Abstract
The cytoplasmic level of flagellin (called Hag) is homeostatically regulated in the Gram-positive bacterium Bacillus subtilis by a partner-switching mechanism between the protein FliW and either the Hag structural protein or CsrA, an RNA binding protein that represses hag translation. Here we show that FliW and the putative secretion chaperone FliS bind to Hag simultaneously but control Hag translation by different mechanisms. While FliW directly inhibits CsrA activity, FliS antagonizes CsrA indirectly by binding to Hag, enhancing Hag secretion, and depleting Hag in the cytoplasm to trigger the FliW partner switch. Consistent with a role for FliS in potentiating Hag secretion, the mutation of fliS crippled both motility and flagellar filament assembly, and both phenotypes could be partially rescued by artificially increasing the concentration of the Hag substrate through the absence of CsrA. Furthermore, the absence of FliS resulted in an approximately 30-fold reduction in extracellular Hag accumulation in cells mutated for CsrA (to relieve homeostatic control) and the filament cap protein FliD (to secrete flagellin into the supernatant). Thus, we mechanistically discriminate between the FliW regulator and the FliS chaperone to show that secretion disrupts flagellin homeostasis and promotes high-level flagellin synthesis during the period of filament assembly in B. subtilis.
INTRODUCTION
Many bacteria swim by assembling helical flagella that are rotated to provide force like a propeller. Flagella are complex machines assembled from over 30 different proteins organized into three structural domains, the basal body, the hook, and the filament (1, 2). The basal body is assembled first into the plasma membrane and houses a type III secretion (T3S) apparatus to secrete a series of proteins that are sequentially polymerized to build the flagellum. First, the rod proteins are assembled to transit the peptidoglycan (and the outer membrane in the case of Gram-negative bacteria). Next, the hook proteins are assembled to form a short extracellular flexible linker domain that acts as a universal joint. Once the hook is assembled, there is a substrate specificity switch that occurs within the secretion apparatus that enables the machine to secrete late-class flagellar proteins. After the specificity switch, two junction proteins are assembled at the end of the hook, followed by a protein called FliD (also known as FlaV and HAP2), which acts as a cap on the end of the extending structure (3–5). Finally, as many as 20,000 flagellin proteins are secreted through the basal body, rod, and hook and assembled underneath the FliD cap to form the long helical filament (6–10). The filament and, thus, flagellin, comprise the bulk of the flagellum, the assembly of which is estimated to consume 2% of the cell's biosynthetic resources (11).
When synthesized, flagellin is among the most abundant proteins made by the cell, and a chaperone, FliS, is thought to promote efficient flagellin export during filament assembly. FliS binds to the disordered C terminus of flagellin subunits in the cytoplasm, and the FliS-flagellin interaction has been reported to have a number of biological consequences (12–14). FliS has been shown to protect flagellin monomers from intracellular proteolysis in vivo and inhibits the polymerization of flagellin in vitro (12, 13). The FliS-flagellin complex has been implicated in the process of flagellin secretion, because FliS structurally resembles type III secretion chaperones and FliS enhances the interaction between flagellin and the secretion apparatus (15, 16). Furthermore, the deletion of the flagellin C terminus, to which FliS binds, reduces or abolishes the secretion of flagellin (17, 18). Finally, the mutation of fliS in Salmonella and Pseudomonas has been shown to impair motility and result in flagellar filaments that are significantly shorter than those of the wild type (19, 20). For Bacillus subtilis, FliS was suggested to be essential for filament assembly due to the inability to raise a lysate for a phage that targets the filament in a strain mutated for fliS (21).
In addition to FliS, B. subtilis encodes a second protein that binds to flagellin called FliW. Residues in the C-terminal region of flagellin (also known as Hag) are required for FliW binding, but FliW is not a chaperone (22). Rather, the FliW-Hag interaction is regulatory and part of a partner-switching mechanism in which Hag homeostatically restricts its own synthesis (23). When Hag stoichiometrically binds to and sequesters FliW, the RNA binding protein CsrA binds to and occludes the Shine-Dalgarno (SD) sequence of the hag transcript and inhibits hag translation (23, 24). When Hag levels are low in the cytoplasm, perhaps as the result of secretion, FliW switches partners and instead binds to CsrA, thereby preventing CsrA-hag mRNA interactions and, hence, allowing high levels of Hag translation. Thus, the Hag-FliW-CsrA system ensures that there is a low threshold level of Hag in the cytoplasm but that Hag translation increases concomitantly with Hag secretion and filament assembly. This mechanism suggests that the Hag protein is not only structural but also regulatory and that Hag expression is coupled to secretion. The homeostatic restriction of flagellin translation may also be functioning by a homologous system in Borrelia burgdorferi and perhaps other organisms (23, 25, 26).
Here we study the relationship of the putative flagellin chaperone FliS to the flagellin homeostatic regulator FliW in B. subtilis. We show that both FliW and FliS bound to flagellin simultaneously and that the mutation of either fliW or fliS abolished motility. The mutation of fliS reduced Hag translation but did not regulate CsrA directly, as is the case for FliW, but indirectly through Hag. Furthermore, the mutation of fliS abolished flagellar filament synthesis, except when csrA was also mutated, suggesting that high levels of flagellin partially bypass the need for FliS. The fliS csrA double mutant produced short filaments like those seen in a Salmonella fliS mutant, perhaps because Salmonella lacks FliW and also likely lacks the flagellin homeostatic feedback found in B. subtilis. Finally, we demonstrate that mutants defective in the FliD filament-capping protein secrete Hag into the supernatant, and we use the fliD mutant to show that the mutation of fliS resulted in a reduced amount of Hag secretion. The data support the model that flagellin cytoplasmic levels are homeostatically autoregulated, governed by the secretion of the flagellin structural subunit.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis strains were grown in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) or on LB plates fortified with 1.5% Bacto agar at 37°C. When appropriate, antibiotics were included at the following concentrations: 10 μg/ml tetracycline, 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 5 μg/ml kanamycin, and 1 μg/ml erythromycin plus 25 μg/ml lincomycin (mls). Isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) was added to the medium at the indicated concentrations when appropriate.
Motility assays. (i) Swarm expansion assay.
Cells were grown to the mid-log phase at 37°C in LB broth and resuspended to 10 optical density at 600 nm (OD600) units in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4 [pH 8.0]) containing 0.5% India ink (Higgins). Freshly prepared LB broth containing 0.7% Bacto agar (25 ml/plate) was dried for 20 min in a laminar flow hood, centrally inoculated with 10 μl of the cell suspension, dried for another 10 min, and incubated at 37°C. The India ink demarks the origin of the colony, and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of the plate, and swarm radius measurements were taken along this transect. For experiments including IPTG, cells were propagated in broth in the presence of IPTG, and IPTG was included in the swarm agar plates.
(ii) Swim expansion assay.
Freshly prepared LB broth containing 0.3% Bacto agar (25 ml/plate) was dried for 10 min in a laminar flow hood, centrally toothpick inoculated, and incubated at 37°C overnight. Plate shots were taken by using a Bio-Rad Gel Doc instrument.
Western blotting.
For the pellet fraction (cytoplasmic and cell associated), B. subtilis strains were grown in LB broth to an OD600 of ∼1.0, and 1 ml of broth culture was harvested by centrifugation, resuspended to 10 OD600 units in lysis buffer (20 mM Tris-HCl [pH 7.0], 10 mM EDTA, 1 mg/ml lysozyme, 10 μg/ml DNase I, 100 μg/ml RNase I, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and incubated for 30 min at 37°C. Ten microliters of lysate was mixed with 2 μl 6× SDS loading dye. For the supernatant fraction (secreted extracellular components), 1 ml of supernatant was collected from B. subtilis strains grown in LB broth to an OD600 of ∼1.0 by centrifugation. The supernatant was clarified by centrifugation at 9,447 × g for 10 min and treated with 100 μl of freshly prepared 0.15% sodium deoxycholate for 10 min at room temperature. Proteins from the supernatant were precipitated by treating the supernatant with 50 μl chilled trichloroacetic acid (TCA) for 2 h, pelleting at 9,447 × g for 10 min at 4°C, and washing with ice-cold acetone. Precipitated protein was resuspended in 50 μl of 0.1 N sodium hydroxide and mixed with 10 μl 6× SDS loading dye. Pellet and supernatant samples were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were electroblotted onto nitrocellulose and probed with a 1:80,000 dilution of anti-SigA primary antibody (a generous gift of Masaya Fujita, University of Houston), a 1:80,000 dilution of anti-Hag primary antibody, and a 1:10,000 dilution of secondary antibody (horseradish peroxidase [HRP]-conjugated goat anti-rabbit immunoglobulin G). The immunoblot was developed by using the Immun-Star HRP Developer kit (Bio-Rad).
For the dot blot, B. subtilis strains were grown in LB broth to an OD600 of ∼0.7, 1 ml of broth culture was harvested by centrifugation, and the supernatant fraction was saved. The supernatant was clarified by centrifugation at 9,447 × g for 10 min. Next, 150 μl of a serial dilution (3-fold at each step) of the supernatant was directly applied onto a nitrocellulose membrane using a dot blot apparatus, and the dot blot was probed with a 1:20,000 dilution of anti-Hag primary antibody (a generous gift of Masaya Fujita, University of Houston) and a 1:100,000 dilution of anti-SigA primary antibody. The dot blot was developed by using the Immun-Star HRP Developer kit (Bio-Rad).
Microscopy.
Fluorescence microscopy was performed with a Nikon 80i microscope with a phase-contrast Nikon Plan Apo 100× objective and an Excite 120 metal halide lamp. FM4-64 was visualized with a C-FL HYQ Texas Red filter cube (excitation filter, 532 to 587 nm; barrier filter, >590 nm), and Alexa Fluor 488 C5 maleimide fluorescent signals were visualized by using a C-FL HYQ fluorescein isothiocyanate (FITC) filter cube (FITC excitation filter, 460 to 500 nm; barrier filter, 515 to 550 nm). Images were captured with a Photometrics Coolsnap HQ2 camera in black and white, false colored, and superimposed by using Metamorph imaging software.
For fluorescence microscopy of flagella, 1 ml of broth culture was harvested at an OD600 of 0.5 to 1.0 and washed once in 1.0 ml of PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4). The suspension was pelleted, resuspended in 50 μl of PBS containing 5 μg/ml Alexa Fluor 488 C5 maleimide (Molecular Probes), and incubated for 5 min at room temperature (27). Cells were then washed twice with 500 μl PBS. When appropriate, membranes were stained by resuspension in 50 μl of PBS containing 5 μg/ml FM4-64 (Molecular Probes) and incubated for 5 min at room temperature. Three microliters of the suspension was placed onto a microscope slide and immobilized with a poly-l-lysine-treated coverslip.
For superresolution microscopy using structured illumination, the OMX 3D-SIM Super-Resolution system at the Indiana University Bloomington Light Microscopy Imaging Center was used. Superresolution microscopy was performed by using a 1.4-numerical-aperture (NA) Olympus 100× oil objective. FM4-64 was visualized by using laser line 561 and emission filter 609 nm to 654 nm, and Alexa Fluor 488 nm was visualized by using laser line 488 nm and emission filter 500 nm to 550 nm. Images were captured by using an Photometrics Cascade II electron-multiplying charge-coupled-device camera, processed by using SoftWorx imaging software, and analyzed by using IMARIS software.
Strain construction.
All constructs were first introduced into domesticated strain PY79 by natural competence and then transferred into the 3610 background using SPP1-mediated generalized phage transduction (28, 29). All strains used in this study are listed in Table 1. All plasmids used in this study are listed in Table S1 in the supplemental material. All primers used in this study are listed in Table S2 in the supplemental material.
Table 1.
Strains
| Strain | Genotype | Reference |
|---|---|---|
| 3610 | Wild type | |
| DK60 | ΔfliW Δhag | |
| DK72 | ΔfliW Δhag amyE::Phagtranslational-lacZ spec | |
| DK159 | ΔfliD ΔfliW | |
| DK374 | srfAC::Tn10 spec epsH::tet | |
| DK375 | ΔcsrA srfAC::Tn10 spec epsH::tet | |
| DK376 | ΔfliW ΔcsrA srfAC::Tn10 spec epsH::tet | |
| DK377 | ΔfliW srfAC::Tn10 spec epsH::tet | |
| DK378 | Δhag srfAC::Tn10 spec epsH::tet | |
| DK396 | ΔfliS ΔcsrA srfAC::Tn10 spec epsH::tet | |
| DK397 | ΔfliS srfAC::Tn10 spec epsH::tet | |
| DS278 | amyE::Phagtranslational-lacZ spec | 23 |
| DS793 | amyE::Phagtranscriptional-lacZ cat | 36 |
| DS1677 | Δhag | |
| DS1916 | amyE::Phag-hagT209C spec | 27 |
| DS6188 | ΔcsrA | 23 |
| DS6189 | ΔfliW ΔcsrA | 23 |
| DS6245 | ΔfliW | 23 |
| DS6772 | ΔcsrA amyE::Phag-hagT209C spec | |
| DS6773 | ΔfliW ΔcsrA amyE::Phag-hagT209C spec | |
| DS6774 | ΔfliW amyE::Phag-hagT209C spec | |
| DS7596 | ΔcsrA amyE::Phagtranslational-lacZ spec | |
| DS7597 | ΔfliW ΔcsrA amyE::Phagtranslational-lacZ spec | |
| DS7598 | ΔfliW amyE::Phagtranslational-lacZ spec | |
| DS7791 | ΔfliD | |
| DS7792 | ΔfliS | |
| DS7812 | ΔfliS amyE::Phag-hagT209C spec | |
| DS7814 | ΔfliS amyE::Phagtranslational-lacZ spec | |
| DS8255 | ΔfliS ΔcsrA | |
| DS8267 | ΔfliS ΔcsrA amyE::Phag-hagT209C spec | |
| DS8323 | ΔcsrA amyE::Phagtranscriptional-lacZ cat | |
| DS8324 | ΔfliW ΔcsrA amyE::Phagtranscriptional-lacZ cat | |
| DS8325 | ΔfliW amyE::Phagtranscriptional-lacZ cat | |
| DS8525 | ΔfliS amyE::Phagtranscriptional-lacZ cat | |
| DS8526 | ΔfliS ΔcsrA amyE::Phagtranslational-lacZ spec | |
| DS8527 | ΔfliS ΔcsrA amyE::Phagtranscriptional-lacZ cat | |
| DS8622 | Δhag amyE::Phagtranslational-lacZ spec | |
| DS9181 | ΔfliS amyE::Physpank-fliS spec | |
| DS9229 | ΔfliD ΔfliS | |
| DS9668 | ΔfliD ΔcsrA | |
| DS9669 | ΔfliD ΔfliS ΔcsrA | |
| DS9733 | ΔfliS ΔcsrA amyE::Physpank-fliS spec | |
| DS9898 | ΔfliD ΔfliW ΔcsrA | |
| DS9902 | ΔfliS Δhag amyE::Phagtranslational-lacZ spec | |
| DS9904 | Δhag amyE::Phagtranscriptional-lacZ cat | |
| DS9905 | ΔfliS Δhag amyE::Phagtranscriptional-lacZ cat |
(i) In-frame deletions.
To generate the ΔfliS in-frame markerless deletion construct, the region upstream of fliS was PCR amplified by using primer pair 2541/2542 and digested with EcoRI and XhoI, and the region downstream of fliS was PCR amplified by using primer pair 2543/2544 and digested with XhoI and BamHI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of pMiniMAD2, which carries a temperature-sensitive origin of replication and an erythromycin resistance cassette, to generate pDP363 (30). Plasmid pDP363 was introduced into PY79 by single-crossover integration by transformation at the restrictive temperature for plasmid replication (37°C), using mls resistance as a selection. The integrated plasmid was then transduced into 3610. To evict the plasmid, the strain was incubated in 3 ml LB broth at a permissive temperature for plasmid replication (22°C) for 14 h, diluted 30-fold in fresh LB broth, and incubated at 22°C for another 8 h. Dilution and outgrowth were repeated two more times. Cells were then serially diluted and plated onto LB agar at 37°C. Individual colonies were patched onto LB plates and LB plates containing mls to identify mls-sensitive colonies that had evicted the plasmid. Chromosomal DNA from colonies that had excised the plasmid was purified and screened by PCR using primer pair 2541/2544 to determine which isolate had retained the ΔfliS allele.
The ΔfliD construct was built in a way similar to ΔfliS by using primer pairs 2537/2538 and 2539/2540 to generate pDP362. Primer pairs 2537/2538 and 2543/2544 were used for the ΔfliD fliS deletion construct pSM36.
The Δhag construct was built in a way similar to ΔfliS by using primer pairs 575/576 and 577/578 to generate pDP200.
(ii) Complementation constructs.
To generate the inducible amyE::Physpank-fliS spec construct pSM34, a PCR product containing fliS was amplified from B. subtilis 3610 chromosomal DNA using primer pair 2906/2907, digested with HindIII and NheI, and cloned into the HindIII and NheI sites of pDR111 containing a spectinomycin resistance cassette, a polylinker downstream of the Physpank promoter, and the gene encoding the LacI repressor between the two arms of the amyE gene (31, 32).
(iii) Protein expression constructs. (a) His-SUMO-FliS fusion protein expression vector.
To generate the translational fusion of FliS to the His-SUMO tag, a fragment containing fliS was amplified by using 3610 as a template and primer pair 2230/2231 and was digested with SapI and XhoI. The fragment was ligated into the SapI and XhoI sites of pTB146 containing an ampicillin resistance cassette (33), to create pSM25.
(b) His-SUMO-Hag fusion protein expression vector.
To generate the translational fusion of FliS to the His-SUMO tag, a fragment containing hag was amplified by using 3610 as a template and primer pair 3317/3331 and was digested with SalI and BamHI. The fragment was ligated into the SalI and BamHI sites of pTB146 containing an ampicillin resistance cassette (33), to create pSM56.
Hag antibody preparation.
One milligram of purified Hag protein was sent to Cocalico Biologicals Inc. for serial injection into a rabbit host for antibody generation. Anti-Hag serum was mixed with Hag-conjugated Affigel-10 beads and incubated overnight at 4°C. The beads were packed onto a 1-cm column (Bio-Rad) and then washed with 100 mM glycine (pH 2.5) to release the antibody and immediately neutralized with 2 M Tris base. The purification of the antibody was verified by SDS-PAGE. Purified anti-Hag antibody was dialyzed into 1× PBS–50% glycerol and stored at −80°C.
SPP1 phage transduction.
Serial dilutions of SPP1 phage stock were added to 0.2 ml of dense culture grown in TY broth (LB broth supplemented after autoclaving with 10 mM MgSO4 and 100 μM MnSO4), and the mixture was statically incubated for 15 min at 37°C. Three milliliters of TYSA (molten TY broth supplemented with 0.5% agar) was added to each mixture and poured atop fresh TY plates, and the mixture was incubated at 37°C overnight. Top agar from the plate containing near-confluent plaques was harvested by scraping into a 50-ml conical tube, vortexed, and centrifuged at 5,000 × g for 10 min. The supernatant was treated with 25 μg/ml DNase (final concentration) before being passed through a 0.45-μm syringe filter and stored at 4°C.
Recipient cells were grown to the stationary phase in 2 ml TY broth at 37°C. Cells (0.9 ml) were mixed with 5 μl of SPP1 donor phage stock. Nine milliliters of TY broth was added to the mixture, and the mixture was allowed to stand at 37°C for 30 min. The transduction mixture was then centrifuged at 5,000 × g for 10 min, the supernatant was discarded, and the pellet was resuspended in the remaining volume. The cell suspension (100 μl) was then plated onto TY medium fortified with 1.5% agar, the appropriate antibiotic, and 10 mM sodium citrate.
β-Galactosidase assay.
One milliliter of cells was harvested from a mid-log-phase (OD600 of ∼0.5) culture grown in LB broth with shaking at 37°C and assayed for β-galactosidase activity, as described previously (23).
Protein purification.
The FliS protein expression vector pSM25 was transformed into Escherichia coli Rosetta Gami cells grown to an OD600 of ∼0.7 in 500 ml of Terrific Broth (12 g tryptone, 24 g yeast extract, 4 ml glycerol, 2.31 g monobasic potassium phosphate, and 12.54 g dibasic potassium phosphate per liter), induced with 1 mM IPTG, and grown overnight at 16°C. Cells were pelleted and resuspended in lysis buffer (50 mM Na2HPO4, 300 mM NaCl, 10 mM imidazole), treated with lysozyme, and lysed by sonication. Lysed cells were ultracentrifuged at 6,339 × g for 30 min. The cleared supernatant was combined with Ni-nitrilotriacetic acid (NTA) resin (Novagen) and incubated for 1 h at 4°C. The bead-lysate mixture was poured onto a 1-cm separation column (Bio-Rad), and the resin was allowed to pack and was washed with wash buffer (50 mM Na2HPO4, 300 mM NaCl, 30 mM imidazole). His-SUMO-FliS bound to the resin was then eluted by using a stepwise elution of wash buffer with 50 to 500 mM imidazole. Elution mixtures were separated by SDS-PAGE and Coomassie stained to verify the purification of the His-SUMO-FliW fusion. Purified His-SUMO-FliS was combined with ubiquitin ligase (protease) and cleavage buffer and incubated overnight at 4°C to cleave the SUMO tag from the FliW protein (34). The cleavage reaction mixture was combined with Ni-NTA beads, incubated for 2 h at 4°C, and centrifuged to pellet the resin. The supernatant was removed and dialyzed in a solution containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% glycerol, and 1 mM dithiothreitol (DTT) and stored at −20°C. The removal of the SUMO tag was verified by SDS-PAGE and Coomassie staining.
His6-Hag, His-SUMO-FliW, and His-SUMO-Hag proteins were purified in a similar way, and the glutathione S-transferase (GST)–CsrA protein was purified as described previously (23).
Protein-protein interaction assays. (i) His-Hag and FliS interactions.
Ni-NTA beads were washed with T(0.1) buffer (25 mM Tris-HCl [pH 8.0], 20% glycerol, 100 mM NaCl, 1 mM DTT, 1× protease inhibitor cocktail [from Roche]) containing 10 mM imidazole. Thirty microliters of washed beads was mixed with 30 μl of 5 μM His-Hag protein and rotated on a Labquake instrument at 4°C for 2 h. Next, the beads bound to the His-Hag protein were centrifuged at 56 × g for 2 min, and the pellet was washed twice with T(1.0) buffer (25 mM Tris-HCl [pH 8.0], 20% glycerol, 1 M NaCl, 1 mM DTT, 1× protease inhibitor cocktail) containing 10 mM imidazole and again washed twice with T(0.1) buffer containing 10 mM imidazole. Thirty microliters of increasing concentrations of FliS (5, 10, and 20 μM) was then added to the washed beads bound to His-Hag and rotated on a Labquake instrument at 4°C for 2 h. The samples were centrifuged at 56 × g for 2 min, and the supernatant was saved. The pellet was washed 4 times with T(0.1) buffer containing 10 mM imidazole. The supernatant and pellet fractions were subjected to SDS-PAGE analysis and stained with Coomassie brilliant blue.
Assays of the His-SUMO-FliW and FliS interaction and the His-SUMO-FliS and Hag or FliW interaction were carried out similarly. The assays of GST-CsrA and FliW or FliS interactions were also done in a similar way by using glutathione-Sepharose beads that bind to the GST tag (23).
(ii) Hag-FliW-FliS interactions.
Ni-NTA beads were washed with T(0.1) buffer containing 10 mM imidazole. Thirty microliters of washed beads was mixed with 30 μl of 5 μM His-Hag protein and rotated on a Labquake instrument at 4°C for 2 h. Next, the beads bound to the His-Hag protein were centrifuged at 56 × g for 2 min, and the pellet was washed twice with T(1.0) buffer containing 10 mM imidazole and again washed twice with T(0.1) buffer containing 10 mM imidazole. Thirty microliters of a mixture of 5 μM FliW and increasing concentrations of FliS (0, 5, 10, and 20 μM) was then added to the washed beads bound to His-Hag and rotated on a Labquake instrument at 4°C for 2 h. The samples were centrifuged at 56 × g for 2 min, and the supernatant was saved. The pellet was washed 4 times with T(0.1) buffer. The supernatant and pellet fractions were subjected to SDS-PAGE analysis and stained with Coomassie brilliant blue. In a similar experiment, 30 μl of a mixture of 5 μM FliS and increasing concentrations of FliW (0, 5, 10, and 20 μM) was added to the washed beads bound to His-Hag and analyzed by SDS-PAGE and Coomassie brilliant blue staining.
RESULTS
FliS and FliW bind to Hag simultaneously.
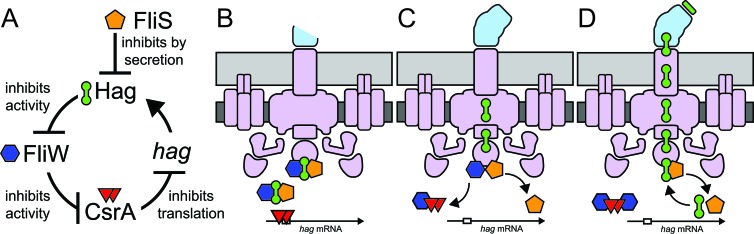
The flagellin protein Hag is reportedly bound by two different proteins in the cytoplasm, FliS and FliW (15, 22, 23). The FliS-Hag interaction is thought to be involved in filament assembly, where FliS acts as a secretion chaperone for flagellin (Fig. 1A) (15). In contrast, the FliW-Hag interaction was recently proposed to be regulatory, where FliW is part of a partner-switching mechanism with the RNA binding protein CsrA to maintain Hag homeostasis inside the cell (23). According to the homeostatic model, when cytoplasmic levels of Hag exceed a threshold, Hag sequesters FliW to liberate CsrA. CsrA in turn limits Hag protein levels by binding to the hag transcript, inhibiting hag translation, and reducing Hag protein levels to restore homeostasis (Fig. 1A). When Hag is secreted during filament assembly, the FliW-CsrA interaction is restored, the hag transcript is relieved from repression, and the Hag protein is translated at a high level. We predicted that the reduced ability to secrete Hag in the absence of a chaperone (e.g., FliS) would have many phenotypes similar to those in the absence of FliW while having additional secretion-specific defects. As FliS and FliW are closely coupled by Hag, we wondered whether the specific roles of the two proteins could be distinguished.
Fig 1.
Model for how FliS governs cytoplasmic flagellin homeostasis through the Hag-FliW-CsrA system. (A) Model for the genetic and biochemical relationships of the FliS, Hag, FliW, and CsrA proteins. T bars indicate inhibition. (B to D) Model for the maintenance of flagellin homeostasis morphogenically coupled to flagellar assembly. Orange pentagons, FliS; green barbells, Hag; dark blue hexagons, FliW; red triangles, CsrA; open box in mRNA, SD sequence; purple, basal body; blue, hook; green, growing filament; dark gray, cell membrane; light gray, peptidoglycan.
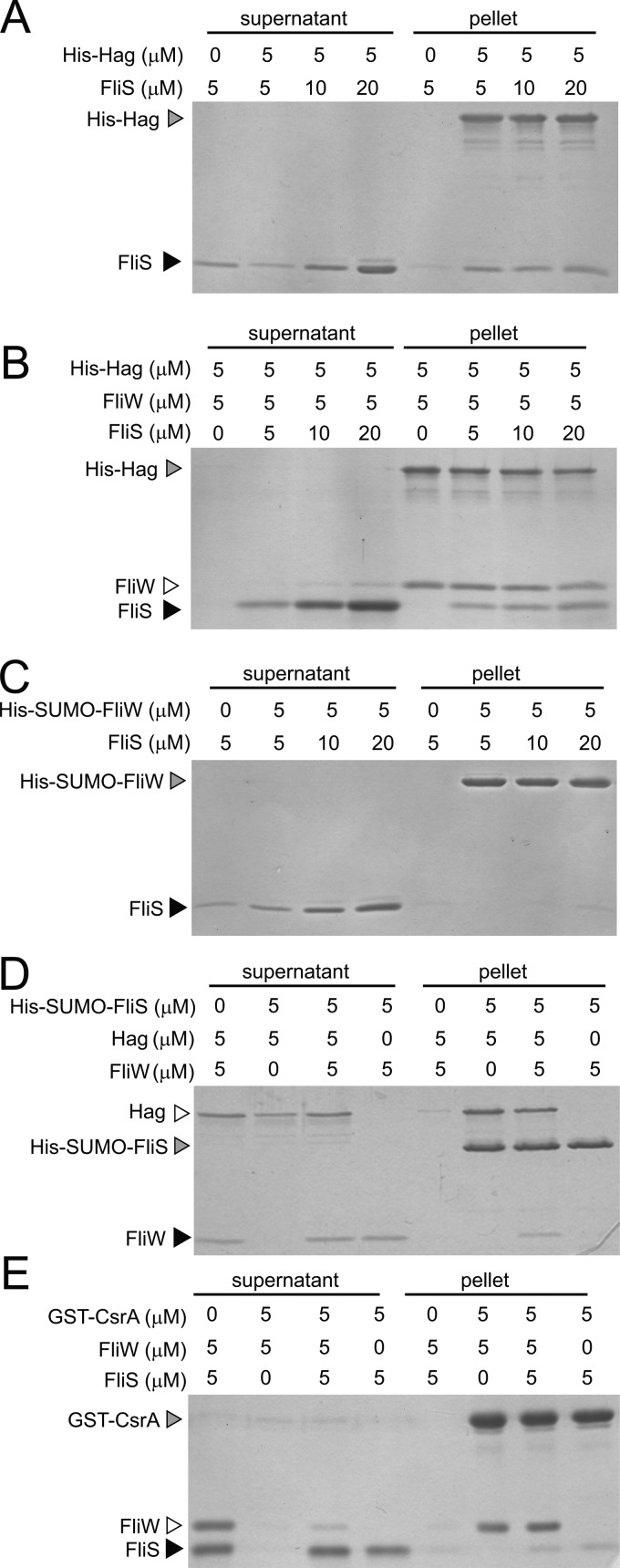
To confirm the Hag-FliS interaction, both proteins were purified for a protein pulldown assay. FliS was previously reported to exist and function in either a monomer or dimer form depending on the bacterial species (12, 16). When the purified B. subtilis FliS protein was subjected to fast pressure liquid chromatography (FPLC) using a Superdex 75 size-exclusion column, FliS eluted predominantly as a dimer (data not shown). For the pulldown assay, various concentrations of FliS dimers were incubated with nickel-NTA agarose beads bound to 5 μM His-Hag fusion protein. The beads were centrifuged, and the supernatant and pellet fractions were resolved by SDS-PAGE and then stained with Coomassie brilliant blue. FliS was retained in the pellet fraction when both FliS and Hag were at roughly equimolar concentrations (Fig. 2A). The retention of FliS in the pellet was dependent on the presence of Hag, as FliS was poorly retained in the pellet when mixed with nickel-NTA agarose beads alone (Fig. 2A). We conclude that FliS and Hag directly interacted in vitro.
Fig 2.
FliS and FliW bind to Hag simultaneously. (A and B) Protein pulldown assays using the indicated amounts of His-Hag (gray carets) loaded onto a nickel column with the indicated amounts of FliS (black carets) and/or FliW proteins (open caret) added. (C) Protein pulldown assay using the indicated amounts of His-SUMO-FliW (gray caret) loaded onto a nickel column with the indicated amounts of FliS (black caret) added. (D) Protein pulldown assay using the indicated amounts of His-SUMO-FliS (gray caret) loaded onto a nickel column with the indicated amounts of FliW (black caret) and/or Hag proteins (open caret) added. (E) Protein pulldown assay using the indicated amounts of GST-CsrA (gray caret) loaded onto a glutathione-Sepharose column with the indicated amounts of FliS (black caret) and/or FliW proteins (open caret) added. Gels were stained with Coomassie brilliant blue. “Supernatant” indicates the proteins that failed to bind to the beads, and “pellet” indicates the proteins that remained bound to the beads following a series of washes.
To determine the effect of FliS on the previously reported FliW-Hag interaction in vitro, various concentrations of FliS were mixed with 5 μM FliW and 5 μM His-Hag fusion protein bound to nickel-NTA agarose beads (23). The beads were centrifuged, and the supernatant and pellet fractions were resolved by SDS-PAGE and then stained with Coomassie brilliant blue. Both FliS and FliW were retained in the pellet fraction at roughly equimolar concentrations, and saturating amounts of FliS did not displace FliW (Fig. 2B). Similar results were obtained when various concentrations of FliW were mixed with 5 μM FliS and 5 μM His-Hag protein bound to nickel-NTA agarose beads: both proteins bound to Hag at roughly equimolar concentrations, and saturating amounts of FliW did not displace FliS (see Fig. S1 in the supplemental material). It appeared that FliW and FliS bound to Hag simultaneously. In addition, FliS did not bind to beads containing only His-SUMO-FliW (Fig. 2C), and FliW did not bind to beads containing only His-SUMO-FliS, unless Hag was also included in the reaction mixture, further supporting a three-protein complex (Fig. 2D). Thus, we conclude that both FliS and FliW bind to Hag simultaneously, that the two proteins do not compete with each other for Hag binding, and that FliS and FliW do not interact directly with one another.
FliS antagonizes Hag negative-feedback inhibition.
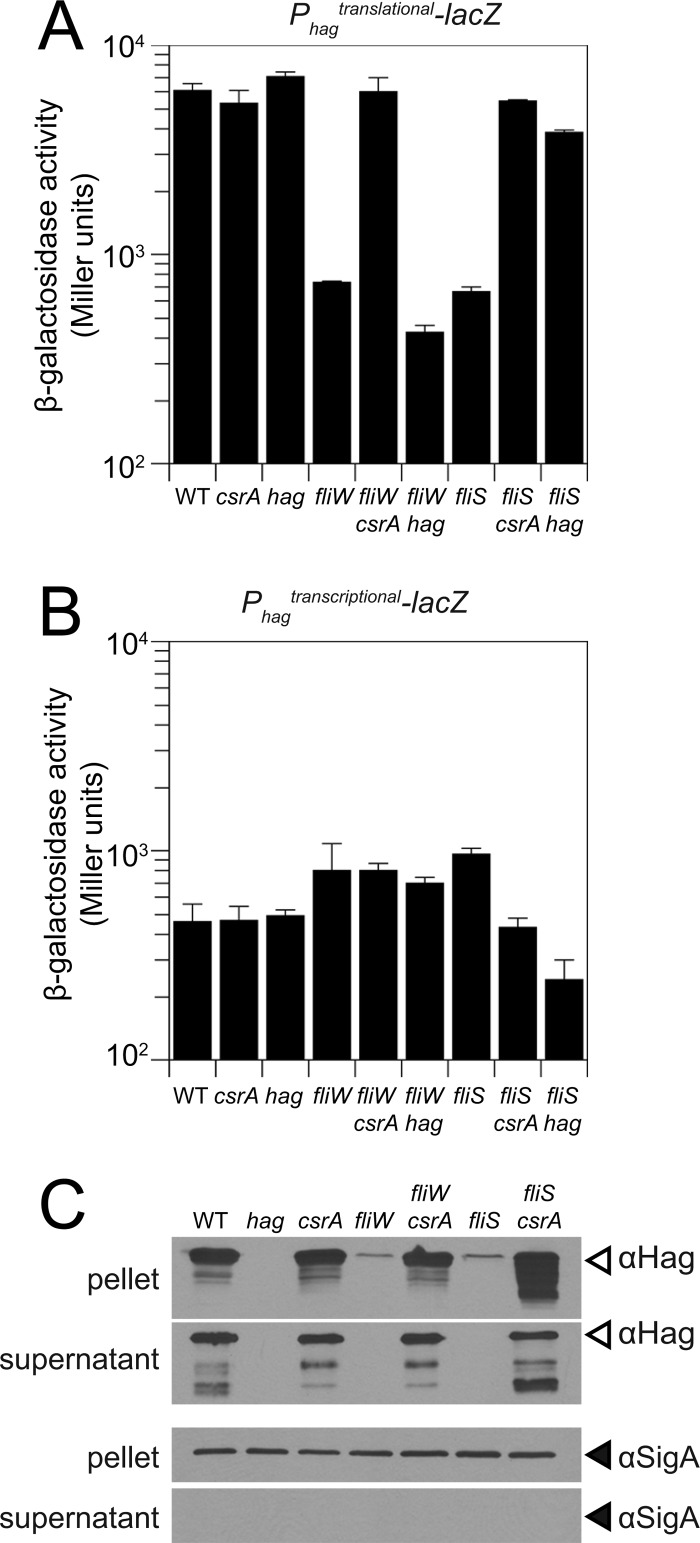
FliW is required for the efficient translation of the hag transcript. The level of expression from a translational reporter in which the lacZ gene encoding β-galactosidase was translationally fused to the Phag promoter, the hag 5′ untranslated region (5′UTR), and the hag start codon (amyE::Phagtranslational-lacZ) was reduced 10-fold in the absence of fliW compared to the wild type (Fig. 3A). The effect of the FliW mutant was mediated by the translational regulator CsrA, as a fliW csrA double mutant restored wild-type expression levels (Fig. 3A). Similarly, the mutation of FliS resulted in a 10-fold reduction in the expression level of the Hag translational reporter, which was restored to wild-type levels in a fliS csrA double mutant (Fig. 3A). Neither the mutation of fliW nor the mutation of fliS significantly decreased the expression level of a transcriptional reporter of lacZ fused to the Phag promoter (amyE::Phagtranscriptional-lacZ) (Fig. 3B). Consistent with the reduction in the level of translation of Hag, the mutation of either fliS or fliW dramatically decreased the amount of Hag protein both associated with the cell and secreted into the supernatant (Fig. 3C). Finally, Hag protein levels were restored to both fliS and fliW background levels by the mutation of csrA (Fig. 3C). We conclude that the mutation of fliS phenocopies the mutation of fliW with respect to Hag translation.
Fig 3.
FliS acts upstream of Hag in the Hag-FliW-CsrA negative-feedback loop. (A and B) β-Galactosidase assays of Phagtranslational-lacZ and Phagtranscriptional-lacZ fusions for strains of the indicated genotypes (raw data are presented in Table S3 in the supplemental material). Error bars are the standard deviations of data from three replicates. (C) Western blot analysis of separate B. subtilis cell lysates (pellet) and TCA-precipitated culture supernatants of the wild-type (WT) (3610), hag (DS1677), csrA (DS6188), fliW (DS6245), fliW csrA (DS6189), fliS (DS7792), and fliS csrA (DS8255) strains, separately probed with anti-Hag and anti-SigA primary antibodies. Note that the pellet represents both the combined Hag pools in the cytoplasm and Hag polymerized into flagellar filaments on the cell surface.
FliW indirectly promotes Hag translation by antagonizing the RNA binding protein CsrA. In the absence of FliW, CsrA binds to two sites in the 5′UTR of the hag transcript and occludes the Shine-Dalgarno (SD) sequence necessary for ribosome binding (24). FliW antagonizes the RNA binding activity of CsrA by direct protein-protein interactions (23). To determine whether FliS and FliW promote the efficient translation of the hag transcript by similar partner-switching mechanisms, we tested whether FliS, like FliW, could bind to CsrA. To determine the interaction with CsrA, a protein pulldown experiment was conducted in which 5 μM FliS or 5 μM FliW was incubated with glutathione-Sepharose beads bound to 5 μM GST-CsrA fusion protein. FliW but not FliS was retained in the pellet fraction (Fig. 2E). We conclude that FliS does not bind to CsrA, and we infer that the mechanism by which FliS antagonizes CsrA is distinct from that of FliW.
One way in which FliS might antagonize CsrA inhibition of hag translation is by antagonizing the cytoplasmic Hag protein level. Hag antagonism would relieve inhibition on FliW such that FliW would in turn bind and sequester CsrA (Fig. 1A). Consistent with FliS antagonizing Hag, a fliS hag double mutant phenocopied a fliS csrA double mutant (Fig. 3A). We note that although there was a 2-fold decrease in the expression level of the hag transcriptional reporter (amyE::Phagtranscriptional-lacZ) in the fliS hag double mutant, the expression level of the hag translational reporter (amyE::Phagtranslational-lacZ) was nonetheless restored to wild-type levels in the fliS hag double mutant (Fig. 3A and B). Thus, the mutation of hag was epistatic to the mutation of fliS. In contrast, the expression level of the hag translation fusion reporter remained low in a fliW hag double mutant (Fig. 3A). Thus, the mutation of fliW was epistatic to the mutation of hag. We conclude that both CsrA and Hag are downstream of FliS, and we infer that FliS antagonizes CsrA indirectly by antagonizing Hag (Fig. 1A).
FliS is required for motility and enhances Hag secretion.
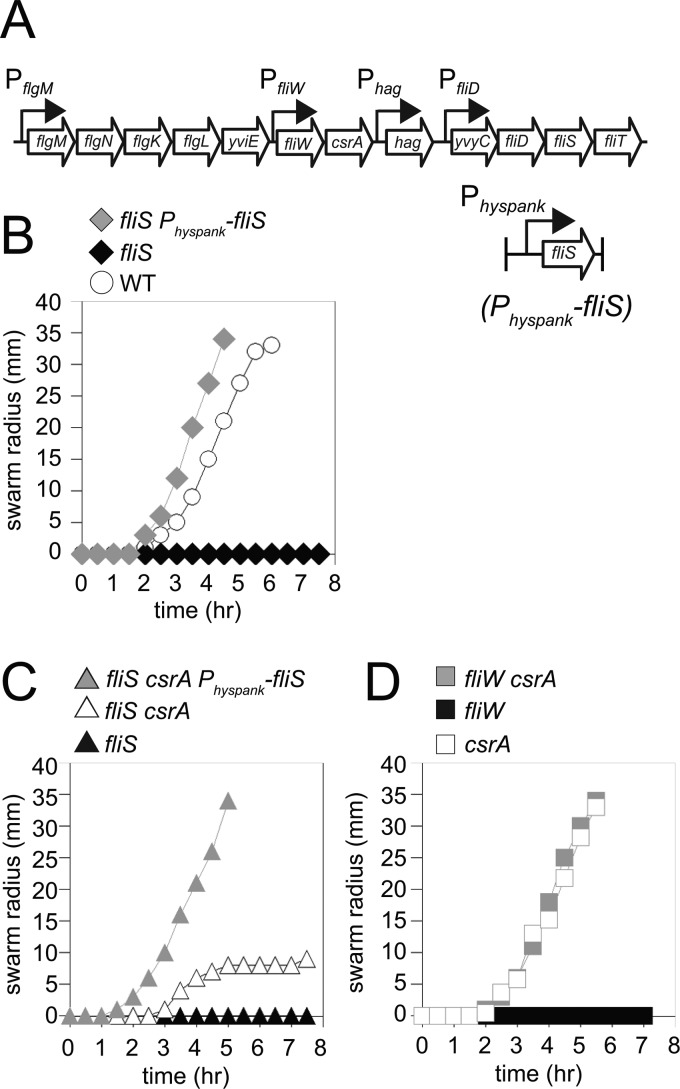
One way in which FliS might antagonize Hag is by enhancing Hag secretion similarly to its reported role in Salmonella enterica. Enhanced Hag secretion would lower the level of the Hag protein in the cytoplasm and thereby disrupt the homeostatic repression of Hag translation. To begin to study the biology of FliS, we first determined the consequence of a fliS mutant on swarming motility (35). Whereas wild-type cells swarmed rapidly atop the surface of an agar petri plate, cells mutated for fliS exhibited a severe defect in motility, which was rescued when fliS was expressed from an IPTG-inducible promoter inserted at an ectopic site in the chromosome (amyE::Physpank-fliS) (Fig. 4A and B). The fliS defect in swarming motility was similar to the severe defect in swarming exhibited by cells mutated for fliW (Fig. 4C and D). Whereas a mutation in csrA was sufficient to restore both wild-type swarming and swimming motility to cells mutated for fliW, the mutation of csrA restored only partial motility to cells mutated for fliS (Fig. 4C and D; see also Fig. S2 in the supplemental material). We conclude that FliS is required to indirectly antagonize CsrA activity but that CsrA antagonism is not sufficient to fully explain the motility defect in a FliS mutant.
Fig 4.
FliS is required for motility. (A) Genetic neighborhood containing the fliW, csrA, hag, fliD, and fliS genes. Large open arrows represent open reading frames (lengths are not to scale). Bent arrows indicate promoters. The promoters PflgM, Phag, and PfliD are dependent on the alternate sigma factor σD, while PfliW is under the control of the vegetative sigma factor σA (24). (B) Quantitative swarm expansion assays for the wild-type (WT) strain (3610), the fliS strain (DS7792), and the fliS Physpank-fliS strain in the presence of 1 mM IPTG (DS9181). (C) Quantitative swarm expansion assays for the fliS csrA strain (DS8255), the fliS strain (DS7792), and the fliS csrA Physpank-fliS strain in the presence of 1 mM IPTG (DS9733). (D) Quantitative swarm expansion assays for the csrA (DS6188), fliW (DS6245), and fliW csrA (DS6189) strains. Each point is the average of data from three replicates.
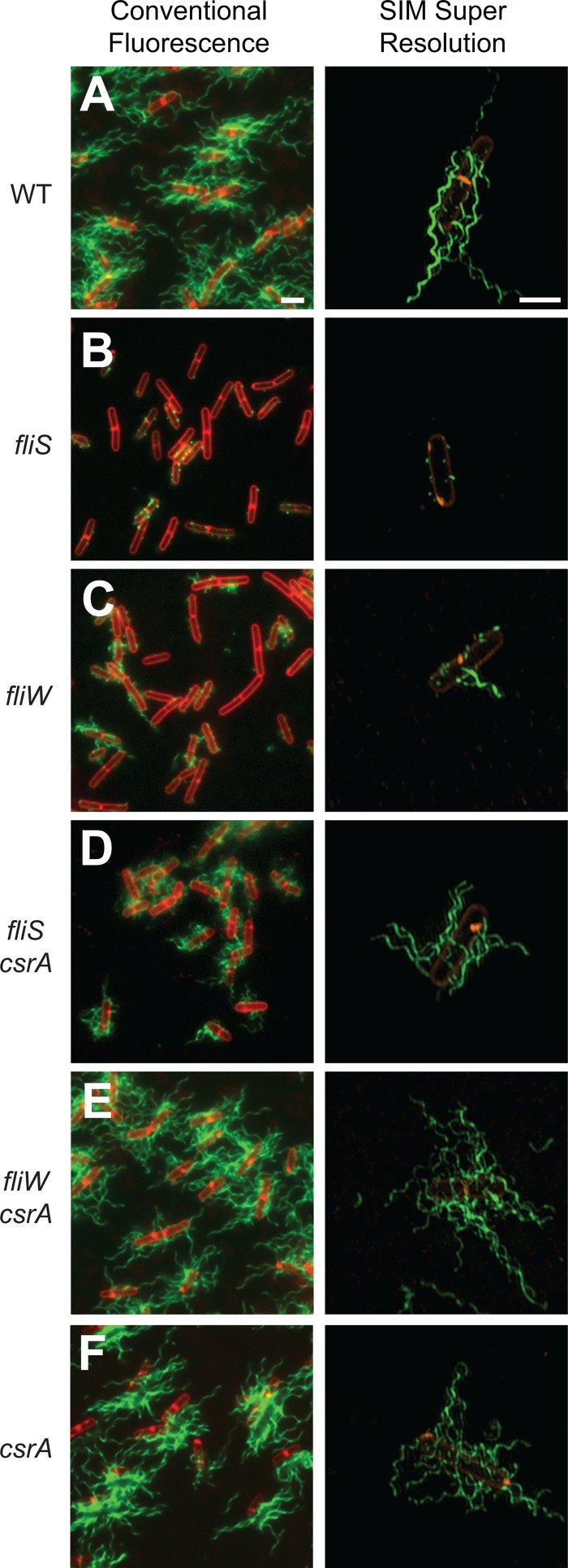
To explore the FliS motility defect further, we fluorescently labeled flagellar filaments in a variety of genetic backgrounds. Wild-type cells produced long filaments, but the mutation of fliS resulted in a defect in filament assembly such that cells exhibited filament stubs that resembled dots on the cell surface (Fig. 5A and B). In contrast, the mutation of fliW resulted in a defect less severe than that of fliS, where cells expressed a mixture of short filaments and filament stubs (Fig. 5C). Whereas the simultaneous mutation of fliW and csrA was sufficient to restore filaments to the wild-type length, the simultaneous mutation of fliS and csrA restored only filaments that remained short (Fig. 5D and E). The mutation of csrA alone produced long filaments, and thus, csrA was fully epistatic to fliW for filament assembly but not to fliS (Fig. 5F). We conclude that FliS is essential for filament assembly except when flagellin is translated at an abnormally high level (such as when cells are mutated for CsrA). We infer that the mutation of CsrA is insufficient, however, to restore wild-type motility to fliS mutants, because the flagellar filaments remain short. We hypothesize that shorter filaments in the fliS csrA double mutant are due to a reduced rate of Hag secretion.
Fig 5.
FliS is required for filament assembly. Shown are fluorescence micrograph overlays from conventional fluorescence microscopy and superresolution SIM fluorescence microscopy of the wild-type (WT) (DS1916) (A), fliS (DS7812) (B), fliW (DS6774) (C), fliS csrA (DS8267) (D), fliW csrA (DS6773) (E), and csrA (DS6772) (F) strains, membrane stained with FM4-64 (false colored red) and flagella stained with Alexa Fluor 488 maleimide (false colored green). Scale bars are 2 μm.
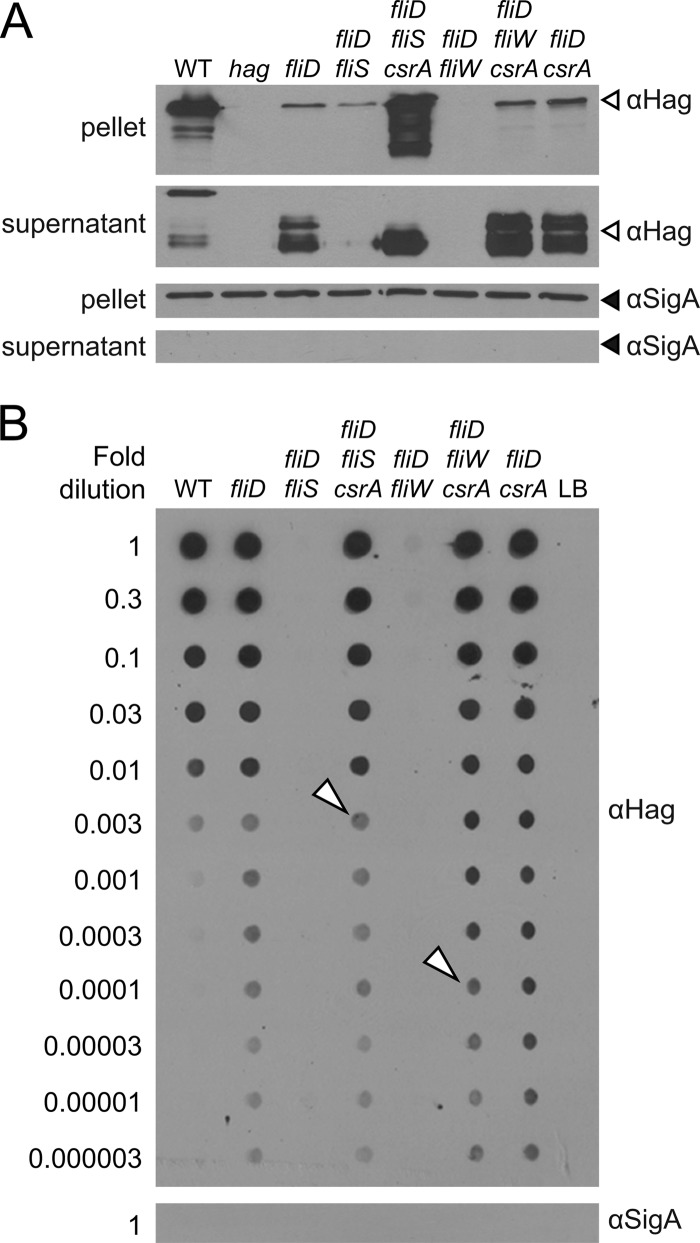
To measure the secretion of Hag, cells were first mutated for the fliD gene, encoding FliD, a protein that serves both as the filament cap and as an extracellular chaperone for the assembly of Hag proteins into the elongating filament (8). When FliD is mutated in S. enterica, the filament is not polymerized, and flagellin monomer units are secreted into the supernatant (3). To confirm that a fliD mutant behaves similarly in B. subtilis, cell pellets and TCA-precipitated culture supernatant fractions were harvested from the wild-type, hag, and fliD mutant strains. Each sample was separately resolved by SDS-PAGE, electroblotted, and probed with either anti-Hag or anti-SigA antibodies for Western blot analysis. Wild-type, but not hag mutant, cells had high levels of Hag in the pellet fraction (from combined cytoplasmic pools and assembled flagella) and high levels of Hag in the supernatant (from Hag secretion and filament shearing) (Fig. 6A). In contrast, cells mutated for fliD had low levels of Hag in the pellet fraction and high levels of Hag in the supernatant. Furthermore, we note that the Hag protein had a lower molecular weight in supernatants of the fliD mutant than in supernatants of the wild type, which is suggestive of proteolytic degradation (Fig. 3C and 6A). We conclude that FliD behaves similarly in B. subtilis and S. enterica such that in the absence of FliD, the Hag protein is not polymerized into the filament and is instead secreted abundantly into the supernatant. We further infer that in the absence of FliD, the inability to correctly fold and polymerize Hag renders the Hag protein susceptible to extracellular proteolytic degradation.
Fig 6.
FliS promotes Hag secretion. (A) Western blot analysis of B. subtilis cell lysates (pellet) and TCA-precipitated culture supernatants of the wild-type (WT) (3610), hag (DS1677), fliD (DS7791), fliS fliD (DS9229), fliS csrA fliD (DS9669), fliW fliD (DK159), fliW csrA fliD (DS9898), and csrA fliD (DS9668) strains, separately probed with anti-Hag and anti-SigA primary antibodies. Note that the pellet represents both the combined Hag pools in the cytoplasm and Hag polymerized into flagellar filaments on the cell surface. (B) Dot blot analysis of serially diluted B. subtilis culture supernatants of the wild-type (3610), fliD (DS7791), fliS fliD (DS9229), fliS csrA fliD (DS9669), fliW fliD (DK159), fliW csrA fliD (DS9898), and csrA fliD (DS9668) strains, separately probed with anti-Hag (all dilutions) and anti-SigA primary (only undiluted culture supernatant) antibodies. Open carets represent the first dilutions of the fliS csrA fliD and fliW csrA fliD samples that leave saturation and represent the two points being qualitatively compared for relative secretion efficiency in the text.
To compare the effects of FliS and FliW on the efficiency of Hag secretion, a fliS fliD double mutant and a fliW fliD double mutant were generated. In the absence of either FliS or FliW, very little Hag protein remained in the pellet, and the Hag protein was undetectable in the supernatant (Fig. 6A). The reduced amount of the Hag protein was due primarily to CsrA, as the amount of the Hag protein increased in both the fliS csrA fliD and fliW csrA fliD triple mutants (Fig. 6A). Importantly, Hag accumulated cytoplasmically in the fliS csrA fliD triple mutant but accumulated extracellularly in the fliW csrA fliD triple mutant (Fig. 6A). We infer that the intracellular Hag accumulation observed in the absence of FliS is consistent with a defect in Hag secretion, but the amount of extracellular Hag in both triple mutants was saturating in the Western blots, rendering subtle qualitative comparisons between the samples difficult. To compare the relative amounts of secreted Hag, the supernatant fractions were serially diluted in a dot blotter and probed with an anti-Hag antibody. The fliS csrA fliD triple mutant accumulated approximately 30- to 100-fold less protein in the supernatant than the fliW csrA fliD triple mutant (Fig. 6B). We conclude that cells lacking FliS have a defect in Hag secretion and that the reduced secretion efficiency likely accounts for the short filaments and reduced motility of the fliS csrA double mutant.
DISCUSSION
An estimated 20,000 flagellin subunits assemble to form a single flagellar filament (2). To compensate for the massive metabolic investment in flagellar assembly, the primary sequence of flagellin has evolved to favor energy-efficient amino acids, and flagellin expression, secretion, and polymerization are highly regulated (11, 12, 23, 37). Flagellin readily oligomerizes into filaments in vitro, but in vivo, polymerization is restricted by intracellular and extracellular chaperones (38, 39). Each flagellin subunit is secreted in an unfolded state through the flagellar rod and hook by the flagellar type III secretion (T3S) apparatus within the basal body. Once flagellin emerges from the secretion conduit, it encounters an extracellular chaperone, FliD, that catalyzes flagellin folding and ushers flagellin assembly at the tip of the nascent filament. Secretion is enhanced by an intracellular chaperone, FliS, that binds to flagellin and may protect flagellin from proteolytic degradation, may maintain flagellin in an unfolded state, and/or may aid in the delivery of flagellin to the secretion apparatus (12, 16, 40, 41). B. subtilis encodes FliS and a second protein that binds to flagellin, FliW, thus raising the possibility of multiple cytoplasmic chaperones (15, 22).
A molecular chaperone can generally be defined as a protein that interacts directly with a target protein and catalyzes the folding state of the target (42). In the case of the putative chaperone FliW, however, the FliW-Hag interaction was found to be regulatory in a manner unrelated to protein folding (23). Instead, FliW acted as a sensor/regulator to control Hag protein levels. When Hag levels are high in the cytoplasm, Hag binds to FliW, and FliW is released from its other binding partner, the RNA binding protein CsrA. Free CsrA, in turn, binds to hag mRNA, represses Hag translation, and causes Hag levels to decrease. When Hag levels are low in the cytoplasm, FliW is free to bind to and antagonize CsrA, liberating the hag transcript and causing Hag levels to rise. Thus, the negative-feedback loop ensures that Hag levels oscillate around a low homeostatic level in the cytoplasm. Although FliW once appeared to be a chaperone, our studies revealed a more complex regulatory system of protein interactions.
FliW is not encoded by Gram-negative gammaproteobacteria (e.g., E. coli and S. enterica), and in these organisms, FliS is the only candidate chaperone. Like other T3S chaperones, FliS is a small, acidic protein (∼133 amino acids) that binds to the C terminus of its secretion substrate and enhances the efficiency of substrate secretion (16, 20). FliS of B. subtilis has a number of properties consistent with a secretion chaperone. FliS and Hag directly interact, and in the absence of FliS, cells are nonmotile due to a severe defect in filament assembly. Furthermore, in vitro, FliS was shown previously to be essential to promote interactions between Hag and the secretion component FlhA (15). In vivo, Hag secretion occurred in the absence of FliS (albeit impaired 30-fold) but only when the intracellular pool of Hag was artificially increased (Fig. 6B). Thus, FliS potentiates the secretion of Hag.
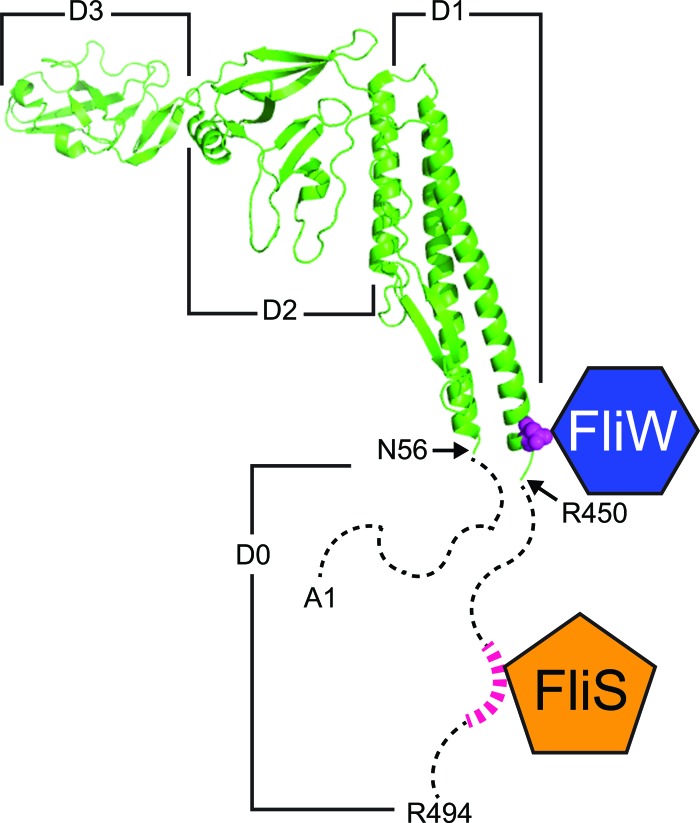
FliS binds to the last 40 amino acids of the Hag C terminus, whereas FliW binding requires residues 33 amino acids upstream of the FliS binding site (Fig. 7) (16, 22, 43). Thus, FliS and FliW bind to two different sites on the Hag protein, and our data indicate that simultaneous binding to Hag results in a FliS-Hag-FliW complex (Fig. 2B and C). We suggest that the simultaneous binding of FliS and FliW to Hag is important for the homeostatic regulatory system because competitive binding for Hag would result in Hag translation that was uncoordinated with flagellar assembly. For example, if the interaction between FliS and Hag displaced FliW, Hag translation would be constitutively activated even in the absence of secretion (Fig. 1A). Conversely, if the interaction between FliW and Hag displaced FliS, Hag secretion and Hag translation would be repressed even during filament synthesis. Instead, both proteins bind simultaneously to Hag, and FliW is displaced only when FliS catalyzes Hag export through the secretion apparatus, ensuring that Hag is translated only at a high level during the assembly of the filament.
Fig 7.
FliW and FliS are predicted to bind to two different regions of the Hag tertiary structure. The structure of the Hag homolog FliC from Salmonella enterica (43) is shown in green and is organized into four structural domains, D0, D1, D2, and D3. The FliW protein (dark blue hexagon) binds to FliC residue N445 (in magenta), with the caveats that S. enterica does not encode FliW and that B. subtilis Hag is much shorter than FliC due to the natural deletion of domain D3. Therefore, the analogous position at which FliW would bind to Hag is residue N255 (22). The FliS protein (orange pentagon) is predicted to bind to FliC in the disordered C terminus of domain D0 at the QAN motif (abstractly highlighted in pink) (16). We note that although the Hag protein is shorter in B. subtilis, the QAN residues are conserved in the C terminus. The N- and C-terminal disordered D0 domains of flagellin are represented by dashed lines (41).
How does FliS potentiate Hag secretion? One model suggests that FliS could protect Hag from cytoplasmic proteolysis (13). In B. subtilis, however, no low-molecular-weight products consistent with proteolytic degradation were observed from cytoplasmic Hag in the fliS mutant (Fig. 3C and 6A). Another model suggests that FliS could prevent the inappropriate polymerization of Hag in the cytoplasm (12). In B. subtilis, however, the homeostatic Hag-FliW-CsrA system may keep Hag below the critical concentration required for polymerization, rendering inhibition by FliS unnecessary. Another model suggests that FliS could maintain Hag in an unfolded, export-competent state prior to secretion (12). In B. subtilis, however, FliW functionally binds to Hag in the cytoplasm, suggesting that perhaps sufficient folding occurs at least at the interaction site, and other work suggests that FliS binding does not inhibit Hag folding in vitro (13, 14). Finally, FliS could catalyze the interaction of Hag with the secretion apparatus and thereby enhance the rate of export. In support of the latter model, work by other laboratories has shown that the secretion apparatus preferentially interacts with the FliS-Hag complex and that FliS governs the rate of filament elongation (15, 20). Based on our results, we favor the model that in B. subtilis, FliS promotes interactions between Hag and the secretion complex, as our data seem to be less supportive of the other options.
We note that there is a final mechanistic possibility for FliS, however, based on our work with FliW. Perhaps, like FliW, FliS is not just a chaperone but is also another, more complicated, regulator of flagellin. If FliS is not strictly a secretion chaperone, then what would FliS regulate, and how would its function be related to a defect in flagellin secretion? One possibility is that FliS could regulate the length of the flagellar filament. Previous data showed that the level of secretion of flagellin decreased proportionally with the length of the extending filament eventually to extinction, and thus, no active mechanism of filament length control needed to be invoked (7). Recent work, however, disagreed with the previous observations and showed that the rate of filament extension did not decrease with length and was in fact constant throughout synthesis (10). Thus, if the filament length is not inherently self-limiting, then how filament length is determined becomes an open question, and FliS could potentially participate by regulating flagellin secretion. We note that there is a precedent for the length control of the flagellar hook, in which the regulator FliK indirectly controls the secretion of the hook structural subunit FlgE (44). While we do not suggest that FliS functions like FliK, the distinction between a chaperone and an allosteric regulator is subtle, and the true function of FliS may be more complicated than we expect.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Powers, Suchetna Mukhopadhyay, and Sarah N. Fontaine for reagents, assistance, and helpful discussions.
We also thank the Indiana University Bloomington Light Microscopy Imaging Center for the OMX 3D-SIM Super-Resolution system, supported by NIH grant S10RR028697-01. This work was supported by NIH grants GM055969 to P.B. and GM093030 to D.B.K.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01654-12.
REFERENCES
- 1. Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6: 455– 465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57: 77– 100 [DOI] [PubMed] [Google Scholar]
- 3. Homma M, Fujita H, Yamaguchi S, Iino T. 1984. Excretion of unassembled flagellin by Salmonella typhimurium mutants defective in hook-associated proteins. J. Bacteriol. 159: 1056– 1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ikeda T, Asakura S, Kamiya R. 1989. Total reconstitution of Salmonella flagellar filaments from hook and purified flagellin and hook-associated proteins in vitro. J. Mol. Biol. 209: 109– 114 [DOI] [PubMed] [Google Scholar]
- 5. Ikeda T, Oosawa K, Hotani H. 1996. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J. Mol. Biol. 259: 679– 686 [DOI] [PubMed] [Google Scholar]
- 6. Emerson SU, Tokuyasu K, Simon MI. 1970. Bacterial flagella: polarity of elongation. Science 169: 190– 192 [DOI] [PubMed] [Google Scholar]
- 7. Iino T. 1969. Polarity of flagellar growth in Salmonella. J. Gen. Microbiol. 56: 227– 239 [DOI] [PubMed] [Google Scholar]
- 8. Ikeda T, Yamaguchi S, Hotani H. 1993. Flagellar growth in a filament-less Salmonella fliD mutant supplemented with hook-associated protein 2. J. Biochem. 114: 39– 44 [DOI] [PubMed] [Google Scholar]
- 9. Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26: 131– 158 [DOI] [PubMed] [Google Scholar]
- 10. Turner L, Stern AS, Berg HC. 2012. Growth of flagellar filaments is independent of flagellar length. J. Bacteriol. 194: 2437– 2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Macnab RM. 1996. Flagella and motility, p 123–145 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 12. Auvray F, Thomas J, Fraser GM, Hughes C. 2001. Flagellin polymerization control by a cytosolic export chaperone. J. Mol. Biol. 308: 221– 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ozin AJ, Claret L, Auvray F, Hughes C. 2003. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerization. FEMS Microbiol. Lett. 219: 219– 224 [DOI] [PubMed] [Google Scholar]
- 14. Papaneophytou CP, Papi RM, Pantazaki AA, Kyriakidis DA. 2012. Flagellin gene (fliC) of Thermus thermophilus HB8: characterization of its product and the involvement to flagella assembly and microbial motility. Appl. Microbiol. Biotechnol. 94: 1265– 1277 [DOI] [PubMed] [Google Scholar]
- 15. Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, Sinning I. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 107: 11295– 11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evdokimov AG, Phan J, Tropea JE, Routzahn KM, Peters HK, III, Pokross M, Waugh DS. 2003. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat. Struct. Biol. 10: 789– 793 [DOI] [PubMed] [Google Scholar]
- 17. Homma M, Fujita H, Yamaguchi S, Iino T. 1987. Regions of Salmonella typhimurium flagellin essential for its polymerization and excretion. J. Bacteriol. 169: 291– 296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. LaVallie ER, Stahl ML. 1989. Cloning of the flagellin gene from Bacillus subtilis and complementation studies of an in vitro-derived deletion mutation. J. Bacteriol. 171: 3085– 3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Capdevila S, Martínez-Granero FM, Sánchez-Contreras M, Rivilla R, Martín M. 2004. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150: 3889– 3897 [DOI] [PubMed] [Google Scholar]
- 20. Yokoseki T, Kutsukake K, Ohnishi K, Iino T. 1995. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology 141: 1715– 1722 [DOI] [PubMed] [Google Scholar]
- 21. Chen L, Helmann JD. 1994. The Bacillus subtilis σD-dependent operon encoding the flagellar proteins FliD, FliS, and FliT. J. Bacteriol. 176: 3093– 3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Titz B, Rajagopala SV, Ester C, Häuser R, Uetz P. 2006. Novel conserved assembly factor of the bacterial flagellum. J. Bacteriol. 188: 7700– 7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. 2011. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 82: 447– 461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 64: 1605– 1620 [DOI] [PubMed] [Google Scholar]
- 25. Sal MS, Chunhao L, Motalab MA, Shibata S, Aizawa S, Charon NW. 2008. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 190: 1912– 1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sze CW, Morado DR, Liu J, Charon NW, Xu H, Li C. 2011. Carbon storage regulator A (CsrABb) is a repressor of Borrelia burgdorferi flagellin protein B. Mol. Microbiol. 82: 851– 864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320: 1636– 1638 [DOI] [PubMed] [Google Scholar]
- 28. Canosi U, Luder G, Trautner TA. 1982. SPP1-mediated plasmid transduction. J. Virol. 44: 431– 436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 14: 1343– 1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70: 1166– 1179 [DOI] [PubMed] [Google Scholar]
- 31. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532– 536 [DOI] [PubMed] [Google Scholar]
- 32. Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57– 61 [DOI] [PubMed] [Google Scholar]
- 33. Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO. J. 28: 193– 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Butt TR, Edavettal SC, Hall JP, Mattern MR. 2005. SUMO fusion technology for difficult to express proteins. Protein Expr. Purif. 43: 1– 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49: 581– 590 [DOI] [PubMed] [Google Scholar]
- 36. Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19: 3083– 3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith DR, Chapman MR. 2010. Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio 1(3):e00131–10 doi:10.1128/mBio.00131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wakabayashi K, Hotani H, Asakura S. 1969. Polymerization of Salmonella flagellin in the presence of high concentrations of salts. Biochim. Biophys. Acta 175: 195– 203 [DOI] [PubMed] [Google Scholar]
- 39. Asakura S, Eguchi G, Iino T. 1964. Reconstitution of bacterial flagella in vitro. J. Mol. Biol. 10: 42– 56 [DOI] [PubMed] [Google Scholar]
- 40. Muskotal A, Kiraly R, Sebestyen A, Gugolya Z, Vegh BM, Vonderviszt F. 2006. Interaction of FliS flagellar chaperone with flagellin. FEBS Lett. 580: 3916– 3920 [DOI] [PubMed] [Google Scholar]
- 41. Aizawa SI, Vonderviszt F, Ishima R, Akasaka K. 1990. Termini of Salmonella flagellin are disordered and become organized upon polymerization into flagellar filament. J. Mol. Biol. 211: 673– 677 [DOI] [PubMed] [Google Scholar]
- 42. Ellis RJ. 1991. Molecular chaperones. Annu. Rev. Biochem. 60: 321– 347 [DOI] [PubMed] [Google Scholar]
- 43. Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410: 331– 337 [DOI] [PubMed] [Google Scholar]
- 44. Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30: 2948– 2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.