Abstract
Fe protein (dinitrogenase reductase) activity is reversibly inactivated by dinitrogenase reductase ADP-ribosyltransferase (DraT) in response to an increase in the ammonium concentration or a decrease in cellular energy in Azospirillum brasilense, Rhodospirillum rubrum, and Rhodobacter capsulatus. The ADP-ribosyl is removed by the dinitrogenase reductase-activating glycohydrolase (DraG), promoting Fe protein reactivation. The signaling pathway leading to DraT activation by ammonium is still not completely understood, but the available evidence shows the involvement of direct interaction between the enzyme and the nitrogen-signaling PII proteins. In A. brasilense, two PII proteins, GlnB and GlnZ, were identified. We used Fe protein from Azotobacter vinelandii as the substrate to assess the activity of A. brasilense DraT in vitro complexed or not with PII proteins. Under our conditions, GlnB was necessary for DraT activity in the presence of Mg-ADP. The PII effector 2-oxoglutarate, in the presence of Mg-ATP, inhibited DraT-GlnB activity, possibly by inducing complex dissociation. DraT was also activated by GlnZ and by both uridylylated PII proteins, but not by a GlnB variant carrying a partial deletion of the T loop. Kinetics studies revealed that the A. brasilense DraT-GlnB complex was at least 18-fold more efficient than DraT purified from R. rubrum, but with a similar Km value for NAD+. Our results showed that ADP-ribosylation of the Fe protein does not affect the electronic state of its metal cluster and prevents association between the Fe and MoFe proteins, thus inhibiting electron transfer.
INTRODUCTION
Biological nitrogen fixation is the reduction of atmospheric N2 to NH3, catalyzed by the enzyme nitrogenase. The most common form of nitrogenase is composed of two component proteins: the molybdenum-iron (MoFe) protein (dinitrogenase, or NifDK), a 240-kDa α2β2 tetramer, and the iron (Fe) protein (dinitrogenase reductase, or NifH), a 64-kDa γ2 homodimer. The Fe protein is responsible for electron transfer to the MoFe protein in a reaction that requires the hydrolysis of two ATP to two ADP molecules and two Pi for each electron transferred. The MoFe protein contains the N2 binding/reduction active site (1). In order to avoid wasteful ATP hydrolysis, several microorganisms have acquired diverse mechanisms to inactivate nitrogenase at the posttranslational level when fixed nitrogen (e.g., ammonium) is available in the environment (2).
The best-described nitrogenase posttranslational regulatory mechanism relies on reversible ADP-ribosylation of the Fe protein. This mechanism has been extensively studied in the alphaproteobacteria Azospirillum brasilense and Rhodospirillum rubrum. Furthermore, bioinformatics analysis supports the idea that a similar mechanism operates in at least 25 bacterial genera (2). Fe protein ADP-ribosylation is catalyzed by dinitrogenase reductase ADP-ribosyltransferase (DraT). This enzyme uses NAD+ as an ADP-ribose donor to modify a conserved arginine (usually Arg101) in one of the subunits of Fe protein (3). The ADP-ribosylation inactivates the Fe protein, presumably by blocking the docking site between the Fe and the MoFe protein (4, 5), preventing the association of the Fe protein with the MoFe protein, and thus electron transfer. The ADP-ribosyl group attached to the Fe protein is removed by dinitrogenase reductase-activating glycohydrolase (DraG), which thereby promotes nitrogenase complex reactivation (6).
The DraT and DraG activities are subject to opposite regulation in vivo (7). In response to a negative stimulus, such as the presence of ammonium or a decrease in the available cell energy, DraT is activated and DraG is inactivated, promoting inactivation of nitrogenase by ADP-ribosylation of the Fe protein. Conversely, when the negative stimulus is removed, DraT is inactivated and DraG is activated, favoring ADP-ribose removal from the Fe protein and nitrogenase activity (2, 7). The signaling pathway linking the cellular nitrogen status to regulated activities of DraT and DraG in A. brasilense and R. rubrum has been extensively studied and relies on the nitrogen signal transduction proteins of the PII protein family. In contrast, the energy-signaling pathway remains unknown (2).
The PII proteins constitute one of the most widely distributed families of signal transduction proteins. These proteins regulate nitrogen metabolism by direct interaction with enzymes, transcriptional regulators, and transporters (8–10). PII proteins are homotrimers, and each monomer carries a flexible loop region, the T loop, that is a crucial element for PII function (11). The signaling status of PII proteins and their capacity to interact with their target proteins are dictated by the structural conformation of the PII T loops (9, 12).
In proteobacteria, the PII T loops are subjected to reversible uridylylation at a conserved residue (Tyr51). Uridylylation reflects the cellular availability of the nitrogen-signaling molecule glutamine, so that under low glutamine, PII is uridylylated (13). PII is also modulated through the allosteric binding of ATP, ADP, and 2-oxoglutarate (2-OG). The PII trimer carries three binding sites for each of these molecules located in the lateral clefts between subunits (11, 14, 15). ATP and ADP bind competitively to the same site on PII (16), whereas 2-OG interacts cooperatively with the bound Mg-ATP (15, 17, 18). Both the occupancy of PII by ATP, ADP, and 2-OG and the uridylylation status of PII alter the T-loop conformation and, hence, the ability of PII to interact with its target proteins (15).
In A. brasilense, two PII proteins sharing 67% identity in their amino acid sequences were detected, namely, GlnB and GlnZ (19). Under nitrogen-fixing conditions, GlnB and GlnZ are fully uridylylated and nitrogenase is active. Upon an ammonium pulse, nitrogenase is inactivated by ADP-ribosylation of the Fe protein. At the same time, GlnB and GlnZ are deuridylylated (20) and interact with DraT and DraG, respectively (21–24). The DraG-GlnZ complex is directed to the membrane by interaction with the ammonium transporter AmtB, resulting in DraG inactivation (20, 22, 24). Given that the DraT-GlnB complex is detected only under conditions where DraT is active, it was postulated that this protein-protein interaction is responsible for DraT activation; however, this has yet to be confirmed (23). The A. brasilense DraT-GlnB complex can be purified after coexpression in Escherichia coli, and the complex exhibits a stoichiometry of 1 DraT monomer to 1 GlnB trimer, is stable in the presence of Mg-ADP, and is destabilized in the presence of Mg-ATP and 2-OG (23).
Here, we used Fe protein from Azotobacter vinelandii as the substrate to analyze the activity of A. brasilense DraT in vitro. The results show that GlnB activates DraT in the presence of Mg-ADP. The DraT-GlnB complex is at least 18-fold more active than DraT alone in inactivating nitrogenase. A GlnB variant carrying a deletion in the T loop failed both to interact with and to activate DraT in vitro. These results suggest a model where DraT can be activated by interaction with the GlnB T loop.
MATERIALS AND METHODS
Materials and reagents.
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) and were used as provided unless otherwise specified.
Bacterial strains, plasmids, and protein purification.
The A. brasilense GlnBΔ42-54 and GlnZ variants were obtained using the Quick-Change site-directed mutagenesis kit (Agilent, Santa Clara, CA) with the pLH25PET (25) or pMSA4 (26) plasmid as a template, respectively. The DNA inserts of the resulting plasmids, pLH25PET H42Q, pMSA4 Q22HS52VS54D, pMSA4 S52VS54D, and pLHGlnB Δ42-54, were completely sequenced to confirm their integrity.
A. brasilense GlnB (25), GlnZ (26), GlnZQ42HS52VS54D, GlnZS52VS54D, GlnBH42Q, GlnBΔ42-54, His-tagged DraT (27), and His-tagged GlnD (28) and recombinant proteins were overexpressed in E. coli strain BL21(DE3) harboring the plasmids listed in Table 1. The PII proteins (GlnB, GlnZ, and GlnBΔ42-54) were purified using a thermal treatment at 70°C for 15 min prior to loading on the 5-ml HiTrap heparin column (GE Healthcare) (26). The PII proteins were eluted in a nonlinear KCl gradient using an Äkta system (GE Healthcare). The samples with purity over 97% as judged by SDS-PAGE were pooled, dialyzed, and stored in aliquots at −80°C until use. His-tagged GlnD was purified essentially as described previously (28) using a 1-ml HiTrap chelating HP column prepared as recommended by the manufacturer (GE Healthcare). Uridylylated GlnB and GlnZ were obtained as described previously (23). His-tagged DraT was purified using a 1-ml HiTrap chelating HP column preequilibrated with buffer A (50 mM Tris-HCl, pH 8.0). The protein was eluted in a stepwise increase of imidazole (50, 100, 200, 300, 400, and 500 mM) in buffer A, using 5 column volumes at each concentration. Soluble fractions with purity over 95% as judged by SDS-PAGE were pooled, kept in an ice bath, and used immediately. For the purification of the A. brasilense His-DraT–GlnB complex, both proteins were coexpressed, and the complex was purified according to the method of Huergo et al. (23) using a 4-ml immobilized metal ion affinity chromatography (IMAC) column. The complex was dialyzed in buffer containing 10% glycerol, followed by gel filtration chromatography on a Superdex 200 column (320 ml) using 50 mM Tris-HCl, pH 8.0, 10% glycerol, 200 mM NaCl, 1 mM MgCl2, 0.5 mM ADP as a buffer. The loaded samples consisted of aggregates of high molecular weight containing both His-tagged DraT and GlnB and a fraction of approximately 64,000 Da, corresponding to the molecular mass of 1 DraT monomer plus 1 GlnB trimer. This low-molecular-weight fraction had higher activity and was used in all subsequent analyses.
Table 1.
Plasmids
| Plasmid | Genotype | Reference |
|---|---|---|
| pLH25 Kmr (pET28a) | Expresses the A. brasilense GlnB | 25 |
| pMSA4 Kmr (pET28a) | Expresses the A. brasilense GlnZ | 26 |
| pLHPETDraT Kmr (pET28a) | Expresses the N-terminally His-tagged A. brasilense DraT | 27 |
| pLHDK5pII Ampr (pDK5) | Expresses the A. brasilense GlnB | 25 |
| pMSA4 Q42HS52VS54D Kmr (pET28a) | Expresses the A. brasilense GlnZ variant Q42HS52VS54D | This work |
| pMSA4 S52VS54D Kmr (pET28a) | Expresses the A. brasilense GlnZ variant S52VS54D | This work |
| pLH25PETGlnB H42Q Kmr (pET28a) | Expresses the A. brasilense GlnB variant H42Q | This work |
| pGlnBΔ42–54 Kmr (pET28a) | Expresses the A. brasilense GlnB carrying a T-loop deletion (Δ42-54) | This work |
| pGlnDHis Kmr (pET28a) | Expresses the N-terminally His-tagged A. brasilense GlnD | 28 |
The nitrogenase MoFe protein was purified from A. vinelandii strain DJ995 and the Fe protein from strain DJ884 based on the methods of Christiansen and Seefeldt (29, 30), respectively. The cells were grown, and both proteins were expressed. Approximately 240 g of cells (wet weight) was processed for each purification, and the crude extracts were prepared by osmotic shock using glycerol. The DJ995 strain encodes a MoFe protein variant carrying a histidine tag on the α-subunit, which allowed protein purification using metal affinity chromatography (29). Cell extract was loaded onto a Zn(II)-charged IMAC column (60 ml of resin) using a peristaltic pump. After loading the extract, the column was washed with 2 column volumes of buffer A (50 mM Tris-HCl, pH 7.9, 500 mM NaCl, 1 mM sodium dithionite) containing 20 mM imidazole. The protein that remained bound to the column was then eluted using a linear gradient of imidazole in an Äkta system (GE Healthcare). The eluted protein was collected at approximately 250 mM NaCl and diluted 8-fold in a degassed buffer (50 mM Tris-HCl, pH 8.0) containing 1 mM sodium dithionite. The diluted protein was then loaded onto a Q-Sepharose column (30 ml of resin) and eluted using a linear NaCl gradient (100 to 300 mM NaCl over 5 column volumes). DJ884 encodes a wild-type Fe protein, which was purified using standard methods (30). Cell extract was loaded onto a Q-Sepharose column (150 ml of resin) using a peristaltic pump. After loading the extract, the column was washed with 1 column volume of no-salt buffer (50 mM Tris-HCl, pH 8.0) containing 1 mM sodium dithionite. The protein that remained bound to the column was then eluted using 4 column volumes of a linear gradient of NaCl in an Äkta system (GE Healthcare). The eluted protein was collected at approximately 350 mM NaCl and concentrated in a Q-Sepharose column (40 ml of resin) by loading the protein fraction, washing with 50 ml of no-salt buffer, reversing the flow, and eluting with salt buffer (50 mM Tris-HCl, pH 8.0, 1 M NaCl) containing 2 mM sodium dithionite. The concentrated protein was then loaded onto a Sephacryl S-200 column (600 ml of resin) and eluted using a salt buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl) containing 2 mM sodium dithionite. Proteins were concentrated using an Amicon concentrator (Beverly, MA) fitted with a YM100 (for MoFe) and YM30 (for Fe) membrane. The proteins were quantified by the biuret method using bovine serum albumin as the standard, and protein purity was monitored by SDS-PAGE. The final purified proteins were pelleted and stored in liquid nitrogen until use. The Fe protein used in these experiments had purity of over 90% and specific activities for acetylene and proton reduction (30, 31) greater than 2,000 nmol of acetylene reduced or H2 evolved min−1 (mg Fe protein)−1. All protein manipulations were conducted in septum-sealed serum vials degassed under an argon atmosphere, and all anaerobic liquid and gas transfers were performed using gas-tight syringes.
DraT-PII complex formation in vitro.
Purified proteins [DraT and GlnB, GlnZ, GlnB-(UMP)3, GlnZ-(UMP)3, or GlnBΔ42-54] were incubated with buffer (50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, and 2 mM ADP) at 4°C overnight using the molar ratio indicated, considering the PII protein a trimer.
Protein pulldown assay.
In vitro protein complex formation was performed using MagneHis-Ni2+ beads (Promega Co., Madison, WI) as described previously (23). Reactions were conducted in buffer containing 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% glycerol, 20 mM imidazole, 0.05% (vol/vol) Tween 20, 5 mM MgCl2, and 1 mM ADP with 5 μg of His-tagged DraT and 10 μg of PII. After washing, loading buffer was added, and the samples were boiled for 5 min prior to 12.5% SDS-PAGE analysis. The gels were stained with Coomassie blue. The data reported were confirmed in at least two independent experiments.
DraT activity assays.
The standard assay for DraT activity was carried out using 20 mM MOPS (morpholinepropanesulfonic acid), pH 7.5, 2 mM NAD+, 1 mM ADP, 1 mM MgCl2, 100 μg of unmodified oxidized Fe protein, and the indicated amounts of DraT or the DraT-PII complex in a 500-μl reaction mixture. The oxidized Fe protein was prepared by passing the reduced Fe protein over a 3-ml G-25 Sephadex column to remove the excess dithionite, which can reduce NAD+ to NADH (32). The samples were incubated for 5 min at 130 rpm at 30°C. The DraT activity was determined indirectly by measuring the Fe protein activity based on H2 evolution, as described previously (31) with modifications. The nitrogenase activity buffer contained 200 mM MOPS, 13.4 mM MgCl2, 60 mM phosphocreatine, 10 mM ATP, 2.3 mg/ml bovine serum albumin (BSA), 0.4 mg/ml creatine phosphokinase, and 24 mM dithionite. This buffer was added to a 1-ml final volume immediately after incubation for DraT activity. The dithionite reduces the NAD+, quenching the DraT reaction. To determine Fe protein activity, MoFe protein was added in a 1:1.4 molar ratio (MoFe/Fe protein), the reaction mixtures were incubated for 8 min at 160 rpm at 30°C, and H2 evolution was measured by gas chromatography. The pattern of ADP-ribosylation was analyzed by 12% low-cross-linker SDS-PAGE as described previously (33).
The DraT assays were conducted in a small vial inside a bigger septum-sealed vial. A dithionite solution (12 mM) was used outside the reaction vial to act as an oxygen scavenger.
EPR spectroscopy.
Resting state and turnover samples of ADP-ribosylated Fe (Fe-ADPR) protein, prepared as described for the DraT activity assay, were monitored by electron paramagnetic resonance (EPR) spectroscopy essentially as described previously (34) and compared with unmodified Fe protein.
Stopped-flow spectrophotometry.
Fe protein was modified using NAD+ in a standard reaction in the presence of the DraT-GlnB complex. To remove the complex, the solution was passed over a 2-ml IMAC column in the presence of buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 0.5 mM ADP). To remove ADP interference, the sample was exchanged with the same buffer in the absence of Mg-ADP and then concentrated in an Amicon cell using a membrane with a 30,000-Da cutoff.
Electron transfer from modified Fe protein to MoFe protein was assessed by absorbance at 430 nm. Observing a change in absorbance at 430 nm is indicative of the redox state change due to one-electron oxidation of the Fe protein metal cluster. This change in absorbance was monitored over time in a Hi-Tech SF61 stopped-flow UV-visible (Vis) spectrophotometer equipped with a data acquisition and curve-fitting system. The reactions were carried out at 25°C under conditions described previously (35).
Statistical analysis.
Statistical analysis was carried out by one-tailed unpaired t test, using the Graph Pad Prism 5 program. The number of experimental replicates was at least three independent experiments. P values of less than 0.05 were considered statistically significant.
RESULTS
DraT is activated by GlnB in vitro.
We have shown previously that A. brasilense DraT interacts with deuridylylated GlnB in vivo and in vitro and that His-tagged DraT has activities comparable to those of the wild-type protein in vivo (20, 21, 23). To assess whether GlnB-DraT complex formation altered DraT activity in vitro, the activity of DraT was compared to that of the DraT-GlnB complex that was obtained either by copurification or by in vitro reconstitution using the purified proteins. DraT activity was determined indirectly by measuring nitrogenase H2 evolution using the modified Fe protein. For these assays, His-tagged DraT or the DraT-GlnB complex was preincubated with the oxidized A. vinelandii Fe protein in the presence of Mg-ADP and NAD+ under anaerobic conditions and in the absence of dithionite. After 5 min at 30°C, the ADP-ribosylation reaction was quenched by adding nitrogenase assay buffer containing excess dithionite to completely reduce NAD+. The purified A. vinelandii MoFe protein was added to the reaction mixture, and the activity was determined by measuring H2 production.
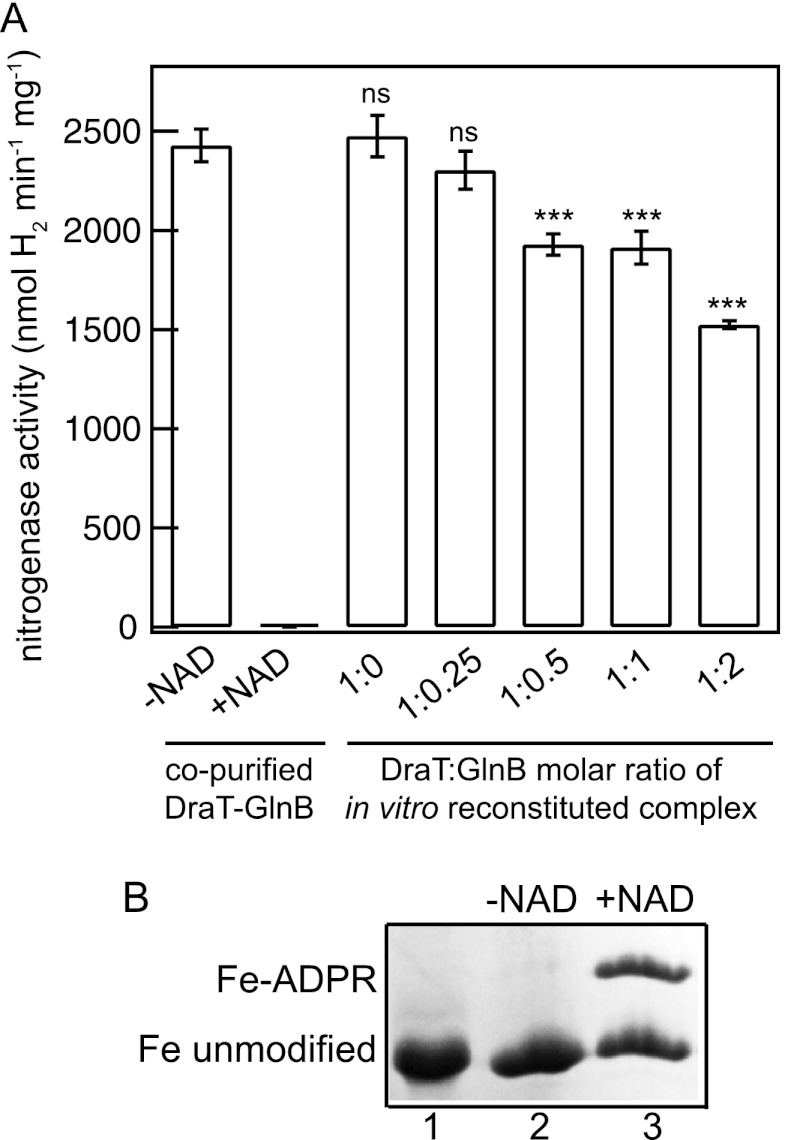
Control reactions in the absence of NAD+ indicated that the Fe protein was fully active and not modified in the presence of the DraT-GlnB complex that was obtained by coexpression and copurification (Fig. 1A and B). Similar activities were observed using DraT alone in the presence of NAD+ (Fig. 1A). Hence, DraT had no significant activity under these conditions. Preincubation of the Fe protein with the DraT-GlnB complex that was obtained by copurification resulted in complete Fe protein inactivation (Fig. 1A) and ADP-ribosylation (Fig. 1B). Furthermore, preincubation of the Fe protein with DraT in the presence of increasing concentrations of GlnB resulted in Fe protein inhibition in a GlnB-dependent manner, with the highest inhibition when DraT-GlnB was used at a 1:2 molar ratio (Fig. 1A). Under similar conditions, the copurified complex was approximately 3-fold more efficient to ADP-ribosylate the Fe protein than the in vitro-reconstituted complex (260 nmol of Fe protein min−1 mg DraT−1 compared to 97 nmol min−1 mg DraT−1) (Fig. 1A). To verify the efficiency of in vitro complex formation, DraT and GlnB were mixed at a 1:2 molar ratio and analyzed by pulldown of the His-tagged DraT using Ni2+ beads. Quantification of the protein bands showed that only 56% of DraT was complexed to GlnB (Fig. 2B). This result may explain the lower efficiency of the in vitro-reconstituted complex in inhibiting the Fe protein in comparison to the DraT-GlnB complex obtained by copurification.
Fig 1.
Fe protein activity and modification after DraT-GlnB complex activity assay. (A) Fe protein activity was determined after incubation at different DraT/GlnB (monomer/trimer) ratios or in the presence of the DraT-GlnB complex obtained by copurification. The reaction mixtures for the DraT assay contained 2 mM NAD+; 1 mM MgCl2; 1 mM ADP; 100 μg of A. vinelandii Fe protein; and DraT, DraT-GlnB complex formed in vitro, or copurified complex and were incubated for 5 min at 130 rpm at 30°C. The reactions were performed using an Fe protein/DraT (dimer/monomer) molar ratio of 42:1. As a control, the reaction was performed in the presence of either copurified complex in the absence of NAD. The DraT-GlnB assay was stopped by reducing NAD+ using dithionite solution, the Fe protein activity was determined by adding 500 μg of the MoFe protein, and the hydrogen evolution was measured. ns, nonsignificant; ***, P < 0.001. The error bars represent the calculated standard deviations. (B) NAD+-dependent modification of A. vinelandii Fe protein catalyzed by the A. brasilense DraT-GlnB complex. Lane 1, control containing purified, unmodified A. vinelandii Fe protein. Lanes 2 and 3, purified A. vinelandii Fe protein was incubated with the A. brasilense copurified DraT-GlnB complex in the presence (lane 3) or absence (lane 2) of NAD+. The unmodified and ADP-ribosylated subunits of the Fe protein were separated using low-cross-link SDS-PAGE, and the gel was stained with Coomassie blue. The percentages of Fe-ADPR and unmodified Fe protein were quantified using Photoshop CS4, resulting in 42 and 58%, respectively.
Fig 2.
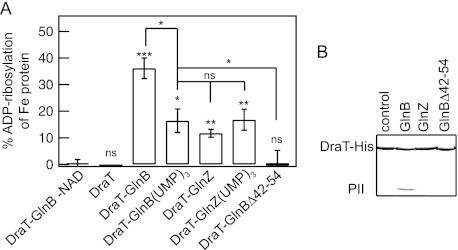
DraT activation and interaction with PII proteins. (A) DraT activity was assessed indirectly from Fe protein activity by measuring hydrogen evolution. A DraT assay was performed using A. vinelandii Fe protein as the substrate in the presence of different PII proteins to form the in vitro complexes. Reaction mixtures containing 2 mM NAD+; 1 mM MgCl2; 1 mM ADP; 100 μg Fe protein; and DraT, DraT-PII complexes formed in vitro, or copurified DraT-GlnB were incubated for 5 min at 130 rpm at 30°C. The reactions were performed using a DraT/PII trimer molar ratio of 1:2 and an Fe protein/DraT molar ratio of 21:1. For a control, the reaction was performed in the presence of either copurified complex, DraT-GlnB (1:2, as shown), or DraT in the absence of NAD+. The DraT activity was stopped by reducing NAD+ using dithionite solution. The Fe protein activity was determined by adding 500 μg of MoFe protein, and the hydrogen production was measured. The activity of the copurified DraT-GlnB complex was 130 ± 2.5 nmol ADP-ribosylated Fe protein min−1 mg DraT−1 under the same conditions, corresponding to 100% ADP-ribosylation of Fe protein. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. The error bars represent the calculated standard deviations. (B) Complex formation between His-tagged DraT and the PII protein GlnB, GlnZ, or GlnBΔ42-54 was assessed by pulldown in the presence of 1 mM ADP. Proteins bound to Ni2+ beads were analyzed by SDS-PAGE, and the gel was stained with Coomassie blue.
DraT activation in vitro requires complex formation with the GlnB T loop.
A. brasilense encodes two highly similar PII proteins, GlnB and GlnZ (19), whereas only GlnB seems able to interact with DraT (21, 23). Unexpectedly, the presence of GlnZ in the DraT assay resulted in significant DraT activation in comparison to DraT alone (Fig. 2A), although this GlnZ-induced activation was less than half of that observed with GlnB (Fig. 2A). Given the high similarity between GlnB and GlnZ (see Fig. S1 in the supplemental material), it is possible that GlnZ interacts with DraT to promote its activation. Likewise, the two PII proteins of Rhodobacter capsulatus were able to interact with DraT using yeast two-hybrid assays (36). However, in A. brasilense, the lower affinity of DraT for GlnZ precluded detection of the protein complex in pulldown assays (Fig. 2B).
The formation of the DraT-GlnB complex is affected by the uridylylation of GlnB (23). When GlnB-(UMP)3 or GlnZ-(UMP)3 was used in the DraT assay, it also resulted in significant DraT activation, though this activation was approximately 2-fold lower than the activation observed using unmodified GlnB (Fig. 2A). Yeast two-hybrid analysis revealed that in R. rubrum the GlnB T-loop mutants failed to interact with DraT (37). In order to further exploit the role of the GlnB T loop, a GlnB variant carrying a partial deletion of the T loop (GlnBΔ42-54) was purified and tested for in vitro interaction with DraT. In contrast to the wild-type GlnB, the GlnBΔ42-54 variant did not coprecipitate with His-tagged DraT in pulldown assays (Fig. 2B) and did not significantly activate DraT in vitro (Fig. 2A). The deletion of the GlnB T loop did not affect the quaternary structure of the GlnBΔ42-54 variant as judged by migration rates on native PAGE (see Fig. S2 in the supplemental material). Furthermore, the deletion of residues 42 to 54 in the highly similar GlnZ protein did not affect binding of ATP and ADP (24), supporting the conclusion that the GlnBΔ42-54 variant retains its overall fold and effector binding properties. Comparison of the GlnB and GlnZ T-loop sequences revealed only 3 nonconserved substitutions at positions 42, 52, and 54 (see Fig. S1 in the supplemental material). When the GlnZ residues at positions 52 and 54 were replaced by the respective residues found in GlnB, interaction between the GlnZ variants (Q42HS52VS54D and S52VS54D) and DraT could be detected by pulldown assay (see Fig. S3 in the supplemental material), reinforcing the involvement of the T loop in the DraT-GlnB interaction. On the other hand, the GlnB T-loop H42 residue seems not to be important for DraT binding, since a GlnB H42Q variant was able to interact with DraT (see Fig. S3 in the supplemental material). Together, these results support a model where the major docking site for DraT is the GlnB T loop and show that this interaction is required for DraT activation.
PII effectors ATP, ADP, and 2-oxoglutarate modulate GlnB-dependent DraT activation.
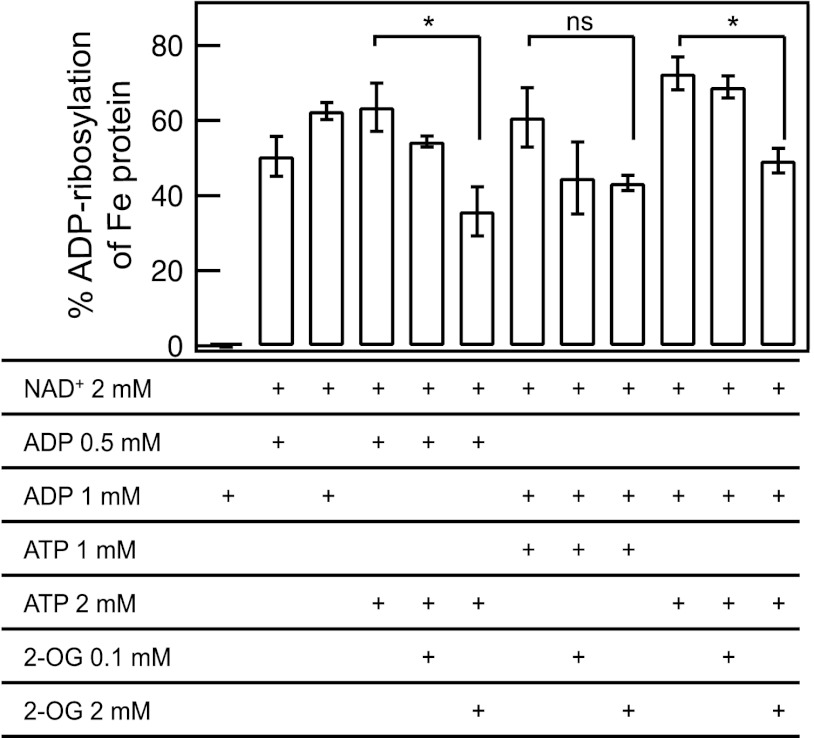
We have previously shown that the A. brasilense DraT-GlnB complex is stable in the presence of Mg-ADP and is destabilized in the presence of Mg-ATP and 2-OG combined (23). In order to exploit the role of the PII effectors ATP, ADP, and 2-OG in GlnB-dependent DraT activation, DraT activity was assayed in the presence of added ATP and ADP combined and under different 2-OG levels reflecting the reported physiological range (38). The DraT-GlnB complex (obtained by copurification) was inactive in the presence of ATP without added ADP (see Fig. S4 in the supplemental material). The activities of the DraT-GlnB complex were similar when using ADP alone or using ATP plus ADP at different ATP/ADP ratios (Fig. 3). However, at high ATP/ADP ratios and in the presence of a high 2-OG concentration (2 mM), the activity was inhibited (Fig. 3). These results are in agreement with our previous results reporting on in vitro complex formation between DraT and GlnB (23) and strongly support the idea that only the ADP-bound conformation of the GlnB T loop is able to activate DraT.
Fig 3.
Effects of ADP, ATP, and 2-oxoglutarate on the activity of the DraT-GlnB complex. DraT activity was assessed indirectly from Fe protein activity by measuring hydrogen evolution. Reaction mixtures for the DraT assay contained 4 mM MgCl2, copurified DraT-GlnB complex, 100 μg A. vinelandii Fe protein, and the effectors, as indicated. For a control, the reaction was performed in the presence of copurified DraT-GlnB complex in the absence of NAD+. The DraT activity was stopped by reducing NAD+ using dithionite solution. The Fe protein activity was determined by adding 500 μg of MoFe protein, and the hydrogen production was measured. ns, nonsignificant; *, P < 0.05. The error bars represent the calculated standard deviations.
GlnB acts by enhancing the kcat and kcat/Km of DraT.
To further investigate the mechanism of DraT activation by GlnB, we determined the Km value for NAD+ of the DraT-GlnB complex (Table 2; see Fig. S5 in the supplemental material). It was not possible to determine the Km for NAD+ using A. brasilense DraT alone, given its extremely low activity. The Km value determined for NAD+ using the A. brasilense DraT-GlnB complex is within the same order of magnitude as the value reported using DraT purified from the related organism R. rubrum (Table 2) (39, 40). Furthermore, the NAD+ Km was well below the reported intracellular concentration of NAD+ in E. coli (about 2 mM) (41). This suggests NAD+ binding by DraT is not affected by interaction with GlnB. On the other hand, both the kcat and the kcat/NAD+ Km ratio for the DraT-GlnB complex are well above the reported values for DraT alone; notably, the DraT-GlnB complex was at least 18-fold more efficient than DraT purified from R. rubrum (Table 2).
Table 2.
Comparison of kinetic constants for reactions catalyzed by DraT from R. rubrum or the DraT-GlnB complex from A. brasilense using A. vinelandii Fe protein as the acceptor
| Fe protein as the acceptor of ADP-ribose | Vmaxa | kcat (s−1)b | Km (μM)c | kcat/Km (s−1 M−1) | Reference |
|---|---|---|---|---|---|
| A. brasilense DraT-GlnB | 6.72 | 0.501 | 166 | 3,018.1 | This work |
| R. rubrum DraT | 0.072 | 0.0022 | 22 | 100 | 39 |
| 1.48 | 0.044 | 268 | 164.2 | 40 |
The values represent nmol of ADP-ribose transferred s−1 mg DraT−1 or nmol ADP-ribosylated Fe protein s−1 mg DraT-GlnB complex−1.
DraT was considered 30,000 Da and DraT-GlnB complex 74,504 Da (calculated based on the sequence using the Expasy program).
Km for NAD+ in the presence of Mg-ADP.
As we did not determine a Km value for the Fe protein, we cannot determine whether GlnB acts to enhance the affinity of DraT for the Fe protein and/or simply affects DraT catalytic efficiency. A previous report indicated that NAD+ binds to the Fe protein-DraT complex but not to DraT alone (42). Thus, given that DraT-GlnB complex formation did not significantly affect the DraT Km for NAD+ (Table 2), it is likely that GlnB acts to enhance the catalytic efficiency of DraT rather than to enhance the affinity for binding the Fe protein.
ADP-ribosylation of Fe protein blocks its ability to transfer electrons to the MoFe protein.
The current model for nitrogenase inactivation by Fe protein ADP-ribosylation is based on studies by Murrel and coauthors (4). They provided indirect evidence that ADP-ribosylation of the Fe protein prevents its interaction with the MoFe protein, thus blocking the electron transfer between these proteins. Structural analysis of the nitrogenase complex revealed that, indeed, the site of Fe protein ADP-ribosylation is located in the docking surface for the MoFe protein (5). To evaluate if the ADP-ribosylation of the Fe protein could somehow affect the [4Fe-4S] cluster, we performed EPR experiments, which showed that this modification does not affect the electronic state of the reduced metal cluster ([4Fe-4S]1+) bound to Mg-ADP (see Fig. S6 in the supplemental material). This result agrees with the midpoint potential being unaffected by the DraT-catalyzed modification (43). Furthermore, the turnover sample prepared in nitrogen gas demonstrated no association and/or no electron transfer from the Fe to the MoFe protein (data not shown).
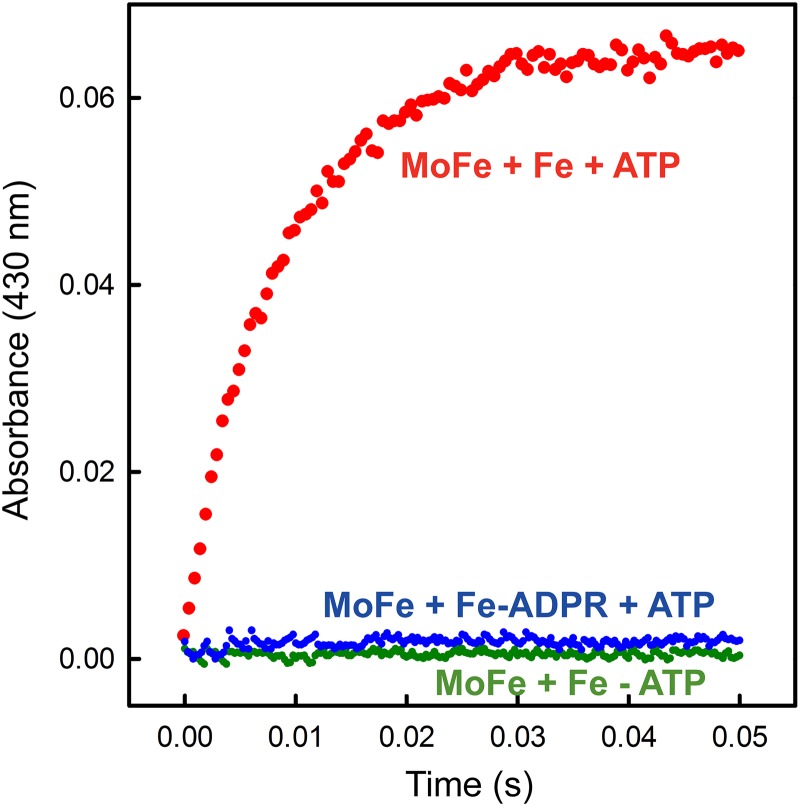
Finally, to determine if ADP-ribosylated Fe protein could be weakly associated with the MoFe protein, which could start the first electron transfer based on the deficit spending mechanism recently described (35), we monitored the electron transfer from the Fe protein to the MoFe protein using stopped-flow spectrophotometry. We monitored the absorbance at 430 nm from 3 ms to 50 ms after mixing to observe the change in the redox state of Fe protein due to its oxidation. There was no change in absorbance for ADP-ribosylated Fe protein (Fig. 4), confirming a previous hypothesis (4) that ADP-ribosylation prevents association of the Fe protein with the MoFe protein.
Fig 4.
Electron transfer from the Fe protein to the MoFe protein monitored by stopped-flow spectroscopy. Shown is the absorbance at 430 nm plotted against the time after mixing ADP-ribosylated or unmodified Fe protein (68 μM) and MoFe protein (18 μM) in one syringe with buffer (100 mM MOPS, pH 7.4, and 18 mM Mg-ATP) or in the absence of nucleotide as a control.
DISCUSSION
The mechanism of nitrogenase regulation through reversible ADP-ribosylation has been the subject of studies since the late 1970s. In the 1980s, the enzymes responsible for Fe protein ADP-ribosylation, DraT and DraG, were isolated and characterized in vitro (3, 6). A series of studies revealed that DraT and DraG have opposite regulation in vivo in the presence of negative effectors, such as ammonium or anaerobiosis in A. brasilense and darkness in R. rubrum (33, 44, 45).
In the last decade, it has become clear that proteins from the PII family are involved in the regulation of DraT in response to ammonium levels (2). The DraT enzyme interacts with GlnB in A. brasilense (21, 23), R. rubrum (37), and R. capsulatus (36). The DraT-GlnB interaction is regulated by the nitrogen status in vivo. Under nitrogen fixation conditions, DraT is inactive, whereas upon an ammonium shock, DraT binds to deuridylylated GlnB and is presumably activated (21).
In this work, we used purified proteins to confirm for the first time that GlnB activates DraT in vitro (Fig. 1). A GlnB variant carrying a deletion in the T loop (GlnBΔ42-54) failed to interact and activate DraT, suggesting that enzyme activation is mediated by interaction between DraT and the GlnB T loop (Fig. 2). Indeed, yeast two-hybrid analysis in R. rubrum showed that amino acid substitutions on the GlnB T loop impaired DraT-GlnB interaction (37). Furthermore, when only 2 amino acids of the GlnZ T loop were replaced with those found in GlnB, the GlnZ variants (Q42HS52VS54D and S52VS54D) were able to interact with DraT at detectable levels (see Fig. S3 in the supplemental material).
The hypothesis that the interaction of DraT with GlnB results in enzyme activation is in accordance with in vivo analysis of PII mutants that failed to modify the Fe protein after an ammonium shock (46–48). However, this is in contrast to a previous model that suggested the presence of a negative regulator of DraT in vivo (7). The early model was based on the fact that R. rubrum DraT was active in vitro after purification (3). However, the authors noted that DraT was only stable when Mg-ADP was present during purification. Furthermore, the DraT preparations used in that work had a contaminant with the molecular weight expected for the GlnB monomer, and attempts to remove this contaminant resulted in loss of DraT activity (3). Hence, it is likely the previously described DraT active in vitro had copurified GlnB.
Our results showed that the ability of GlnB to activate DraT was positively influenced by ADP and negatively influenced by 2-OG in the presence of high ATP/ADP ratios (Fig. 3), agreeing with our previous protein interaction studies (23). In prokaryotes, the 2-OG levels report the nitrogen status: the molecule accumulates under nitrogen limitation and decreases in response to an ammonium upshift (38, 49, 50). Furthermore, the binding of ATP and ADP to GlnB is influenced by 2-OG so that GlnB is presumably saturated with Mg-ATP and 2-OG under nitrogen-fixing conditions and with Mg-ADP after an ammonium upshift (18, 38, 51). Hence, an ammonium upshift would increase the affinity of DraT for GlnB bound to ADP, resulting in DraT activation. In addition, under nitrogen-fixing conditions, GlnB is fully uridylylated, and this form is a weaker activator of DraT (Fig. 3). The concerted action of GlnB effectors and uridylylation would ensure that GlnB will interact with DraT to activate the enzyme only when ammonium is abundant.
Comparison between the kinetic constants of the A. brasilense DraT-GlnB complex and those published for DraT from R. rubrum strongly suggests that GlnB acts to enhance the enzyme's catalytic efficiency, although our data cannot precisely define the mechanism of DraT activation by GlnB. Previous in vitro analysis showed that some of the properties of His-tagged DraT, including solubility and affinity for Ni2+, changed in response to interaction with GlnB, suggesting conformational changes in DraT upon GlnB binding (23). Such conformational changes could result in a DraT structure with improved catalytic efficiency.
Finally, we noted that despite the proximity of the Fe protein ADP-ribosylation site to the [4Fe-4S] cluster ligand Cys-98, the EPR signal of iron in this cluster is not affected by ADP-ribosylation. Furthermore, ADP-ribosylation of the Fe protein completely abolished its ability to transfer electrons to the MoFe protein, as observed by stopped-flow spectrophotometric analysis, confirming that ADP-ribosylation of the Fe protein prevented its association with the MoFe protein, thus inhibiting nitrogenase activity.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Luiza M. Araujo for kindly providing the plasmid to express GlnD protein.
V.R.M. was the recipient of a mobility fellowship from CAPES (process number 0388/11-4). This work was supported by CNPq, INCT da Fixação de Nitrogênio/MCT, Fundação Araucária, and CAPES.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01517-12.
REFERENCES
- 1. Seefeldt LC, Hoffman BM, Dean DR. 2009. Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 78:701–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huergo LF, Pedrosa FO, Muller-Santos M, Chubatsu LS, Monteiro RA, Merrick M, Souza EM. 2012. PII signal transduction proteins: pivotal players in post-translational control of nitrogenase activity. Microbiology 158:176–190 [DOI] [PubMed] [Google Scholar]
- 3. Lowery RG, Ludden PW. 1988. Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J. Biol. Chem. 263:16714–16719 [PubMed] [Google Scholar]
- 4. Murrell SA, Lowery RG, Ludden PW. 1988. ADP-ribosylation of dinitrogenase reductase from Clostridium pasteurianum prevents its inhibition of nitrogenase from Azotobacter vinelandii. Biochem. J. 251:609–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. 1997. Structure of ADP. AIF4-stabilized nitrogenase complex and its implications for signal transduction.Nature 387:370–376 [DOI] [PubMed] [Google Scholar]
- 6. Saari LL, Triplett EW, Ludden PW. 1984. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J. Biol. Chem. 259:15502–15508 [PubMed] [Google Scholar]
- 7. Zhang Y, Burris RH, Ludden PW, Roberts GP. 1997. Regulation of nitrogen fixation in Azospirillum brasilense. FEMS Microbiol. Lett. 152:195–204 [DOI] [PubMed] [Google Scholar]
- 8. Forchhammer K. 2008. PII signal transducers: novel functional and structural insights. Trends Microbiol. 16:65–72 [DOI] [PubMed] [Google Scholar]
- 9. Huergo LF, Chandra G, Merrick M. 3 August 2012. PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. doi:10.1111/j.1574–6976.2012.00351.x. [DOI] [PubMed]
- 10. Ninfa AJ, Atkinson MR. 2000. PII signal transduction proteins. Trends Microbiol. 8:172–179 [DOI] [PubMed] [Google Scholar]
- 11. Xu Y, Cheah E, Carr PD, van Heeswijk WC, Westerhoff HV, Vasudevan SG, Ollis DL. 1998. GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J. Mol. Biol. 282:149–165 [DOI] [PubMed] [Google Scholar]
- 12. Radchenko M, Merrick M. 2011. The role of effector molecules in signal transduction by PII proteins. Biochem. Soc. Trans. 39:189–194 [DOI] [PubMed] [Google Scholar]
- 13. Jiang P, Peliska JA, Ninfa AJ. 1998. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37:12782–12794 [DOI] [PubMed] [Google Scholar]
- 14. Conroy MJ, Durand A, Lupo D, Li X- D, Bullough PA, Winkler FA, Merrick M. 2007. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. U. S. A. 104:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Truan D, Huergo LF, Chubatsu LS, Merrick M, Li XD, Winkler FK. 2010. A new PII protein structure identifies the 2-oxoglutarate binding site. J. Mol. Biol. 400:531–539 [DOI] [PubMed] [Google Scholar]
- 16. Jiang P, Ninfa AJ. 2007. Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in vitro. Biochemistry 46:12979–12996 [DOI] [PubMed] [Google Scholar]
- 17. Fokina O, Chellamuthu V-R, Forchhammer K, Zeth K. 2010. Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc. Natl. Acad. Sci. U. S. A. 107:19760–19765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang P, Ninfa AJ. 2009. Sensation and signaling of α-ketoglutarate and adenylylate energy charge by the Escherichia coli PII signal transduction protein require cooperation of the three ligand-binding sites within the PII trimer. Biochemistry 48:11522–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Zamaroczy M. 1998. Structural homologues PII and Pz of Azospirillum brasilense provide intracellular signalling for selective regulation of various nitrogen-dependent functions. Mol. Microbiol. 29:449–463 [DOI] [PubMed] [Google Scholar]
- 20. Huergo LF, Souza EM, Araujo MS, Pedrosa FO, Chubatsu LS, Steffens MBR, Merrick M. 2006. ADP-ribosylation of dinitrogenase reductase in Azospirillum brasilense is regulated by AmtB-dependent membrane sequestration of DraG. Mol. Microbiol. 59:326–337 [DOI] [PubMed] [Google Scholar]
- 21. Huergo LF, Chubatsu LS, Souza EM, Pedrosa FO, Steffens MBR, Merrick M. 2006. Interactions between PII proteins and the nitrogenase regulatory enzymes DraT and DraG in Azospirillum brasilense. FEBS Lett. 580:5232–5236 [DOI] [PubMed] [Google Scholar]
- 22. Huergo LF, Merrick M, Pedrosa FO, Chubatsu LS, Araujo LM, Souza EM. 2007. Ternary complex formation between AmtB, GlnZ and the nitrogenase regulatory enzyme DraG reveals a novel facet of nitrogen regulation in bacteria. Mol. Microbiol. 66:1523–1535 [DOI] [PubMed] [Google Scholar]
- 23. Huergo LF, Merrick M, Monteiro RA, Chubatsu LS, Steffens MBR, Pedrosa FO, Souza EM. 2009. In vitro interactions between the PII proteins and the nitrogenase regulatory enzymes dinitrogenase reductase ADP-ribosyltransferase (DraT) and dinitrogenase reductase-activating glycohydrolase (DraG) in Azospirillum brasilense. J. Biol. Chem. 284:6674–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajendran C, Gerhardt ECM, Bjelic S, Gasperina A, Scarduelli M, Pedrosa FO, Chubatsu LS, Merrick M, Souza EM, Winkler FK, Huergo LF, Li XD. 2011. Crystal structure of the GlnZ-DraG complex reveals a different form of PII-target interaction. Proc. Natl. Acad. Sci. U. S. A. 108:18972–18976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huergo LF, Filipaki A, Chubatsu LS, Yates MG, Steffens MB, Pedrosa FO, Souza EM. 2005. Effect of the over-expression of PII and PZ proteins on the nitrogenase activity of Azospirillum brasilense. FEMS Microbiol. Lett. 253:47–54 [DOI] [PubMed] [Google Scholar]
- 26. Moure VR, Razzera G, Araújo LM, Oliveira MAS, Gerhardt ECM, Müller-Santos M, Almeida F, Pedrosa FO, Valente AP, Souza EM, Huergo LF. 2012. Heat stability of proteobacterial PII protein facilitates purification using a single chromatography step. Protein Expr. Purif. 81:83–88 [DOI] [PubMed] [Google Scholar]
- 27. Huergo LF, Souza EM, Steffens MBR, Yates MG, Pedrosa FO, Chubatsu LS. 2005. Effects of over-expression of the regulatory enzymes DraT and DraG on the ammonium-dependent post-translational regulation of nitrogenase reductase in Azospirillum brasilense. Arch. Microbiol. 183:209–217 [DOI] [PubMed] [Google Scholar]
- 28. Araujo ML, Huergo LF, Invitti AL, Gimenes CI, Bonatto AC, Monteiro RA, Souza EM, Pedrosa FO, Chubatsu LS. 2008. Different responses of the GlnB and GlnZ proteins upon in vitro uridylylation by the Azospirillum brasilense GlnD protein. Braz. J. Med. Biol. Res. 41:289–294 [DOI] [PubMed] [Google Scholar]
- 29. Christiansen J, Goodwin PJ, Lanzilotta WN, Seefeldt LC, Dean DR. 1998. Catalytic and biophysical properties of a nitrogenase apo-MoFe protein produced by a nifB-deletion mutant of Azotobacter vinelandii. Biochemistry 37:12611–12623 [DOI] [PubMed] [Google Scholar]
- 30. Seefeldt LC, Morgan TV, Dean DR, Mortenson LE. 1992. Mapping the site(s) of MgATP and MgADP interaction with the nitrogenase of Azotobacter vinelandii. Lysine 15 of the iron protein plays a major role in MgATP interaction. J. Biol. Chem. 267:6680–6688 [PubMed] [Google Scholar]
- 31. Barney BM, Igarashi RY, Dos Santos PC, Dean DR, Seefeldt LC. 2004. Substrate interaction at an iron-sulfur face of the FeMo-cofactor during nitrogenase catalysis. J. Biol. Chem. 279:53621–53624 [DOI] [PubMed] [Google Scholar]
- 32. Lowery RG, Saari LL, Ludden PW. 1986. Reversible regulation of the nitrogenase iron protein from Rhodospirillum rubrum by ADP-ribosylation in vitro. J. Bacteriol. 166:513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanemoto RH, Ludden PW. 1984. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J. Bacteriol. 158:713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanzilotta WN, Fisher K, Seefeldt LC. 1996. Evidence for electron transfer from the nitrogenase iron protein to the molybdenum-iron protein without MgATP hydrolysis: characterization of a tight protein-protein complex. Biochemistry 35:7188–7196 [DOI] [PubMed] [Google Scholar]
- 35. Danyal K, Dean DR, Hoffman BM, Seefeldt LC. 2011. Electron transfer within nitrogenase: evidence for a deficit-spending mechanism. Biochemistry 50:9255–9263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pawlowski A, Riedel Klipp K-UW, Dreiskemper P, Groß S, Drepper T, Masepohl B. 2003. Yeast two-hybrid studies on interaction of proteins involved in regulation of nitrogen fixation in the phototrophic bacterium Rhodobacter capsulatus. J. Bacteriol. 185:5240–5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y, Conrad MC, Zhang Y, Roberts GP. 2006. Identification of Rhodospirillum rubrum GlnB variants that are altered in their ability to interact with different targets in response to nitrogen status signals. J. Bacteriol. 188:1866–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radchenko MV, Thornton J, Merrick M. 2010. Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J. Biol. Chem. 285:31037–31045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lowery RG, Ludden PW. 1989. Effect of nucleotides on the activity of dinitrogenase reductase ADP-ribosyltransferase from Rhodospirillum rubrum. Biochemistry 28:4956–4961 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y, Kim K, Ludden PW, Roberts GP. 2001. Isolation and characterization of draT mutants that have altered regulatory properties of dinitrogenase reductase ADP-ribosyltransferase in Rhodospirillum rubrum. Microbiology 147:193. [DOI] [PubMed] [Google Scholar]
- 41. Bennett BD, Kimball EH, Gao M, Osterhout R, Dien SJV, Rabinowitz DJ. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grunwald SK, Ludden PW. 1997. NAD-dependent cross-linking of dinitrogenase reductase and dinitrogenase reductase ADP-ribosyltransferase from Rhodospirillum rubrum. J. Bacteriol. 179:3277–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Halbleib CM, Zhang Y, Ludden PW. 2000. Regulation of dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase by a redox-dependent conformational change of nitrogenase Fe protein. J. Biol. Chem. 275:3493–3500 [DOI] [PubMed] [Google Scholar]
- 44. Liang JH, Nielsen GM, Lies DP, Burris RH, Roberts GP, Ludden PW. 1991. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J. Bacteriol. 173:6903–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Y, Burris RH, Ludden PW, Roberts GP. 1993. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J. Bacteriol. 175:6781–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Drepper T, Gross S, Yakunin AF, Hallenbeck PC, Masepohl B, Klipp W. 2003. Role of GlnB and GlnK in ammonium control of both nitrogenase systems in the phototrophic bacterium Rhodobacter capsulatus. Microbiology 149:2203–2212 [DOI] [PubMed] [Google Scholar]
- 47. Klassen G, Souza EM, Yates MG, Rigo LU, Costa RM, Inaba J, Pedrosa FO. 2005. Nitrogenase switch-off by ammonium ions in Azospirillum brasilense requires the GlnB nitrogen signal-transducing protein. Appl. Environ. Microbiol. 71:5637–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y, Pohlmann EL, Ludden PW, Roberts GP. 2001. Functional characterization of three GlnB homologs in the photosynthetic bacterium Rhodospirillum rubrum: roles in sensing ammonium and energy status. J. Bacteriol. 183:6159–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dodsworth JA, Leigh JA. 2006. Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc. Natl. Acad. Sci. U. S. A. 103:9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan J, Doucette CD, Fowler WU, Feng Piazza X-JM, Rabitz HA, Wingreen NS, Rabinowitz JD. 2009. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol. Syst. Biol. 5:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gerhardt ECM, Araujo LM, Ribeiro RR, Chubatsu LS, Scarduelli M, Rodrigues TE, Monteiro RA, Pedrosa FO, Souza EM, Huergo LF. 2012. Influence of the ADP/ATP ratio, 2-oxoglutarate and divalent ions on Azospirillum brasilense PII protein signalling. Microbiology 158:1656–1663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.