again, these results were confirmed using whole cells, where purified LOS was replaced with 108 CFU of C. jejuni (Fig. 5B). For all experiments, endotoxin-free water and LPS from E. coli were used as negative and positive controls for activation of mTLR4-MD2, respectively (Fig. 5A). LPS from R. sphaeroides is a known mTLR4-MD2 agonist and therefore was used as a negative control (15). The ability of mTLR4-MD2 to recognize a wide range of lipid A substrates (i.e., penta- and tetra-acylated lipid A) compared to hTLR4-MD2 is well documented in the literature (15, 16), but increased recognition of pEtN-modified LOS was unexpected. Considering the important roles these molecules play during infection and the reliance on the murine host as a model for human infection, investigation of both hTLR4-MD2 and mTLR4-MD2 is necessary.
Fig 5.
Activation of murine TLR4-MD2 by C. jejuni LOS and whole cells. Activation of TLR4 was monitored using HEK-293 cells stably transfected with murine TLR4-MD2-CD14. TLR activation was monitored colorimetrically using a SEAP reporter gene placed under the control of an NF-κB-inducible promoter. (A) HEK-293 cells were stimulated for 24 h with the indicated ligands. Highly purified LOS from the indicated strains of C. jejuni was added to each well in triplicate at the indicated concentrations. The values are the means of results from triplicate wells ± standard deviations. Compared to the wild type, LOS from the eptC mutant showed a significant increase in mTLR4-MD2 activation, requiring 100-fold less LOS for significant activation than the wild type, 1 ng/ml for the wild type versus 0.01 ng/ml for eptC. mTLR4-MD2 activation by LOS prepared from the eptC eptC+ complemented strain was identical to that of the wild type. (B) These results were confirmed using whole cells, where purified LOS was replaced with C. jejuni cells (1 × 108). Compared to the C. jejuni wild-type or eptC eptC+ strain, the eptC mutant cells showed a significant increase in mTLR4-MD2 activation. Titration curves for whole-cell experiments were performed and revealed results similar to those for purified LOS (data not shown). (A) For all experiments, medium and LPS from E. coli were used as negative and positive controls for activation of mTLR4-MD2, respectively. Unlike hTLR4-MD2, LPS from R. sphaeroides is a known mTLR4-MD2 agonist and was not used as a negative control. *, P ≤ 0.001. Statistical comparisons of mean activation were made between the wild type and select mutants (LOS/whole cells). The data were generated from three experimental replicates of a single biological replicate. However, all results agreed between at least two biological replicates.
hTLR2, known to recognize several conserved bacterial structures, including lipoteichoic acid and lipoproteins, has also been shown to recognize atypical and heavily modified forms of LPS (36, 37). However, the ability of aberrant forms of LPS to activate hTLR2 is controversial in the literature. Some of these discrepancies may arise because of contaminating lipoproteins in LPS preparations. We felt it was important to determine if pEtN-modified LOS from C. jejuni was able to activate hTLR2. Activation of hTLR2 was monitored using HEK-293 cells stably transfected with hTLR2 and quantified using a SEAP reporter method described above. No significant activation of hTLR2 was seen in LOS prepared from all C. jejuni strains (see Fig. S4 in the supplemental material). For each experiment, Pam3CSK4, a synthetic lipopeptide, and LPS from E. coli were used as positive and negative controls for activation of hTLR2 (see Fig. S4 in the supplemental material). Identical results were seen for activation of murine TLR2 (data not shown).
Phosphoethanolamine modifications by EptC in C. jejuni are required for efficient host colonization.
To date, nothing is known regarding participation of C. jejuni LOS modifications in host colonization. Presumably, the addition of pEtN to LOS by EptC provides an advantage through resistance to CAMPs encountered in the host environment and attenuated hTLR4-MD2 activation. Moreover, the reduction in motility in EptC mutants most likely would translate into reduced colonization, as motility and flagellum production are required for colonization of a host (22, 38). Considering our current and past findings, we felt it was important to proceed with an in vivo animal study to determine the role played by EptC in host colonization.
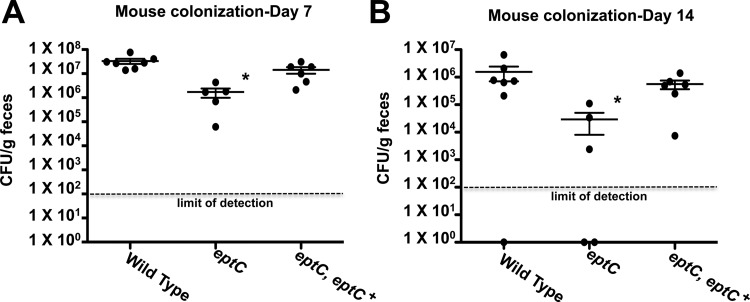
To examine the abilities of select C. jejuni strains to colonize a mammalian host, we used the BALB/cByJ intestinal-colonization model previously shown to identify colonization differences between wild-type and colonization-defective mutant strains (39, 40). Briefly, C. jejuni 81-176 wild-type, eptC, and eptC eptC+ strains were administered by oral gavage to BALB/cByJ mice (Jackson Laboratory) at a dose of 1 × 109 CFU. After 7 and 14 days, C. jejuni cells shed in fecal pellets were enumerated on agar plates and reported as CFU per gram of fecal material. The eptC-deficient mutant showed a significant decrease in colonization compared to the wild type at both 7 (Fig. 6A) and 14 (Fig. 6B) days (P < 0.05). The colonization defect was restored to wild-type levels in the eptC eptC+ complemented strain for both time points postinfection. Most interestingly, the colonization defect in the eptC mutant strain compared to the wild type is enhanced at 14 days postinfection, with 19- and 53-fold decreases in colonization at day 7 and day 14, respectively, suggesting that EptC plays an important role in persistent infection.
Fig 6.
Colonization of BALB/cByJ mice with select strains of C. jejuni. BALB/cByJ mice were infected orogastrically with the indicated strains at doses of approximately 1 × 109 CFU. After 7 (A) and 14 (B) days, C. jejuni shed in fecal pellets was determined by counting the CFU per gram of feces. Each dot represents the amount of C. jejuni recovered from a single mouse, and the mean colonization for each cohort is indicated by a horizontal bar with standard deviations. The eptC-deficient mutant showed a significant decrease in colonization compared to the wild type at both 7 and 14 days. This colonization defect was restored to wild-type levels in the eptC eptC+ complemented strain. Statistical analysis was performed using the Mann-Whitney test. *, P < 0.05. The data were generated from a single independent experiment.
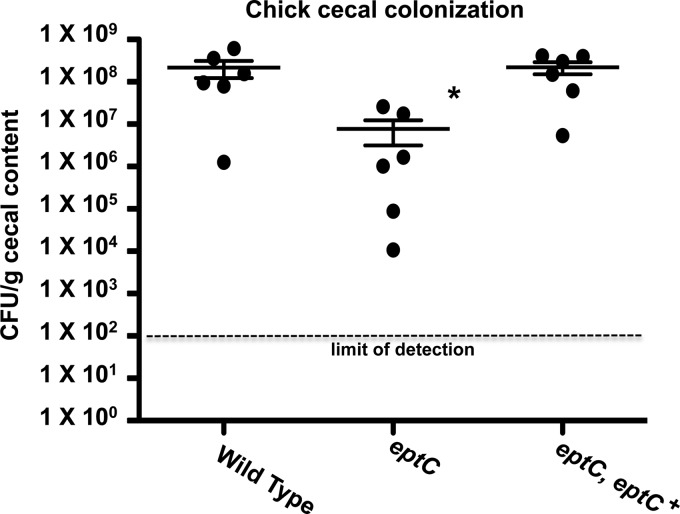
To determine whether the murine colonization defect of the eptC mutant also occurred in an avian host, the abilities of select C. jejuni 81-176 strains to colonize the ceca of chicks after oral inoculation were determined, as previously described (22). Briefly, 1-day-old chicks were orally inoculated with approximately 1 × 102 CFU of select C. jejuni strains. At 7 days postinfection, the chicks were sacrificed and the cecal contents were recovered and plated to determine the number of CFU per gram of cecal contents. The eptC-deficient mutant showed a 28-fold decrease in colonization compared to the wild type (P < 0.05) (Fig. 7). This colonization defect was restored to wild-type levels in the eptC eptC+ complemented strain (Fig. 8). Chicks colonized with EptC-deficient strains show greatly reduced commensal capacity, suggesting an important role for a pEtN-modified surface in the establishment of a C. jejuni-chicken commensal relationship.
Fig 7.
Chick commensal colonization capacities of select C. jejuni strains. One-day-old chicks were orally inoculated with approximately 1 × 102 CFU of the indicated strains (background, 81-176). Each dot represents the amount of C. jejuni recovered from the cecum of a single chick 7 days postinfection, and the mean for each cohort is indicated by a horizontal bar with standard deviations. The eptC-deficient mutant showed a significant decrease in colonization compared to the wild type. This colonization defect was restored to wild-type levels in the eptC eptC+ complemented strain. Statistical analysis was performed using the Mann-Whitney test. *, P < 0.05. The data were generated from a single independent experiment.
Fig 8.
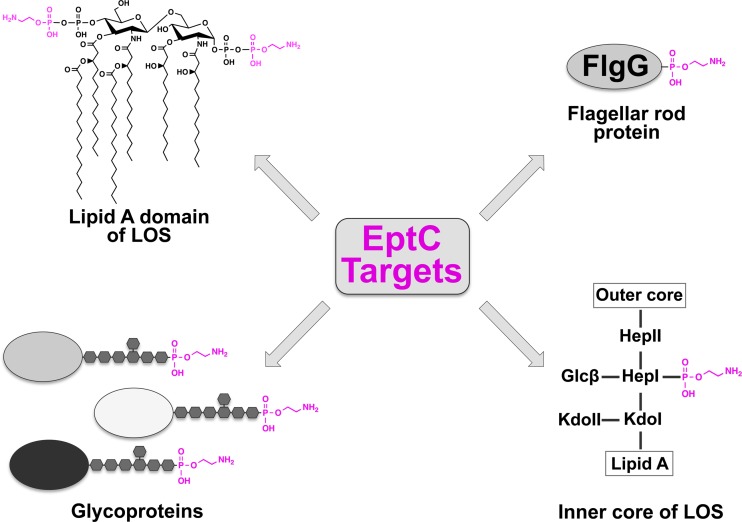
Proposed model for pEtN modifications catalyzed by EptC. The model illustrates the promiscuous enzymatic nature of EptC in the periplasmic modification of target substrates with pEtN (magenta). Glc, glucose.
DISCUSSION
Despite modern sanitation, improved food-handling procedures, and increased surveillance, C. jejuni continues to cause significant food-borne morbidity worldwide. A recent study monitoring C. jejuni contamination in retail meat samples (i.e., chicken) found contamination rates as high as 49.9% (41). To persist as a commensal organism within its avian reservoir, survive the environmental stressors encountered from farm to consumption, and proliferate within a new host, C. jejuni must adapt to an ever-changing environment. The bacterial cell surface and appendages protruding from it interface with the surrounding environment, bringing them into contact with the environment and host immunological defenses, making bacterial surface structures important for survival. To prevent detection and survive in adverse environments, bacteria have evolved elaborate systems and surface structures, many of which undergo phase variation (4, 7, 42). To date, we have examined multiple isolates of C. jejuni, and all produce pEtN-modified lipid A, suggesting that EptC is not phase variable (9). Furthermore, no variable homopolymeric tracts, a mechanism of phase variation shown to control gene expression (43), are found upstream or within the coding sequence of eptC. The mechanisms used by C. jejuni to alter its surface and circumvent detection/killing by host defenses warrant further investigation.
C. jejuni EptC is a member of a large family of proteins (COG2194) found in a number of pathogenic bacteria (10), many of which have not been characterized. Those with known functions appear to mostly target LPS (i.e., PmrC, CptA, and Lpt3) and are thought to provide resistance to CAMPs (26, 27, 44). However, several members stand out because of their enzymatic targets (i.e., EptC and PptA) (9, 42). For example, PptA, found in Neisseria spp., was shown to catalyze the transfer of pEtN and phosphocholine to serine residues of PilE, a structural subunit of the type IV pili. These multisite modifications provide structural and antigenic diversity and are involved in dissemination of N. meningitidis upon cell contact (45). Having multiple enzymatic targets, C. jejuni EptC also stands out and is quite unusual. To date, four different enzymatic targets have been identified: (i) 1- and 4′-phospate groups of lipid A, (ii) Hep I of the core oligosaccharide of LOS, (iii) the terminal sugar of N-linked heptasaccharides on several protein targets, and (iv) the flagellar rod protein FlgG at position Thr75 (Fig. 8). How EptC recognizes four different substrates and catalyzes the addition of pEtN to all of them is unknown. Furthermore, modification of lipid A and FlgG has been linked to CAMP resistance, modulation of hTLR4-MD2 activation, and efficient motility (9, 10). However, the purpose of N-linked glycan and LOS core pEtN modification is unknown and is under investigation.
LPS serves as a powerful activator of the innate immune system via TLR4-MD2. It is well documented that bacteria modify the lipid A component of LPS, attempting to modulate recognition by the TLR4-MD2 receptor. One of the best examples is the gastric pathogen H. pylori, which modifies de novo lipid A from a hexa-acylated bis-phosphorylated form to a tetra-acylated form bearing a single pEtN residue (31–33), rendering its LPS immunologically silent. Interestingly, interruption of the lipid A modification pathway in H. pylori results in increased recognition by TLR4-MD2 and the inability to colonize a murine host (17). C. jejuni LOS shows limited potential to activate hTLR4-MD2 compared to LPS purified from E. coli (Fig. 4A). This has been attributed, in part, to additional amide-linked acyl chains incorporated into the lipid A of C. jejuni (Fig. 1B) (14). Here, we show that the addition of pEtN to LOS contributes to an increase in hTLR4-MD2 activation. This finding agrees with a previous report that found a positive correlation between hTLR4-MD2 activation and the number of phosphoryl substituents (i.e., phosphate groups and/or pEtN) on the lipid A of Neisseria spp. (46). However, these experiments used LPS purified from bacteria producing mixed species of lipid A and never directly demonstrated a role for pEtN modification in hTLR4-MD2 activation. To date, core modifications have not been shown to influence recognition of and/or binding to the TLR4-MD2 receptor. Based upon structural studies of both mTLR4-MD2 and hTLR4-MD2 (16, 47), the lipid A portion of LPS/LOS is clearly the domain of the molecule important for immune recognition. However, our findings do not rule out the possibility that pEtN-modified core structures influence mTLR4-MD2 or hTLR4-MD2 signaling. The role that increased hTLR4-MD2 activation by pEtN-modified LOS plays in the pathogenesis of C. jejuni is unknown. Recently, it was demonstrated that chickens, the commensal reservoir for C. jejuni, have TLR orthologs similar to those found in humans (13). Like mTLR4-MD2, chicken TLR4-MD2 showed differential recognition of LOS purified from C. jejuni (13) compared to the recognition profile of hTLR4-MD2. Considering the commensal relationship formed between C. jejuni and chickens, the role pEtN-modified LOS plays in the recognition by chicken TLR4-MD2 is of interest and is under investigation.
The exact reason for decreased colonization in both a murine and an avian model is difficult to determine due to the number of targets modified with pEtN by EptC. Moreover, the number of phenotypes influenced by the activity of EptC (i.e., CAMP resistance, motility, and TLR4-MD2 modulation) limits our ability to properly pinpoint the exact cause of decreased colonization. For example, motility and flagellum production are required for colonization of murine and avian hosts by C. jejuni (22, 48). Furthermore, loss of CAMP resistance in C. jejuni strains resulting from LOS core truncations also resulted in a reduction in colonization (18). EptC-deficient strains show a reduction in motility and impaired flagellum production, as well as loss of CAMP resistance, which likely act in concert to influence reduced colonization rates. A role for increased hTLR4-MD2 activation by pEtN-modified LOS in the colonization of a host is even more complicated due to the differential (i.e., decreased) activation of mTLR4-MD2. Given that all C. jejuni strains we have examined produce pEtN-modified lipid A (9), the need for resistance to CAMP is most likely of greater importance than increased detection by hTLR4-MD2 within the host. Adding to the complexity of interpretation is the loss of pEtN-modified N-linked glycans and Hep I of core LOS, both of which could influence a variety of host cellular processes and virulence phenotypes. In conclusion, this study shows that EptC influences several surface structures and virulence phenotypes involved in host-pathogen interactions important for survival in a host. In light of these findings, more research is needed to identify the functional and regulatory roles that EptC and orthologous enzymes in a variety of pathogens play in bacterial pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) (grants AI064184 and AI76322 to M.S.T., AI058284 and AI055715 to S.A.T., GM103655 to J.S.B., and AI065539 to D.R.H.), the Welch Foundation (grant F1155 to J.S.B.), the Army Research Office (grant 61789-MA-MUR to M.S.T.), and the USDA (National Research Initiative Grant 2009-35201-05039 to D.R.H.).
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01046-12.
REFERENCES
- 1. Samuel MC, Vugia DJ, Shallow S, Marcus R, Segler S, McGivern T, Kassenborg H, Reilly K, Kennedy M, Angulo F, Tauxe RV. 2004. Epidemiology of sporadic Campylobacter infection in the United States and declining trend in incidence, FoodNet 1996–1999. Clin. Infect. Dis. 38(Suppl. 3):S165–S174 [DOI] [PubMed] [Google Scholar]
- 2. Ang CW, Laman JD, Willison HJ, Wagner ER, Endtz HP, De Klerk MA, Tio-Gillen AP, Van den Braak N, Jacobs BC, Van Doorn PA. 2002. Structure of Campylobacter jejuni lipopolysaccharides determines antiganglioside specificity and clinical features of Guillain-Barre and Miller Fisher patients. Infect. Immun. 70:1202–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Szymanski CM, Michael FS, Jarrell HC, Li J, Gilbert M, Larocque S, Vinogradov E, Brisson JR. 2003. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278:24509–24520 [DOI] [PubMed] [Google Scholar]
- 4. Szymanski CM, Logan SM, Linton D, Wren BW. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11:233–238 [DOI] [PubMed] [Google Scholar]
- 5. Ewing CP, Andreishcheva E, Guerry P. 2009. Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191:7086–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76:295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trent MS, Stead CM, Tran AX, Hankins JV. 2006. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12:205–223 [DOI] [PubMed] [Google Scholar]
- 9. Cullen TW, Trent MS. 2010. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U. S. A. 107:5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cullen TW, Madsen JA, Ivanov PL, Brodbelt JS, Trent MS. 2012. Characterization of unique modification of flagellar rod protein FlgG by Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J. Biol. Chem. 287:3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott NE, Nothaft H, Edwards AV, Labbate M, Djordjevic SP, Larsen MR, Szymanski CM, Cordwell SJ. 2012. Modification of the Campylobacter jejuni N-linked glycan by EptC-mediated addition of phosphoethanolamine. J. Biol. Chem. 287:29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Zoete MR, Keestra AM, Roszczenko P, van Putten JP. 2010. Activation of human and chicken Toll-like receptors by Campylobacter spp. Infect. Immun. 78:1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Mourik A, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, van Putten JP, Wosten MM. 2010. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J. Biol. Chem. 285:15828–15836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. 2002. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 3:354–359 [DOI] [PubMed] [Google Scholar]
- 16. Ohto U, Fukase K, Miyake K, Shimizu T. 2012. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl. Acad. Sci. U. S. A. 109:7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. 2011. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 7:e1002454 doi:10.1371/journal.ppat.1002454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naito M, Frirdich E, Fields JA, Pryjma M, Li J, Cameron A, Gilbert M, Thompson SA, Gaynor EC. 2010. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192:2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westphal O, Jann K. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure, p 83–91 In Whistler RL. (ed), Methods in carbohydrate chemistry, vol 5 Academic Press, Inc., New York, NY [Google Scholar]
- 20. Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J. Immunol. 165:618–622. [DOI] [PubMed] [Google Scholar]
- 21. Hancock REW. 19 September 1999, posting date. Hancock Laboratory methods: modified MIC method for cationic antimicrobial peptides. Department of Microbiology and Immunology, University of British Columbia, Vancouver, Canada: http://www.cmdr.ubc.ca/bobh/methods.htm [Google Scholar]
- 22. Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471–484 [DOI] [PubMed] [Google Scholar]
- 23. Baqar S, Bourgeois AL, Schultheiss PJ, Walker RI, Rollins DM, Haberberger RL, Pavlovskis OR. 1995. Safety and immunogenicity of a prototype oral whole-cell killed Campylobacter vaccine administered with a mucosal adjuvant in non-human primates. Vaccine 13:22–28 [DOI] [PubMed] [Google Scholar]
- 24. Pajaniappan M, Hall JE, Cawthraw SA, Newell DG, Gaynor EC, Fields JA, Rathbun KM, Agee WA, Burns CM, Hall SJ, Kelly DJ, Thompson SA. 2008. A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol. Microbiol. 68:474–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reynolds CM, Kalb SR, Cotter RJ, Raetz CR. 2005. A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem. 280:21202–21211 [DOI] [PubMed] [Google Scholar]
- 26. Tamayo R, Choudhury B, Septer A, Merighi M, Carlson R, Gunn JS. 2005. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J. Bacteriol. 187:3391–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mackinnon FG, Cox AD, Plested JS, Tang CM, Makepeace K, Coull PA, Wright JC, Chalmers R, Hood DW, Richards JC, Moxon ER. 2002. Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol. 43:931–943 [DOI] [PubMed] [Google Scholar]
- 28. Wright JC, Hood DW, Randle GA, Makepeace K, Cox AD, Li J, Chalmers R, Richards JC, Moxon ER. 2004. lpt6, a gene required for addition of phosphoethanolamine to inner-core lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae. J. Bacteriol. 186:6970–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Diamond G, Beckloff N, Weinberg A, Kisich KO. 2009. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 15:2377–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Dijk A, Veldhuizen EJ, Kalkhove SI, Tjeerdsma-van Bokhoven JL, Romijn RA, Haagsman HP. 2007. The beta-defensin gallinacin-6 is expressed in the chicken digestive tract and has antimicrobial activity against food-borne pathogens. Antimicrob. Agents Chemother. 51:912–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tran AX, Karbarz MJ, Wang X, Raetz CR, McGrath SC, Cotter RJ, Trent MS. 2004. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J. Biol. Chem. 279:55780–55791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stead CM, Zhao J, Raetz CR, Trent MS. 2010. Removal of the outer Kdo from Helicobacter pylori lipopolysaccharide and its impact on the bacterial surface. Mol. Microbiol. 78:837–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stead CM, Beasley A, Cotter RJ, Trent MS. 2008. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. J. Bacteriol. 190:7012–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. 2006. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J. Bacteriol. 188:4531–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qureshi N, Takayama K, Kurtz R. 1991. Diphosphoryl lipid A obtained from the nontoxic lipopolysaccharide of Rhodopseudomonas sphaeroides is an endotoxin antagonist in mice. Infect. Immun. 59:441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Erridge C. 2010. Endogenous ligands of TLR2 and TLR4: agonists or assistants? J. Leukoc. Biol. 87:989–999 [DOI] [PubMed] [Google Scholar]
- 37. Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, Saint Girons I, Haake DA, Godowski PJ, Hayashi F, Ozinsky A, Underhill DM, Kirschning CJ, Wagner H, Aderem A, Tobias PS, Ulevitch RJ. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat. Immunol. 2:346–352 [DOI] [PubMed] [Google Scholar]
- 38. Wassenaar TM, van der Zeijst BA, Ayling R, Newell DG. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171–1175 [DOI] [PubMed] [Google Scholar]
- 39. Pei Z, Burucoa C, Grignon B, Baqar S, Huang XZ, Kopecko DJ, Bourgeois AL, Fauchere JL, Blaser MJ. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66:938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baqar S, Applebee LA, Bourgeois AL. 1995. Immunogenicity and protective efficacy of a prototype Campylobacter killed whole-cell vaccine in mice. Infect. Immun. 63:3731–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao S, Young SR, Tong E, Abbott JW, Womack N, Friedman SL, McDermott PF. 2010. Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl. Environ. Microbiol. 76:7949–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hegge FT, Hitchen PG, Aas FE, Kristiansen H, Lovold C, Egge-Jacobsen W, Panico M, Leong WY, Bull Virji VM, Morris HR, Dell A, Koomey M. 2004. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. U. S. A. 101:10798–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 44. Lee H, Hsu FF, Turk J, Groisman EA. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, Imhaus AF, Martin P, Trellet M, Clary G, Chafey P, Camoin L, Nilges M, Nassif X, Dumenil G. 2011. Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331:778–782 [DOI] [PubMed] [Google Scholar]
- 46. Liu M, John CM, Jarvis GA. 2010. Phosphoryl moieties of lipid A from Neisseria meningitidis and N. gonorrhoeae lipooligosaccharides play an important role in activation of both MyD88- and TRIF-dependent TLR4-MD-2 signaling pathways. J. Immunol. 185:6974–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191–1195 [DOI] [PubMed] [Google Scholar]
- 48. Newell DG, McBride H, Dolby JM. 1985. Investigations on the role of flagella in the colonization of infant mice with Campylobacter jejuni and attachment of Campylobacter jejuni to human epithelial cell lines. J. Hyg. 95:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. 2008. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol. Microbiol. 67:264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.