Abstract
Gram-negative bacteria produce outer membrane vesicles (OMVs) that package and deliver proteins, small molecules, and DNA to prokaryotic and eukaryotic cells. The molecular details of OMV biogenesis have not been fully elucidated, but peptidoglycan-associated outer membrane proteins that tether the outer membrane to the underlying peptidoglycan have been shown to be critical for OMV formation in multiple Enterobacteriaceae. In this study, we demonstrate that the peptidoglycan-associated outer membrane proteins OprF and OprI, but not OprL, impact production of OMVs by the opportunistic pathogen Pseudomonas aeruginosa. Interestingly, OprF does not appear to be important for tethering the outer membrane to peptidoglycan but instead impacts OMV formation through modulation of the levels of the Pseudomonas quinolone signal (PQS), a quorum signal previously shown by our laboratory to be critical for OMV formation. Thus, the mechanism by which OprF impacts OMV formation is distinct from that for other peptidoglycan-associated outer membrane proteins, including OprI.
INTRODUCTION
Vesiculation is a highly conserved process occurring in all domains of life (1–4). Among prokaryotes, vesicle formation has been reported in both Gram-negative and Gram-positive bacteria (1, 2, 4, 5). Gram-negative bacteria produce spherical, bilayered vesicles derived from the outer membrane that range in size from 20 to 500 nm (6–10). Similar to the outer membrane, outer membrane vesicles (OMVs) possess an outer leaflet of lipopolysaccharide (LPS) and an inner leaflet of phospholipid (11–15). OMVs also contain outer membrane proteins and entrap periplasmic components as they are released (16–18). OMVs have been found to be associated with Gram-negative bacteria growing planktonically and in surface-attached biofilm communities as well as natural environments (11, 19–21).
Despite their biological importance, the molecular mechanism of OMV formation has not been fully elucidated, though multiple factors have been reported to affect the process (13, 22–28) and numerous models encompassing these factors have been proposed (9, 19, 28–31). A primary hurdle to elucidating the mechanism of OMV formation has been the inability to identify factors that contribute to OMV production. Using the model opportunistic pathogen Pseudomonas aeruginosa, our laboratory demonstrated that the quorum-sensing signal 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal [PQS]) stimulates P. aeruginosa OMV biogenesis (10, 32). Surprisingly, signaling by PQS was not required for OMV formation (10); instead, OMV formation proceeds through direct interaction of PQS with the LPS component of the outer membrane (29). Based on these results, we recently proposed a detailed P. aeruginosa OMV biogenesis model, dubbed the bilayer-couple model, in which PQS induces membrane curvature by stably inserting and expanding the outer leaflet of the outer membrane relative to the inner leaflet (31), resulting in localized membrane curvature and ultimately vesiculation.
One question regarding the bilayer-couple model that remains is the role that peptidoglycan-associated outer membrane proteins play in P. aeruginosa OMV biogenesis. Multiple studies in bacteria other than P. aeruginosa have suggested that OMV formation is localized to regions of the outer membrane not tethered to the underlying peptidoglycan layer (6, 8, 33). Loss of the peptidoglycan-associated outer membrane protein OmpA, Pal, or Lpp significantly increases OMV formation in Escherichia coli, Salmonella enterica serovar Typhimurium, and Vibrio cholerae (6, 34–37). Homologs of OmpA, Pal, and Lpp exist in P. aeruginosa, although their involvement in OMV biogenesis is not known (38). OprF is a 38-kDa OmpA homolog that serves both as a porin and as a tether that noncovalently links the outer membrane to peptidoglycan (39). OprF exists in two conformations: when closed, the C terminus anchors the outer membrane to the peptidoglycan layer, and when open, the C terminus inserts into the outer membrane, forming a functional porin (40). OprL is an 18-kDa Pal homolog that also tethers the outer membrane to peptidoglycan (41–44). Finally, OprI is an 8-kDa homolog of Braun's lipoprotein (Lpp) and is proposed to covalently interact with the peptidoglycan layer (45), though this interaction has been reported to differ among P. aeruginosa strains (39, 46). OprI is highly abundant in the outer membrane (46) and, similar to E. coli Lpp, can exist in a free and a peptidoglycan-bound form (46).
The goal of this study was to assess the involvement of these three peptidoglycan-associated outer membrane proteins in P. aeruginosa OMV biogenesis. Here we demonstrate that deletion of oprF and oprI induces P. aeruginosa vesiculation through two distinct mechanisms. The absence of OprF increases OMV production via increased PQS production, while loss of OprI presumably decreases tethering of the outer membrane to peptidoglycan. These findings are presented in the context of the membrane bilayer-couple model to provide a working model for P. aeruginosa OMV biogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) broth or tryptic soy broth (TSB) with ampicillin (100 μg/ml) or tetracycline (10 μg/ml), when appropriate. P. aeruginosa strains were grown in brain heart infusion (BHI) broth with carbenicillin (150 μg/ml), gentamicin (50 μg/ml), or tetracycline (50 μg/ml), when appropriate.
Table 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 Δ(lacZYA-argF)U169 deoR ϕ80dlacZΔM15 | 47 |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 48 |
| P. aeruginosa | ||
| PA14 | Wild type | 49 |
| oprL mutant | PA14 ΔoprL | This study |
| oprI mutant | PA14 ΔoprI | This study |
| oprF mutant | PA14 oprF::Mar2XT7 (Gmr) | 49 |
| pqsH mutant | PA14 ΔpqsH | This study |
| oprF pqsH mutant | PA14 oprF::Mar2XT7 ΔpqsH (Gmr) | This study |
| oprI pqsH mutant | PA14 ΔoprI ΔpqsH | This study |
| Plasmids | ||
| pGEM-T Easy | Sequencing vector | Promega |
| pEX18Tc | Gene replacement vector (oriT+ sacB+ Tcr) | 50 |
| pEX18Tc-oprL | pEX18Tc containing 1-kb sequences flanking oprL | This study |
| pEX18Tc-oprI | pEX18Tc containing 1-kb sequences flanking oprI | This study |
| pEX18Tc-pqsH | pEX18Tc containing 1-kb sequences flanking pqsH | This study |
| pEX1.8 | Broad-host-range expression vector, IPTG inducible (Apr) | 51 |
| pEX1.8-oprF | pEX1.8 carrying oprF | This study |
| pEX1.8-oprI | pEX1.8 carrying oprI | This study |
DNA manipulations.
DNA manipulations were performed using standard procedures (52). PCR was performed using an Expand Long Template PCR system (Roche). A QIAprep spin miniprep kit (Qiagen) or GeneJET plasmid miniprep kit (Fermentas) was used for plasmid purification. Restriction endonucleases and buffers were purchased from New England BioLabs or Fermentas Life Sciences. A DNeasy tissue kit (Qiagen) was used to extract chromosomal DNA. DNA sequencing was performed at the DNA Core Facility at the University of Texas Institute for Cell and Molecular Biology.
Construction of P. aeruginosa deletion strains.
Unmarked deletions in oprL, oprI, and pqsH were made via allelic exchange as previously described (50) with some modifications. Deletion plasmids were constructed using the primer pairs listed in Table 2. The two amplicons were combined using overlap extension PCR and digested using BamHI (for oprL and oprI deletions) or EcoRI and XbaI (for pqsH deletion) and ligated into pEX18Tc. Each deletion plasmid was transformed into E. coli SM10 and conjugated into P. aeruginosa PA14. For the P. aeruginosa oprF pqsH double mutant, the pqsH deletion plasmid was conjugated into the oprF mutant. Mutant selection was performed as previously described (50), with some modifications. To select the pEX18Tc-oprL, pEX18Tc-oprI, and pEX18Tc-pqsH transconjugants, conjugations were spread onto LB plates with 50 μg/ml tetracycline and 25 μg/ml nalidixic acid. To select for the pEX18Tc-pqsH transconjugant in the oprF mutant background, conjugations were spread on a morpholinepropanesulfonic acid (MOPS)-buffered defined medium (25 mM MOPS [pH 7.2], 93 mM NH4Cl, 43 mM NaCl, 3.7 mM KH2PO4, 1 mM MgSO4, 3.5 μM FeSO4 · 7H2O) supplemented with ∼1.25% agarose, 20 mM succinate, and 20 μg/ml tetracycline. To select for the oprF pqsH double mutant, transconjugants were grown overnight in MOPS supplemented with 20 mM succinate and 20 μg/ml tetracycline, diluted into antibiotic-free medium, and spread onto LB plates supplemented with 10% sucrose. Mutants were confirmed by PCR and sequencing.
Table 2.
Primer sequences
| Name | Sequencea |
|---|---|
| oprL-flanking regions | |
| oprL-P1 | CCGGATCCGAGAAGCTCACCGGTATCAAG |
| oprL-P2 | GTGCTTGGGCATAACGACTTCCATGTAACTCCTAATGAACCC |
| oprL-P3 | GAAGTCGTTATGCCCAAGCAC |
| oprL-P4 | CAGGATCCGTACTGGGAAATGACCTGCTG |
| oprI-flanking regions | |
| oprI-P1 | 5′-CCGGATCCAGGTACTCCAGGTTCAGCCAC |
| oprI-P2 | 5′-GTTTTCAACAGGTCGTGAGACCGGTGGACATTTCCATAACAGCAATC |
| oprI-P3 | 5′-GGTCTCACGACCTGTTGAAAAC-3′ |
| oprI-P4 | 5′-CCGGATCC AGGTGATCAAGGCCAAGTAC-3′ |
| pqsH-flanking regions | |
| pqsH-P1 | 5′-CTGAATTCCTTGTCCTGCAGGTCGATATC-3′ |
| pqsH-P2 | 5′-CATCGCCGAACTCGAAAACAGGATAAGAACGGTCATCCGTTGC-3′ |
| pqsH-P3 | 5′-GCAACGGATGACCGTTCTTATCCTGTTTTCGAGTTCGGCGATG-3′ |
| pqsH-P4 | 5′-CTTCTAGAGATTGCTACAGGTAGCGAGG-3′ |
| Complementation | |
| oprF-for | 5′-CTAACTGACCATCAAGATGGG-3′ |
| oprF-rev | 5′-CCCAAGCTTTTTTCCTTAGAGGCTCA-3′ |
| oprI-for | 5′-CGGAATTCGTCCACCTTAAGGGGAAC-3′ |
| oprI--rev | 5′-CCCAAGCTTCAGGTCGTGAGACCTAT-3′ |
Underlined sequences represent recognition sites for restriction endonucleases.
Complementation of the P. aeruginosa oprF and oprI mutants.
oprF and oprI were PCR amplified from PA14 chromosomal DNA using the primer pairs indicated in Table 2. The oprF PCR product was cloned into the pGEM-T Easy vector (Promega) and digested with PstI and HindIII. The oprI PCR product was purified and digested using EcoRI and HindIII. Purified digested products were separately ligated into PstI/HindIII- or EcoRI/HindIII-digested pEX1.8, and the resulting plasmids (pEX1.8-oprF and pEX1.8-oprI) were verified via DNA sequencing. It should be noted that the oprI gene amplified and cloned in this study contained 3 base pair differences from the published PA14 genome, resulting in codon changes H36D, X47E, and K79N. Plasmids were electroporated into the oprF mutant and oprI mutant (52). Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to cultures at 500 μM to induce gene expression.
OMV preparation.
For OMV preparation, P. aeruginosa overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.001 to 0.05 in BHI broth. Cells were grown to an OD600 of 2.7 to 3.9 with shaking at 250 rpm in a 1:10 culture volume/flask volume ratio. When adding exogenous PQS, synthetic PQS resuspended in 500 μl methanol was added to 25 ml BHI before adding cells, such that the final concentrations of PQS in the culture were 0.5, 10, 20, and 40 μM. OMVs were purified using methods described previously (13). Briefly, cells were removed by centrifugation (5,000 × g for 15 min), and the resulting supernatant was filtered through a 0.45-μm-pore-size membrane (Whatman PuraDisc 25-mm-diameter syringe filters, polyethersulfone [PES]). OMVs were pelleted from cell-free supernatants using an ultracentrifuge with a Beckman 70Ti rotor at 265,000 × g for 1 h and resuspended in MV buffer (50 mM Tris, 5 mM NaCl, 1 mM MgSO4, pH 7.4).
OMV quantification.
OMV production was quantified using a previously described phospholipid assay of purified vesicles (53, 54) with some modifications. Purified OMV pellets were extracted with 2 volumes of chloroform, dried under N2 gas, and resuspended in chloroform (500 μl or 1 ml chloroform). The absorbance was measured at 470 nm and normalized by the OD600 of the extracted culture. To determine the linear range of detection for the assay, commercially available phosphatidylethanolamine (PE; Fluka Biochemika) was used to generate a standard curve with concentrations ranging from 7.8 to 250 μg/ml. Measurements made below the limit of detection were assigned a value equal to the lowest limit of the standard curve.
PQS and HHQ extraction and quantification.
PQS and HHQ were extracted from cultures using 2 volumes of acidified ethyl acetate (acidified with 0.1 ml acetic acid/liter ethyl acetate). The organic phase was removed and dried under a continuous stream of N2 gas. PQS was quantified using thin-layer chromatography (TLC) (53). For TLC, dried samples were resuspended in methanol (Optima grade; Fisher), and 5 μl was spotted onto a dried straight-phase phosphate-impregnated TLC plate. Samples were separated using a 95:5 dichloromethane-methanol mobile phase. Synthetic PQS standards were used to generate a standard curve. PQS spots were measured via photography with excitation by long-wave UV light. HHQ was quantified using high-performance liquid chromatography (HPLC) as previously described (66).
Proteomics.
Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) was performed as described previously (55). Briefly, OMVs were isolated as described above and resuspended in lysis buffer (25 mM Tris-HCl [pH 7.5], 5 mM dithiothreitol [DTT], 1.0 mM EDTA, 1× Calbiochem protease inhibitor cocktail [CPICPS]). Fifty microliters of diluted OMV lysate was incubated at 55°C for 45 min with 50 μl of trifluoroethanol (TFE) and 15 mM DTT, followed by incubation with 55 mM iodoacetamide (IAM) in the dark for 30 min. The sample volume was adjusted to 1 ml with buffer (50 mM Tris, pH 8.0), followed by a 1:50 (wt/wt) trypsin digestion for 4.5 h. The reaction was halted by adding 2% (vol/vol; 20 μl) formic acid. The sample was lyophilized, resuspended with buffer C (95% H2O, 5% acetonitrile, 0.01% formic acid), and cleaned using a C18 tip (Thermo Fisher Scientific). The eluted sample was again lyophilized, resuspended with 120 μl buffer C, and filtered through an Amicon Ultra-0.5 filter (for 12 min at 14,000 × g at 4°C). Each sample was injected 2 times into an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), and data were collected in a 0 to 90% acetonitrile gradient over 5 h. The raw files from LC-MS/MS experiments are available at http://www.marcottelab.org/index.php/PSEAE_oprF.2012.
LC-MS/MS raw files were searched against the P. aeruginosa PA14 protein sequence database (downloaded from the PseudoCAP database, 23 November 2009 version) (56) with randomly shuffled protein sequences as a decoy. Four different search engines, Crux (57), X!Tandem with k score (58, 59), InsPecT (60), and MS-GFDB (61), were used with default options. The results were then integrated with the MSblender program (55). APEX scores (62, 63) estimating absolute protein abundance were calculated using the number of peptide-spectrum matches assigned by MSblender with a false discovery rate with a cutoff of <0.01 and APEX observability score (Oi values) trained by whole-cell lysate proteomics data. Protein localization information was also downloaded from PseudoCAP (56). To simplify the localization data, cellular compartments were prioritized in the following order: outer membrane, extracellular, periplasmic, cytoplasmic membrane, and cytoplasmic. For example, a protein annotated as both periplasmic and cytoplasmic would be considered a periplasmic protein in this analysis. Proteins not localized to one of these five compartments on the basis of annotation were considered unknown. All search results and detailed parameters are also available at http://www.marcottelab.org/index.php/PSEAE_oprF.2012. A summary of the results is available in Table S1 in the supplemental material.
RESULTS AND DISCUSSION
Several OMV biogenesis models hypothesize that loss of outer membrane connections to the underlying peptidoglycan is required for OMV release (6, 8, 33). Supporting this model, deletion of the peptidoglycan-associated outer membrane proteins OmpA, Pal, and Lpp has been shown to significantly increase OMV formation in E. coli, S. Typhimurium, and V. cholerae (6, 34–37). Based on these findings, we predicted that inactivation of peptidoglycan-associated outer membrane proteins in P. aeruginosa would increase OMV formation. To test this hypothesis, OMV formation of the P. aeruginosa PA14 oprF, oprI, and oprL mutants was assessed as previously described using a spectrophotometric lipid assay (31). It is important to note that in strain PA14, oprI is reported to have a premature stop codon (TAA) at position +139 relative to the ATG start codon (64); however, when we sequenced oprI from P. aeruginosa PA14, it was found that the codon encompassing position +139 instead encodes glutamic acid (139T → G), indicating that the open reading frame is intact. This was confirmed by LC-MS/MS data, which showed that OprI is translated and encodes glutamic acid at amino acid 47.
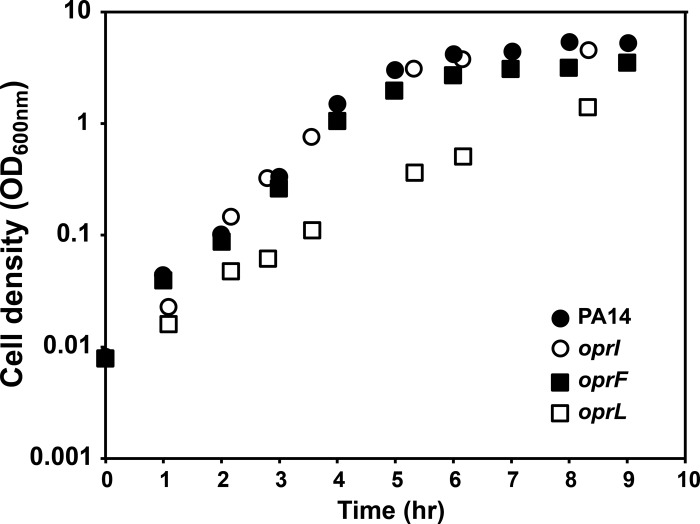
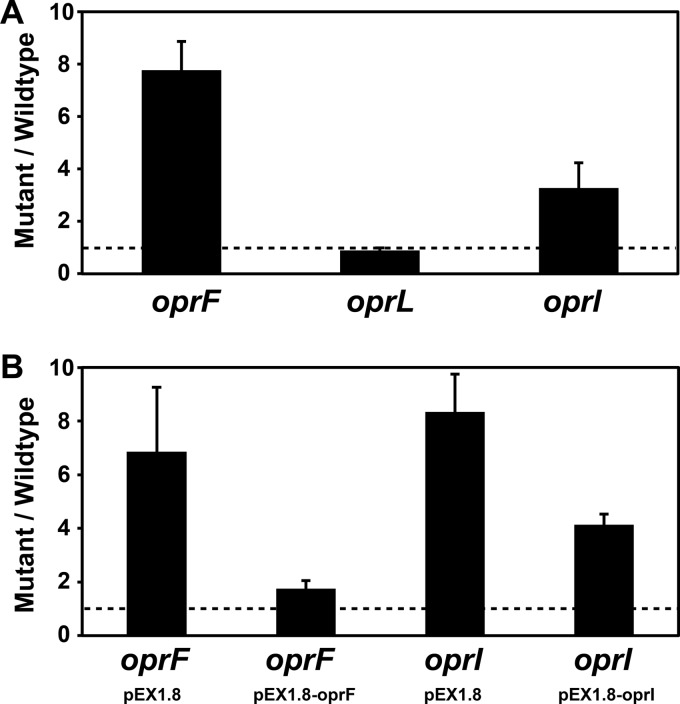
While the oprI and oprF mutants grew at rates equivalent to the rate of wild-type (wt) P. aeruginosa (Fig. 1), they produced ∼3-fold and ∼8-fold more OMVs, respectively (Fig. 2A). Expression of oprI and oprF in trans in the corresponding mutants reduced OMV levels (Fig. 2B), indicating that increased OMV production was due to the loss of OprF and OprI. The oprL mutant showed a slight decrease in growth rate and growth yield (Fig. 1), although it produced OMVs at levels equivalent to those for the wt (Fig. 2A). While the growth rates of wt P. aeruginosa and the oprF mutant were equivalent, the oprF mutant reached slightly lower cell yields (Fig. 1). On the basis of these lower cell yields (OD600 of ∼5 for wt P. aeruginosa and ∼3.5 for the oprF mutant) and the observation that P. aeruginosa can undergo autolysis (65), it was possible that the increase in OMV production in the oprF mutant was due to the presence of cytoplasmic membrane components (arising from lysis) in our OMV preparations. While we did not think that this was likely since the growth yield differences were small, it was critical to examine this possibility experimentally.
Fig 1.
Growth characteristics of wt P. aeruginosa and the oprI, oprF, and oprL mutants. Bacteria were grown with shaking (250 rpm) at 37°C in BHI. Representative growth curves are shown.
Fig 2.
Inactivation of oprF and oprI increases P. aeruginosa OMV production. (A) Fold change in OMV production by the P. aeruginosa oprF, oprL, and oprI mutants. Bacteria were grown with shaking (250 rpm) at 37°C to an OD600 of ∼3.5, and OMVs were quantified by measuring OMV lipid. All lipid measurements were normalized to cell number. For each replicate, the fold change in OMV production was calculated by dividing mutant lipid levels by wt lipid levels. The dotted line represents no change in OMV production. Error bars represent standard errors of the means (n ≥ 4). (B) Complementation of the oprF and oprI mutants. The fold change in OMV production by the oprF and oprI mutants carrying either vector alone (pEX1.8) or the complementation plasmids (pEX1.8-oprF or pEX1.8-oprI) is shown. Bacteria were grown with shaking (250 rpm) at 37°C to an OD600 of ∼3.5 with 500 μM IPTG. OMVs were quantified and compared to those of wt P. aeruginosa carrying pEX1.8 as described for panel A. Error bars represent standard errors of the means (n ≥ 4).
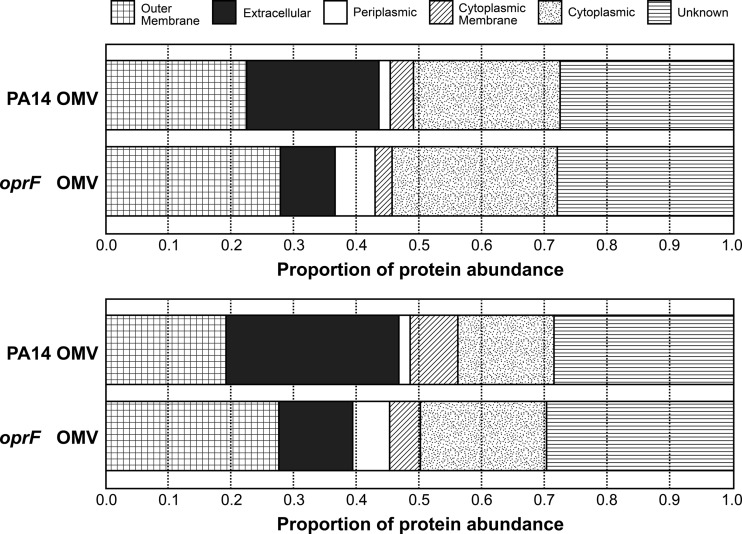
If the OMV preparations from the oprF mutant were contaminated with cytoplasmic membranes, we reasoned that these preparations would be enriched in cytoplasmic membrane proteins. To examine this, the proteome of OMV samples from wt P. aeruginosa and the oprF mutant were determined using LC-MS/MS. Raw files, results, and details of the analyses are available at http://www.marcottelab.org/index.php/PSEAE_oprF.2012 and in Table S1 in the supplemental material. As was observed by several other groups, OMV preparations in wt P. aeruginosa are enriched for outer membrane and periplasmic proteins, although some cytoplasmic and cytoplasmic membrane proteins are also present. The relative abundances indicate that the oprF mutant OMV sample was also enriched for outer membrane proteins and not inner membrane proteins (Fig. 3), indicating that the increase in OMV production in the oprF mutant is not due to cell lysis and contamination by cytoplasmic membrane proteins.
Fig 3.
OMVs from the P. aeruginosa oprF mutant are not enriched for cytoplasmic membrane proteins. Two biological replicates displaying the proportion of protein abundance from each cellular compartment in wt P. aeruginosa OMVs and P. aeruginosa oprF mutant OMVs. The protein abundance of each compartment was estimated by dividing the sum of APEX scores of proteins identified in each compartment by the total APEX score for each sample. Protein localization predictions were obtained from the Pseudomonas Genome Database. In wt PA14 samples, 159 (first replicate) and 533 (second replicate) proteins were identified. In the oprF mutant samples, 504 (first replicate) and 1,140 (second replicate) proteins were identified.
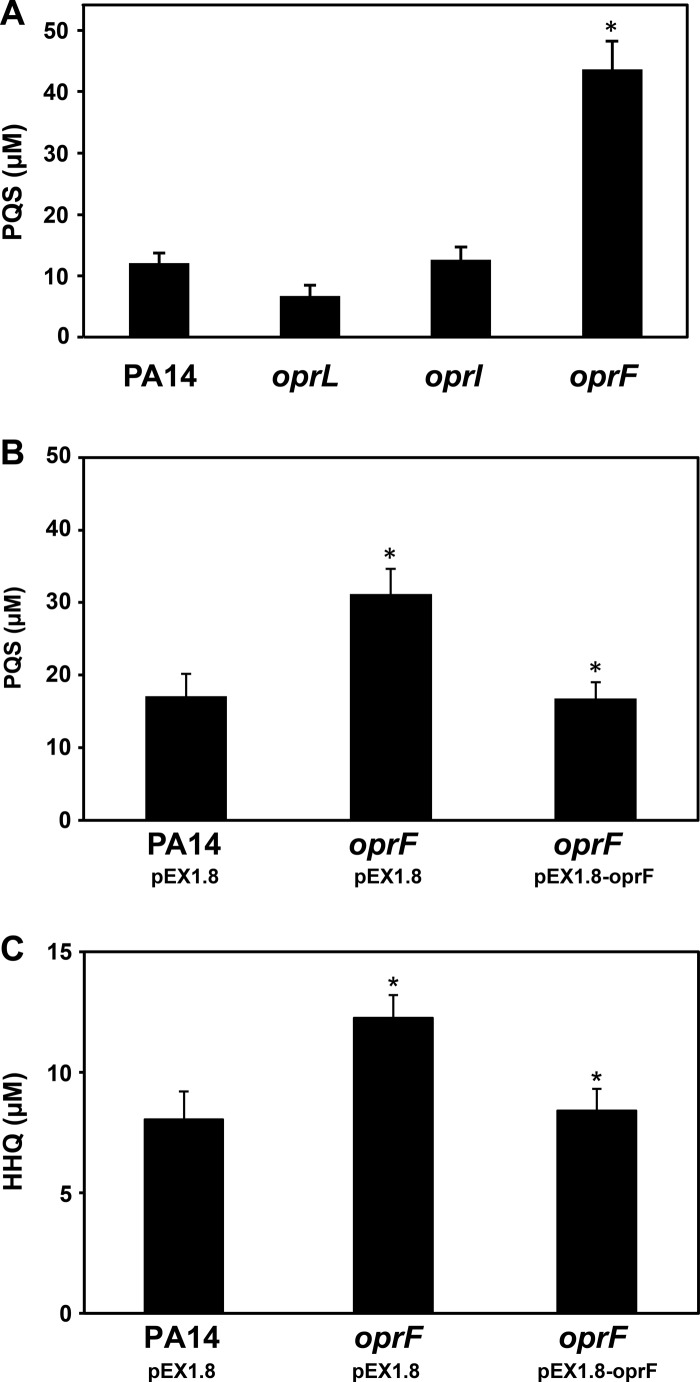
Based on work in other bacteria, the increase in OMV levels in the oprF and oprI mutants was presumably due to detachment of the outer membrane from the underlying peptidoglycan layer (6, 34–37). However, another possibility is that inactivation of these proteins altered the levels of PQS, thus leading to increased OMVs. To test whether production of PQS and its direct precursor, 2-heptyl-4-quinolone (HHQ), were affected in the oprF and oprI mutants, PQS was measured using TLC and HHQ was measured using high-performance liquid chromatography (66). The oprF mutant produced ∼4-fold more PQS and ∼1.5-fold more HHQ than wt P. aeruginosa (Fig. 4A and C), while the oprI mutant produced PQS levels equivalent to those for the wt (Fig. 4A). Importantly, PQS and HHQ production could be genetically complemented by expression of oprF in trans in the oprF mutant (Fig. 4B and C). Recent evidence partially conflicts with these results, determining that a P. aeruginosa oprF mutant produces lower levels of PQS (67). Our study likely contradicts this study due to the fact that different quantification methods were used: Fito-Boncompte et al. (67) used an LC/MS method (68) to quantify PQS, and this study used TLC. In contrast to Fito-Boncompte et al., we and others have found that in the absence of a chelator in the mobile phase, PQS is difficult to quantify using liquid chromatography (66, 69) due to poor peak resolution; thus, TLC provides a more quantifiable approach.
Fig 4.
PQS production by wt P. aeruginosa and the oprL, oprI, and oprF mutants. (A) PQS was extracted from whole cultures and quantified using TLC. The oprF mutant produces ∼4-fold more PQS than wt. (B) Complementation of the P. aeruginosa oprF mutant with pEX1.8-oprF restores PQS to wt levels. (C) The oprF mutant produces slightly more HHQ than the wt, and complementation of the oprF mutant restores HHQ to wt levels. *, P < 0.02 via two-tailed Student's t test, assuming equal variance (n ≥ 4).
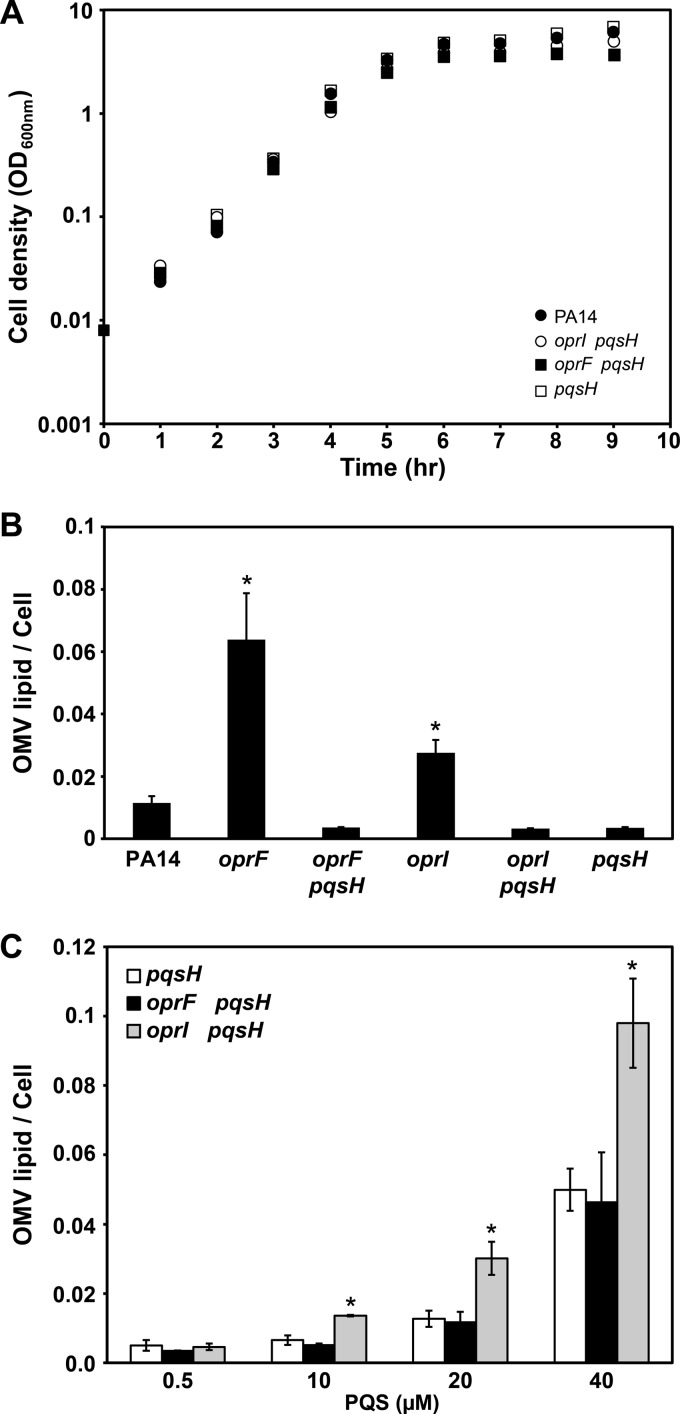
To determine if the increase in OMV production by the P. aeruginosa oprF mutant was due to increased PQS production, the gene (pqsH) encoding the enzyme responsible for the terminal step in PQS production was deleted in the P. aeruginosa oprF mutant. Since this strain is unable to produce PQS, assessment of OMV formation by this strain allows the determination of the importance of PQS for enhanced OMV formation in the oprF mutant. The P. aeruginosa oprF pqsH double mutant grew similarly to wt P. aeruginosa (Fig. 5A) and produced extremely low levels of OMVs (Fig. 5B). In fact, OMVs were not detectable in over half of the OMV preparations. These data support the hypothesis that increased OMV production in the oprF mutant is a result of increased PQS production. To further test this hypothesis, we examined OMV production by the P. aeruginosa oprF pqsH double mutant following supplementation with increasing amounts of PQS (Fig. 5C). OMV production in this strain increased with increasing amounts of PQS (Fig. 5C). Interestingly, addition of PQS at levels produced by the P. aeruginosa oprF mutant (40 μM) resulted in production of very high levels of OMVs equivalent to those observed in the oprF mutant (Fig. 2A and 5B and C). In addition, the PQS-induced OMV production by the P. aeruginosa oprF pqsH double mutant was similar to that observed upon addition of PQS to the P. aeruginosa pqsH mutant (Fig. 5C). These data again support a model in which the increase in PQS production and not simply the lack of OprF is responsible for the increase in OMV formation by the P. aeruginosa oprF mutant.
Fig 5.
Enhanced OMV production by the oprF mutant, but not the oprI mutant, is due to increased PQS production. (A) Representative growth curves of wt P. aeruginosa (PA14), the oprI pqsH double mutant, the oprF pqsH double mutant, and the pqsH mutant (pqsH) grown with shaking (250 rpm) at 37°C in BHI. (B) OMV production by wt P. aeruginosa, the oprF mutant, the oprF pqsH double mutant, the oprI mutant, the oprI pqsH double mutant, and the pqsH mutant. The majority of samples from strains lacking pqsH did not produce detectable amounts of OMVs. (C) OMV production upon addition of increasing levels of PQS. Synthetic PQS was added exogenously to cultures to a final concentration of 0.5, 10, 20, or 40 μM, and OMV levels were quantified. All cultures were grown with shaking (250 rpm) at 37°C to an OD600 of ∼3.5, and OMVs were quantified using the lipid assay. *, P ≤ 0.01 compared to wild type (B) or the pqsH mutant (C) via two-tailed Student's t test, assuming equal variance (n ≥ 3).
The oprI mutant produced more OMVs than the wt; however, unlike the oprF mutant, it produced wt levels of PQS (Fig. 4A). For this reason, we hypothesized that the increased OMV production by the oprI mutant was not due to increased PQS production but instead was due to loss of peptidoglycan tethering. To test this hypothesis, we constructed an oprI pqsH double mutant and examined OMV production in the presence and absence of exogenous PQS. Similar to the pqsH mutant, the oprI pqsH double mutant did not produce detectable levels of OMVs (Fig. 5B); however, the oprI pqsH double mutant produced 2-fold more OMVs than the pqsH mutant upon addition of exogenous PQS (Fig. 5C). These experiments indicate that PQS is necessary for production of detectable OMVs in the absence of OprI; however, loss of OprI leads to increased production of OMVs in the presence of PQS. These data, combined with the fact that OprI is the only P. aeruginosa outer membrane protein known to covalently bind to peptidoglycan, suggest that this protein limits PQS-mediated production of OMVs through tethering to peptidoglycan.
This work provides additional insight into the mechanism of OMV formation in P. aeruginosa. While the absence of the OmpA homolog OprF increases OMV production, we showed that unlike other bacterial species, this increase is not directly attributable to the loss of peptidoglycan binding but instead is attributable to increased production of PQS. As demonstrated for many other bacterial species (6, 8, 33), deletion of Braun's lipoprotein homolog oprI resulted in an increase in OMV production most likely through the loss of the major peptidoglycan-associated lipoprotein. Several models, which are not necessarily mutually exclusive, describe the molecular mechanisms of OMV formation (8, 13, 26–29, 31, 33), but few studies have clarified which models apply to different species and/or growth conditions. These data have allowed us to refine the P. aeruginosa bilayer-couple model (31) for OMV biogenesis through demonstration that OprI reduces PQS-mediated OMV formation.
Supplementary Material
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01253-12.
REFERENCES
- 1. Dorward DW, Garon CF. 1990. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl. Environ. Microbiol. 56: 1960–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knox KW, Vesk M, Work E. 1966. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 92: 1206–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thery C, Zitvogel L, Amigorena S. 2002. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2: 569–579 [DOI] [PubMed] [Google Scholar]
- 4. Work E, Knox KW, Vesk M. 1966. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann. N. Y. Acad. Sci. 133: 438–449 [DOI] [PubMed] [Google Scholar]
- 5. Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107: 19002–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72: 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dorward DW, Garon CF, Judd RC. 1989. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 171: 2499–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoekstra D, van der Laan JW, de Leij L, Witholt B. 1976. Release of outer membrane fragments from normally growing Escherichia coli. Biochim. Biophys. Acta 455: 889–899 [DOI] [PubMed] [Google Scholar]
- 9. Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64: 163–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437: 422–425 [DOI] [PubMed] [Google Scholar]
- 11. Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20: 291–303 [DOI] [PubMed] [Google Scholar]
- 12. Chatterjee SN, Das J. 1967. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J. Gen. Microbiol. 49: 1–11 [DOI] [PubMed] [Google Scholar]
- 13. Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177: 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kahn ME, Maul G, Goodgal SH. 1982. Possible mechanism for donor DNA binding and transport in Haemophilus. Proc. Natl. Acad. Sci. U. S. A. 79: 6370–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pettit RK, Judd RC. 1992. The interaction of naturally elaborated blebs from serum-susceptible and serum-resistant strains of Neisseria gonorrhoeae with normal human serum. Mol. Microbiol. 6: 729–734 [DOI] [PubMed] [Google Scholar]
- 16. Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS. 2011. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11: 3424–3429 [DOI] [PubMed] [Google Scholar]
- 17. Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45: 9–13 [DOI] [PubMed] [Google Scholar]
- 18. Kesty NC, Kuehn MJ. 2004. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 279: 2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19: 2645–2655 [DOI] [PubMed] [Google Scholar]
- 20. Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188: 5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schooling SR, Hubley A, Beveridge TJ. 2009. Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 191: 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burdett ID, Murray RG. 1974. Electron microscope study of septum formation in Escherichia coli strains B and B-r during synchronous growth. J. Bacteriol. 119: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burdett ID, Murray RG. 1974. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J. Bacteriol. 119: 303–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 178: 2767–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Clarke AJ, Beveridge TJ. 1996. A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J. Bacteriol. 178: 2479–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188: 5385–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63: 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tashiro Y, Sakai R, Toyofuku M, Sawada I, Nakajima-Kambe T, Uchiyama H, Nomura N. 2009. Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa. J. Bacteriol. 191: 7509–7519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 69: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61: 839–846 [DOI] [PubMed] [Google Scholar]
- 31. Schertzer JW, Whiteley M. 2012. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3(2): e00297–11 doi:10.1128/mBio.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mashburn-Warren L, Howe J, Brandenburg K, Whiteley M. 2009. Structural requirements of the Pseudomonas quinolone signal for membrane vesicle stimulation. J. Bacteriol. 191: 3411–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wensink J, Witholt B. 1981. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur. J. Biochem. 116: 331–335 [DOI] [PubMed] [Google Scholar]
- 34. Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180: 4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184: 754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sonntag I, Schwarz H, Hirota Y, Henning U. 1978. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 136: 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yem DW, Wu HC. 1978. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J. Bacteriol. 133: 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hancock RE, Siehnel R, Martin N. 1990. Outer membrane proteins of Pseudomonas. Mol. Microbiol. 4: 1069–1075 [DOI] [PubMed] [Google Scholar]
- 39. Hancock RE, Irvin RT, Costerton JW, Carey AM. 1981. Pseudomonas aeruginosa outer membrane: peptidoglycan-associated proteins. J. Bacteriol. 145: 628–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. 2006. Pseudomonas aeruginosa porin OprF exists in two different conformations. J. Biol. Chem. 281: 16220–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koebnik R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16: 1269–1270 [DOI] [PubMed] [Google Scholar]
- 42. Lim A, Jr, De Vos D, Brauns M, Mossialos D, Gaballa A, Qing D, Cornelis P. 1997. Molecular and immunological characterization of OprL, the 18 kDa outer-membrane peptidoglycan-associated lipoprotein (PAL) of Pseudomonas aeruginosa. Microbiology 143(Pt 5): 1709–1716 [DOI] [PubMed] [Google Scholar]
- 43. Mizuno T. 1981. A novel peptidoglycan-associated lipoprotein (PAL) found in the outer membrane of Proteus mirabilis and other Gram-negative bacteria. J. Biochem. 89: 1039–1049 [PubMed] [Google Scholar]
- 44. Mizuno T. 1979. A novel peptidoglycan-associated lipoprotein found in the cell envelope of Pseudomonas aeruginosa and Escherichia coli. J. Biochem. 86: 991–1000 [DOI] [PubMed] [Google Scholar]
- 45. Duchene M, Barron C, Schweizer A, von Specht BU, Domdey H. 1989. Pseudomonas aeruginosa outer membrane lipoprotein I gene: molecular cloning, sequence, and expression in Escherichia coli. J. Bacteriol. 171: 4130–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mizuno T, Kageyama M. 1979. Isolation of characterization of a major outer membrane protein of Pseudomonas aeruginosa. Evidence for the occurrence of a lipoprotein. J. Biochem. 85: 115–122 [DOI] [PubMed] [Google Scholar]
- 47. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 48. de Lorenzo V, Timmis KN. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235: 386–405 [DOI] [PubMed] [Google Scholar]
- 49. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103: 2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212: 77–86 [DOI] [PubMed] [Google Scholar]
- 51. Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179: 5756–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1997. Short protocols in molecular biology, 3rd ed John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 53. Schertzer JW, Brown SA, Whiteley M. 2010. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol. Microbiol. 77: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stewart JC. 1980. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 104: 10–14 [DOI] [PubMed] [Google Scholar]
- 55. Kwon T, Choi H, Vogel C, Nesvizhskii AI, Marcotte EM. 2011. MSblender: a probabilistic approach for integrating peptide identifications from multiple database search engines. J. Proteome Res. 10: 2949–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39: D596–D600 doi:10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Park CY, Klammer AA, Kall L, MacCoss MJ, Noble WS. 2008. Rapid and accurate peptide identification from tandem mass spectra. J. Proteome Res. 7: 3022–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Craig R, Beavis RC. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- 59. Keller A, Eng J, Zhang N, Li XJ, Aebersold R. 2005. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1: 2005.0017 doi:10.1038/msb4100024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. 2005. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77: 4626–4639 [DOI] [PubMed] [Google Scholar]
- 61. Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJ, Pevzner PA. 2010. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 9: 2840–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu P, Vogel C, Wang R, Yao X, Marcotte EM. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25: 117–124 [DOI] [PubMed] [Google Scholar]
- 63. Vogel C, Marcotte EM. 2008. Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat. Protoc. 3: 1444–1451 [DOI] [PubMed] [Google Scholar]
- 64. Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7: R90 doi:10.1186/gb-2006-7-10-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184: 6481–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Palmer GC, Schertzer JW, Mashburn-Warren L, Whiteley M. 2011. Quantifying Pseudomonas aeruginosa quinolones and examining their interactions with lipids. Methods Mol. Biol. 692: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fito-Boncompte L, Chapalain A, Bouffartigues E, Chaker H, Lesouhaitier O, Gicquel G, Bazire A, Madi A, Connil N, Veron W, Taupin L, Toussaint B, Cornelis P, Wei Q, Shioya K, Deziel E, Feuilloley MG, Orange N, Dufour A, Chevalier S. 2011. Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immun. 79: 1176–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lepine F, Deziel E, Milot S, Rahme LG. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 1622: 36–41 [DOI] [PubMed] [Google Scholar]
- 69. Niewerth H, Bergander K, Chhabra SR, Williams P, Fetzner S. 2011. Synthesis and biotransformation of 2-alkyl-4(1H)-quinolones by recombinant Pseudomonas putida KT2440. Appl. Microbiol. Biotechnol. 91: 1399–1408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.