Abstract
Francisella tularensis is classified as a category A priority pathogen and causes fatal disseminated disease in humans upon inhalation of less than 50 bacteria. Although drugs are available for treatment, they are not ideal because of toxicity and route of delivery, and in some cases patients relapse upon withdrawal. We have an ongoing program to develop novel FAS-II FabI enoyl-ACP reductase enzyme inhibitors for Francisella and other select agents. To establish F. tularensis FabI (FtFabI) as a clinically relevant drug target, we demonstrated that fatty acid biosynthesis and FabI activity are essential for growth even in the presence of exogenous long-chain lipids and that FtfabI is not transcriptionally altered in the presence of exogenous long-chain lipids. Inhibition of FtFabI or fatty acid synthesis results in loss of viability that is not rescued by exogenous long-chain lipid supplementation. Importantly, whole-genome transcriptional profiling of F. tularensis with DNA microarrays from infected tissues revealed that FtfabI and de novo fatty acid biosynthetic genes are transcriptionally active during infection. This is the first demonstration that the FabI enoyl-ACP-reductase enzyme encoded by F. tularensis is essential and not bypassed by exogenous fatty acids and that de novo fatty acid biosynthetic components encoded in F. tularensis are transcriptionally active during infection in the mouse model of tularemia.
INTRODUCTION
Although the incidence of the disease tularemia is low and although drugs are available for treatment, interest in the causative agent Francisella tularensis remains due to its highly infectious nature and because of the possibility that F. tularensis could be used deliberately as a bioterror agent. Currently, treatment of tularemia consists of a single course of streptomycin augmented with a fluoroquinolone if needed (1). While the emergence of naturally resistant strains is rare, there is a need for new chemotherapeutics to treat acute disseminated infections due to the fact that current therapies must be administered early during infection for positive disease outcome. Further, due to the low 50% lethal dose (LD50; <50 CFU) of F. tularensis, antibiotic treatments that result in 100% clearance of the bacteria are required (1–3).
The bacterial type II fatty acid synthesis (FAS-II) pathway is distinct from the mammalian fatty acid biosynthesis pathway and therefore has emerged as an attractive pathway for novel therapeutic intervention. As a rate-limiting step in the FAS-II system, the enoyl-ACP reductase enzyme FabI has been a prime example of an enzyme that can be targeted for clinically relevant drug discovery. As part of our broad-spectrum drug discovery program, we have developed a series of lead compounds with activity against FAS-II enoyl-ACP reductases of Mycobacterium tuberculosis and priority pathogens F. tularensis, Burkholderia pseudomallei, and Yersinia pestis (4–6). Lead compounds have been shown to inhibit F. tularensis FabI (FtFabI) enoyl-ACP reductase activity in vitro and demonstrate efficacy against F. tularensis in the mouse model of infection (4, 6). Since drug efficacy in host models only indirectly demonstrates the essentiality of the FtfabI gene product because of possible off-target effects, there is a need to directly determine the requirement for FtFabI for de novo synthesis of fatty acids and activity during infection.
Recently it has been reported that there is a significant decrease in expression and production of FAS-II enzymes encoded by Streptococcus agalactiae in the presence of exogenous fatty acids. This observation indicates that this bacterium can utilize host lipids to circumvent de novo fatty acid biosynthesis (7). Based on this observation, it has been argued that the FAS-II pathway is not a suitable antimicrobial target for Gram-positive organisms because fatty acid synthesis is not essential in the host environment. However, it has also been demonstrated that fatty acid synthesis is required to maintain viability of the Gram-positive organism Staphylococcus aureus, and the differences observed in the essentiality of the enoyl-reductase step of FAS-II may be dependent on the ability of pathogens to subsist on exogenously supplied fatty acids (8). The heterogeneity of these observations suggests that there are organism-specific capabilities in terms of nutrient uptake and utilization that may rely heavily on the evolutionary background, the metabolic capacity, and the pathobiological niche that an organism occupies (7, 9, 10).
While the utilization of host lipids and the extent to which host lipids can be used appear to be more relevant to Gram-positive organisms and applicable to specific species of Gram-positive bacteria, it is important to evaluate FAS-II essentiality on a pathogen-by-pathogen basis. Gram-negative bacteria are known to be able to take up exogenous lipids and utilize them as intermediate substrates for the synthesis of phospholipids; however, unique bacterial lipids cannot be obtained directly from host sources, thus requiring biosynthesis by the bacterium. Notably, F. tularensis is characterized by hydroxyl fatty acids (11), and the major lipid A form is tetraacylated with 3-hydroxyoctadecanoic acid, 3-hydroxyhexadecanoic acid, hexadecanoic acid, and tetradecanoic acid (12). Although the unique lipid components of F. tularensis are unlikely to be supplied by the host and incorporated as intermediates without biosynthesis, as part of a rigorous academic drug discovery program there is a need to determine if FtFabI is essential under infectious conditions in order to efficiently translate in vitro potency to in vivo efficacy and to substantiate this protein as a clinically relevant drug target.
To experimentally substantiate FtFabI as a clinically relevant drug target for the treatment of tularemia, we set out to verify that FtFabI activity is essential for growth, that FtfabI expression is not influenced by the presence of an exogenously supplied mixture of long-chain lipids, and that FtfabI is expressed during infection. We report here that de novo biosynthesis of fatty acids is essential in vitro based on the inability to obtain an auxotrophic strain in the presence of exogenous fatty acids. The presence of exogenous fatty acids does not alter the transcriptional activity of fatty acid biosynthetic genes or FtfabI, and whole-genome transcriptional profiling of F. tularemia in infected tissues showed that FtfabI and other fatty acid biosynthetic genes were active during infection. In addition, the MICs of known FtFabI lead inhibitors and fatty acid biosynthesis inhibitors were unaffected in vitro by supplementation with a mixture of long-chain lipids to the culture medium. Together, the inability to engineer an FtfabI conditional mutant and the transcriptional profile observed during infection demonstrate the essentiality of FtfabI to bacterial viability and confirm the FAS-II pathway as a clinically relevant drug target for the priority pathogen F. tularensis.
MATERIALS AND METHODS
Bacterial strains and growth.
F. tularensis Schu4 and F. tularensis LVS were provided by J. Petersen (Centers for Disease Control, Fort Collins, CO). In general, F. tularensis Schu4 and LVS strains were cultured in Mueller-Hinton broth (BD) modified with 0.025% ferric pyrophosphate (Sigma-Aldrich), 2% IsoVitaleX (BD), and 0.1% glucose (Sigma-Aldrich) at 37°C with constant shaking overnight. For the fatty acid supplementation studies, F. tularensis was grown in modified Mueller-Hinton broth supplemented with 1% lipid mixture containing 2 μg/ml arachidonic and 10 μg/ml each linoleic, linolenic, myristic, oleic, palmitic, and stearic acid (Invitrogen).
Drug treatment studies.
The MIC of ketoacyl-ACP synthase (FabF) inhibitors thiolactomycin and cerulenin and enoyl-ACP reductase (FabI) inhibitors triclosan and SBPT04 were assessed, as previously described (4), against bacteria grown either in the presence or the absence of the exogenous lipid mixture.
The effect of exogenous long-chain lipids on the recovery of treated F. tularensis was assessed by treating F. tularensis with either triclosan at 1× MIC (0.062 mg/liter) or SBPT04 at 1× MIC (0.25 mg/liter) for 18 h. Then, the concentration of each drug was reduced by a 1:1,000 dilution into Mueller-Hinton broth with or without 1% lipid mixture supplement. Viable bacterial growth was determined by plating serial 10-fold dilutions at 0, 2, 4, 6, and 8 h after antibiotic exposure.
Construction and manipulation of fabI merodiploid, deletion, and conditional alleles.
Plasmids used in this study were recombinant derivatives of F. tularensis chromosomal integration vectors pMP812 and pMP815 (13), provided by H. P. Schweitzer, Colorado State University. Cloning and strain maintenance were carried out in Escherichia coli DH5α (Life Technologies) in Luria-Bertani broth (Becton, Dickinson). Phusion High-Fidelity DNA polymerase, restriction endonucleases, site-directed mutagenesis, and other DNA modifying enzymes were supplied by New England BioLabs, and genomic, plasmid, and amplicon DNAs were purified with reagents manufactured by Qiagen. Oligonucleotides were synthesized by Integrated DNA Technologies.
All electroporations, transformations of sucrose-competent F. tularensis LVS and Schu4 with pMP815- or pMP812-based allelic exchange constructs, and subsequent generation of primary and secondary recombinants were performed as described by LoVullo et al. (13). The FtfabI merodiploid strain was constructed as follows. Two fragments, a 202-bp element containing the rpsL promoter and the 783-bp FtfabI coding sequence (CDS) with 126 bp of downstream sequence, were amplified from F. tularensis Schu4 DNA and cloned into the blaB integration vector pMP815. A 5×His tag was engineered into the construct by site-directed mutagenesis. The ΔFtfabI mutant was created using two nonsequential fragments with extensions providing overlapping ends amplified from the FtfabI genomic region of LVS DNA. The two fragments were combined in a second amplification and were cloned into a sacB suicide plasmid pMP812 (13) to form a 356-bp deletion of the fabI promoter and 5′ CDS sequences flanked by 535 bp upstream and 759 bp downstream. The FtfabI conditional mutation allele created from a segment of the Schu4 FtfabI genomic region including 435 bp upstream and 437 bp downstream of sequence coding residue T241 was cloned into pMP812. The T241 codon was converted from ACT to TTT by site-directed mutagenesis to create T241F, orthologous to the fabI S241F mutation in temperature-sensitive E. coli JP1111 (14).
F. tularensis mouse model of infection and isolation of bacterial RNA from infected tissues.
Six-week-old female C57BL/6 mice were purchased from Jackson Laboratories, Bar Harbor, ME. All mice were housed in sterile microisolator cages in the laboratory animal resources facility or in the Regional Biocontainment Laboratory biosafety level 3 (BSL-3) facility at Colorado State University (Fort Collins, CO) and provided water and food ad libitum. All research involving animals was conducted in accordance with the Animal Care and Use Committee-approved animal guidelines and protocols. Mice were infected with F. tularensis Schu4 via the intranasal (i.n.) route. Mice were anesthetized with ketamine-xylazine, and 10 μl of inoculum (50 to 100 CFU) was administered to each of the nares in sequential droplets, allowing mice to inhale the fluid (20 μl total). Infected mice were monitored for morbidity twice daily and were euthanized at predetermined endpoints.
Lungs and spleen from infected mice were homogenized in TRIzol (Invitrogen), and total RNA was extracted using phenol-chloroform and ethanol precipitation. Samples were DNase I (Fermentas) treated to remove genomic DNA contaminants. Total RNA was depleted for rRNA and polyadenylated RNA using Ribominus Mouse and Bacterial Modules and a poly(T) Dynabead module (Invitrogen). The resulting samples, enriched for F. tularensis RNA, were pooled and converted to cDNA (Superscipt III; Invitrogen) and amplified using Klenow fragment (Fermentas) and random hexamer primers (3 μg/μl; Invitrogen, Carlsbad, CA), in the presence of Cy3-coupled dUTPs (GE), at 37°C for 2 h. Amplified cDNA was purified using a PCR purification kit (Qiagen).
In vivo transcriptional analysis.
Custom 70-mer, cDNA oligonucleotide sets designed from the fully assembled genome of F. tularensis Schu4 (Operon Biotechnologies) were printed in-house using a Bio-Rad Chipwriter Pro microarray printer on polyamine-coated glass microarray slides (Array-it). Slides were postprocessed by UV cross-linking, blocked with blocking buffer consisting of 10% bovine serum albumin (BSA; Sigma-Aldrich) and 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), rinsed with water, and dried. Labeled cDNA was combined with 1:1:0.2 formamide–20× SSC–10% SDS hybridization buffer and 10 μg of Saccharomyces cerevisiae tRNA (Sigma), heated to 98°C for 2 min, cooled, and applied under a coverslip placed above the array on the glass slide. Arrays were hybridized at 42°C for 16 h in the dark. Arrays were washed in a decreasing SSC gradient and dried.
Arrays were scanned using a Genepix Pro 4000B scanner and Genepix software. Raw intensity values were normalized across technical triplicates and analyzed for presence or absence based on fluorescence intensity. Genes were considered present if the fluorescence intensity was greater than two times the background fluorescence intensity (signal-to-noise ratio [SNR] of 2), corresponding to the lowest detectable signal (Gene Expression Omnibus [GEO] accession number GSE39871). Gene lists were grouped by functional annotation to determine biochemical pathways that were overrepresented in the data set.
Genes of interest that were observed as expressed during infection using arrays were confirmed using quantitative real-time PCR. Briefly, cDNA synthesis from total RNA was carried out using a First Strand cDNA Synthesis Kit (Invitrogen). One microgram of total RNA was combined with random hexamer and heated for 5 min. Ten microliters of buffered enzyme mix (2 μl of 10× buffer, 4 μl of MgCl2 [6 mM], 2 μl of 0.1 M dithiothreitol [DTT], 1 μl of RNase Out, and 1 μl of Superscript) was added and incubated at 25°C for 10 min, at 50°C for 50 min, and at 85°C for 5 min. Platinum SYBR green quantitative PCR (qPCR) Supermix-UDG (Invitrogen) was combined with gene-specific primers (5 nmol) and 50 ng of template (cDNA) and run in triplicate on an IQ5 thermocycler (Bio-Rad). Resulting data from each condition were compared to values for uninfected controls in an independent fashion using the ΔCT (where CT is threshold cycle) method.
RESULTS
Exogenous long-chain lipids do not alter transcriptional response of fatty acid biosynthetic components.
It is known that the transcription and production of components within a biosynthetic pathway are responsive relative to their utilization and requirement for growth. Therefore, the transcriptional profile resulting from growth under different nutrient conditions and environments can be used for comparison as well as for assessing the involvement of specific metabolic processes (e.g., fatty acid synthesis and β-oxidation) that respond if fatty acids are utilized as intermediate substrates for macromolecular synthesis. Accordingly, the transcriptional activity of F. tularensis grown in defined medium and in medium supplemented with a mixture of long-chain lipids was assessed using F. tularensis whole-genome microarrays. Analyses of the global transcriptional responses revealed a concordance of 84% between bacteria grown in fatty acid-supplemented medium and bacteria grown in control medium (variance of < 0.05; differentially expressed greater than 1.5-fold). Specifically, fatty acid biosynthesis genes accA, accB, accC, and accD encoding the malonyl-coenzyme A (CoA) synthase and fabF, fabG, fabH, fabI, and fabZ encoding FAS-II components were not differentially expressed, nor were FTT0910 encoding a putative phospholipase, FTT0941c encoding a putative esterase/lipase, and fadD1, fadD2, and acbP, which encode the β-oxidation components annotated in the F. tularensis genome (Fig. 1A and B). The resulting 94% concordance in the transcriptional responses of 23 open reading frames (ORFs) involved in fatty acid metabolism between bacteria grown with exogenous fatty acid supplementation and control bacteria indicates that exogenous host lipids do not drastically affect the metabolic flux of F. tularensis FAS-II and β-oxidation.
Fig 1.
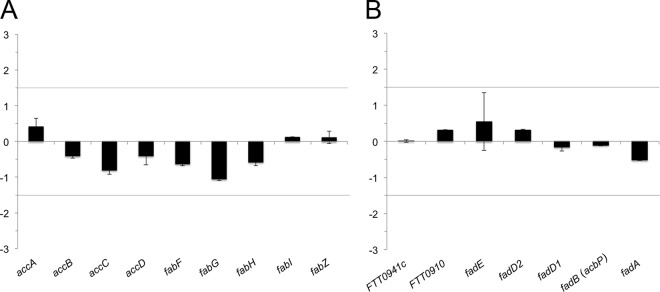
F. tularensis FAS-II and β-oxidation pathways are unaffected by supplementation of exogenous fatty acids in vitro. F. tularensis LVS was grown in standard medium and in the presence of 3% fatty acid supplement. RNA from three biological replicates was labeled and hybridized to a total of nine individual F. tularensis full-genome microarrays. Transcriptional analysis reveals that the fatty acid (A) and β-oxidation (B) pathways are not differentially regulated by fatty acid supplementation. Horizontal lines indicate the minimum threshold cutoff for genes to be considered differentially regulated. Values on the y axes represent fold change in expression.
FtfabI is essential for growth even in the presence of exogenous fatty acids.
Although the fatty acid biosynthetic pathway is attractive for drug discovery and is the target for the antituberculosis drug isoniazid that targets the enoyl-ACP reductase InhA in M. tuberculosis (15) and although inhibitors of the Staphylococcus aureus FabI enzyme have in vivo activity (16–19), we wanted to establish that FtFabI was essential for growth. We have developed long-residence-time inhibitors of InhA (20, 21), saFabI (22), and the F. tularensis FabI enzyme (FtFabI) (6), one of which (SBPT04) demonstrates 100% efficacy, controls dissemination, and prevents relapse in an animal model of tularemia (4). However, a recent report raised the possibility that at least in some bacteria, de novo fatty acid synthesis may not be required for growth in the presence of fatty acids, which are thought to be present in the host environment (7). Therefore, to confirm that FtfabI is essential for growth even when supplied with exogenous fatty acids, we employed molecular approaches to assess the essentiality of FtfabI and de novo fatty acid biosynthesis in the presence of exogenous long-chain lipids. Based on the rationale that F. tularensis utilizes exogenous lipids, it should be possible to create a fatty acid-auxotrophic strain with lipid supplementation. After a population screen was performed in the absence and presence of exogenously supplied fatty acids, it was determined that availability of fatty acids cannot rescue a native FtfabI mutant. After multiple attempts, we were unable to engineer a stable FtfabI knockout strain even in the presence of a second copy of FtfabI, and we were not able to create a temperature-sensitive conditional mutant strain. The inability to develop a stable merodiploid-complemented knockout strain or a conditional mutant is consistent with our observations with other bacterial pathogens and those reported in M. tuberculosis that expression and production of enoyl-ACP reductases are tightly controlled and that the bacterium does not readily tolerate manipulation of FtfabI (23). Specifically, the fact that we were unable to engineer a fatty acid-auxotrophic strain or a stable merodiploid-complement strain substantiates that FtfabI is essential and that its activity is not bypassed by the availability of exogenous long-chain lipids.
Inhibition of FtFabI is gene dosage dependent.
To confirm that the molecular target for triclosan and SBPT04 is FtFabI, a gene dosage drug sensitivity study was performed. Since high-level expression of FtfabI is not well tolerated, low-level expression was used to demonstrate a correlation between FtfabI expression and an increase in MIC. Expression of FtfabI at approximately 3.5-fold (Fig. 2A) resulted in 2.4-fold and 2.3-fold increases in the MICs of triclosan and SBPT04, respectively (Fig. 2B). Importantly, the gene dosage-associated increase in MIC demonstrates that the molecular target for triclosan and SBPT04 in F. tularensis is FtFabI, which further substantiates that F. tularensis cannot utilize exogenous fatty acids to bypass FtFabI inhibition.
Fig 2.
Gene dosage drug sensitivity study. Low-level overexpression of FtfabI correlates with an increase in MIC of both triclosan and SBPT04. (A) The transcriptional activity of the fabI merodiploid strain of F. tularensis in comparison to the wild type. (B) Evaluation of MIC90 of wild-type (wt) F. tularensis and a fabI merodiploid F. tularensis strain (FabI+) in the presence of PT04, triclosan (TRC), and doxycycline (DOXY).
Inhibition of FtFabI and fatty acid biosynthesis are unaffected by exogenous long-chain lipids.
To directly assess whether exogenous long-chain lipids alter the sensitivity of FtFabI-inhibitory compounds, we tested the MICs of the known FabI inhibitors, triclosan and SBPT04 (4), along with known FabF inhibitors thiolactomycin and cerulenin as positive controls. The ability of F. tularensis to utilize exogenous long-chain lipids to circumvent FtFabI inhibition was evaluated by determining the MIC and posttreatment rescue.
The MICs of the FtFabI inhibitors triclosan and SBPT04 were determined in the presence of long-chain lipids. FabF inhibitors thiolactomycin and cerulenin were tested as positive controls for inhibition of fatty acid synthesis to ensure that the observed effects were not due to off-target effects or drug binding by exogenous lipids. There was no difference in inhibitory activities of FabI or FabF inhibitors when they were tested with exogenous long-chain lipid supplementation compared to results in unsupplemented controls (Table 1).
Table 1.
Supplementation of exogenous fatty acids does not affect inhibition of FabF or FabI
| Inhibitor | MIC (mg/liter) against the indicated F. tularensis strain and treatmenta |
|||
|---|---|---|---|---|
| LVS |
Schu4 |
|||
| −Lipids | + Lipids | −Lipids | + Lipids | |
| FabF inhibitors | ||||
| TLM | 4 | 4 | 16 | 16 |
| Cerulinin | 2 | 2 | 1 | 1 |
| FabI inhibitors | ||||
| SBPT04 | 0.25 | 0.5 | 1 | 1 |
| Triclosan | 0.06 | 0.12 | 0.25 | 0.25 |
Data represent MICs of known FabI inhibitors triclosan and SBPT04 and FabF inhibitors thiolactomycin and cerulenin against F. tularensis strains LVS and Schu4 grown in the presence (+) or absence (−) of a mixture of long-chain lipids.
Similarly, exogenous long-chain lipids did not reverse the inhibitory activity of triclosan or SBPT04. Supplying exogenous long-chain lipids for up to 8 h after drug treatment did not alter the recovery and growth of bacteria treated for 18 h. These results are consistent with the inability to obtain an auxotrophic strain of F. tularensis and indicate that F. tularensis requires de novo fatty acid biosynthesis and cannot subsist solely on freely available lipids.
The F. tularensis FAS-II pathway is transcriptionally active in infected tissues.
Although bacteria grown in defined culture medium displayed a transcriptional response that indicated that they could not utilize exogenously supplied fatty acids for fatty acid synthesis, it is still necessary to assess whether the transcriptional profile of bacteria from infected tissues shows a pattern of fatty acid gene expression consistent with active de novo fatty acid biosynthesis. Accordingly, the whole bacterial genome transcriptional response of F. tularensis obtained from infected host tissues was assessed using F. tularensis whole-genome DNA microarrays. We have studied the host response to infection with F. tularensis and dissemination to secondary sites and have determined that infection of the spleen is an important indicator of disease progression and mortality (24) and thus is an important consideration for drug discovery and development of clinically relevant agents. Accordingly, total RNA from spleen samples was harvested at 120 h postinfection and pooled from two mice for microarray analysis. Array data were subjected to a threshold cutoff based on signal-to-noise ratios to identify F. tularensis Schu4 ORFs that were present, thus discriminating between F. tularensis ORFs that are transcriptionally active in infected tissue.
Genes that had a transcriptional signal-to-noise ratio of 2 or greater were deemed to be present, thus implicating activity of a metabolic pathway. In addition, triplicate arrays allowed the elimination of any feature that was flagged as poor quality on any of the three arrays to increase stringency of calling active metabolism. Overall, a total of 1,022 F. tularensis Schu4 ORFs were identified as transcriptionally active in the spleens of infected mice (data not shown). Gene lists were subjected to functional enrichment analysis to determine pathways that had a disproportionally high number of genes with a particular ontological classification. Overall, the transcriptional activity of F. tularensis from infected spleens included basic metabolic pathways of amino acid, protein biosynthesis, carbon metabolism via the tricarboxylic acid cycle (TCA) cycle, and fatty acid biosynthesis (Table 2).
Table 2.
Expression profiling of the F. tularensis transcriptome during infection reveals the metabolic requirements for growth in the hosta
| Gene function and locus | Gene name | Annotation |
|---|---|---|
| Amino acid biosynthesis | ||
| FTT0588 | aroA | 3-Phosphoshikimate 1-carboxyvinyltransferase |
| FTT1154c | aroB | 3-Dehydroquinate synthetase |
| FTT0876c | aroC | Chorismate synthase |
| FTT1155c | aroK | Shikimate kinase I |
| FTT1287 | cbs | Cystathionine beta-synthase (cysteine synthase) |
| FTT0640 | ilvD | Dihydroxy-acid dehydratase |
| FTT0560c | serC | Phosphoserine aminotransferase |
| FTT1772c | trpA | Tryptophan synthase alpha chain |
| FTT1773c | trpB | Tryptophan synthase beta chain |
| Fatty acid biosynthesis | ||
| FTT1498c | accA | Acetyl-CoA carboxylase carboxyl transferase subunit alpha |
| FTT0472 | accB | Acetyl-CoA carboxylase, biotin carboxyl carrier protein subunit |
| FTT0372c | accD | Acetyl-CoA carboxylase beta subunit |
| FTT1377 | fabF | 3-Oxoacyl-(acyl-carrier-protein) synthase II |
| FTT1375 | fabG | 3-Oxoacyl-(acyl-carrier-protein) reductase |
| FTT1373 | fabH | 3-Oxoacyl-(acyl carrier protein) synthase III |
| FTT0782 | fabI | Enoyl-(acyl-carrier-protein) reductase (NADH) |
| FTT0148 | FTT0148 | Fatty acid desaturase |
| FTT1552 | ole1 | Delta 9 acyl-lipid fatty acid desaturase |
| FTT1372 | plsX | Fatty acid/phospholipid synthesis protein lsX |
| Stress response | ||
| FTT1769c | clpB | ClpB protein |
| FTT1269c | dnaK | Chaperone protein dnaK (heat shock protein family 70 protein) |
| FTT0666c | FTT0666c | Methylpurine-DNA glycosylase family protein |
| FTT0733 | FTT0733 | Glutathione peroxidase |
| FTT0846 | FTT0846 | Deoxyribodipyrimidine photolyase |
| FTT0886 | FTT0886 | DNA repair protein RecN |
| FTT0986 | FTT0986 | Conserved hypothetical protein |
| FTT0356 | htpG | Chaperone Hsp90, heat shock protein HtpG |
| FTT0862c | htpX | Heat shock protein HtpX |
| FTT0721c | katG | Peroxidase/catalase |
| FTT1736c | kdpD | Two-component sensor protein kdpD |
| FTT1387c | ligN | DNA ligase |
| FTT0644c | mfd | Transcription-repair coupling factor, ATP-dependent |
| FTT0486 | mutL | DNA mismatch repair protein |
| FTT0648c | nth | Endonuclease III |
| FTT1518 | ogt | Methylated-DNA–protein-cysteine methyltransferase |
| FTT0111 | polA | DNA polymerase I |
| FTT0873c | radA | DNA repair protein RadA |
| FTT0478c | recJ | Single-stranded DNA-specific exonuclease |
| FTT1087c | rep | ATP-dependent DNA helicase |
| FTT1112c | rpoH | RNA polymerase sigma-32 factor |
| FTT0658 | ruvA | Holliday junction DNA helicase, subunit A |
| FTT1013c | ruvB | Holliday junction DNA helicase, subunit B |
| FTT0656 | ruvC | Holliday junction endodeoxyribonuclease |
| FTT1578c | ung | Uracil-DNA glycosylase |
| FTT0245 | usp | Universal stress protein |
| FTT1312c | uvrA | DNA excision repair enzyme, subunit A |
| FTT0777 | uvrC | DNA excision repair enzyme, subunit C |
| FTT0121 | uvrD | DNA helicase II |
| FTT0959c | xthA | Exodeoxyribonuclease III |
| Protein biosynthesis | ||
| FTT1096c | alaS | Alanyl-tRNA synthetase |
| FTT1616 | cysS | Cysteinyl-tRNA synthetase |
| FTT0925 | fmt | Methionyl-tRNA formyltransferase |
| FTT0316 | frr | Ribosome recycling factor |
| FTT0323 | fusA | Elongation factor G |
| FTT0307 | gltX | Glutamyl-tRNA synthetase |
| FTT0419 | glyQ | Glycyl-tRNA synthetase alpha chain |
| FTT0052 | hisS | Histidyl-tRNA synthetase |
| FTT0915c | ileS | Isoleucyl-tRNA synthetase |
| FTT0050 | infB | Translation initiation factor IF-2 |
| FTT0818 | infC | Translation initiation factor IF-3 |
| FTT0990 | leuS | Leucyl-tRNA synthetase |
| FTT0192 | lysU | Lysyl-tRNA synthetase |
| FTT1290 | metG | Methionyl-tRNA synthetase |
| FTT1002c | pheT | Phenylalanyl-tRNA synthetase, beta subunit |
| FTT0476c | poxA | Lysyl-tRNA synthetase |
| FTT0168 | prfA | Peptide chain release factor 1 |
| FTT0191 | prfB | Peptide chain release factor 2 |
| FTT0118 | prfC | Peptide chain release factor 3 |
| FTT1412 | proS | Prolyl-tRNA synthetase |
| FTT0817 | thrS | Threonyl-tRNA synthetase |
| FTT0314 | tsf | Protein chain elongation factor EF-Ts |
| FTT0137 | tufA | Elongation factor Tu |
| FTT0691 | tyrS | Tyrosyl-tRNA synthetase |
| FTT0299 | valS | Valyl-tRNA synthetase |
| TCA cycle | ||
| FTT0306 | fumC | Fumarate hydratase, class II |
| FTT0071c | gltA | Citrate synthase |
| FTT1526c | idh | Isocitrate dehydrogenase |
| FTT0074 | sdhA | Succinate dehydrogenase, catalytic and NAD/flavoprotein subunit |
| FTT0075 | sdhB | Succinate dehydrogenase iron-sulfur protein |
| FTT0072 | sdhC | Succinate dehydrogenase, cytochrome b556 |
| FTT0077 | sucB | Dihydrolipoamide succinyltransferase (2-oxoglutarate dehydrogenase complex) |
Total RNA was isolated from the spleens of mice at 120 h postinfection with F. tularensis Schu4. Material from two mice was pooled and enriched for bacterial transcripts, converted to labeled cDNA, and hybridized to Francisella full-genome cDNA microarrays. Gene lists were interrogated for pathways with an overabundance of representative genes.
Although transcriptional activity identified by microarray provided an overall picture of the metabolic activities of F. tularensis in infected tissues, it was necessary to validate the microarray analysis and, specifically, the expression of the F. tularensis-encoded FAS-II system because of the nature of measuring transcriptional activity in complex biological samples. Accordingly, FAS-II gene expression was assessed using quantitative real-time reverse transcription-PCR (RT-PCR) at 96 h postinfection in both the lungs and spleen of F. tularensis Schu4-infected mice for confirmation. Analysis of quantitative reverse transcription (qRT) data revealed expression of all F. tularensis annotated FAS-II pathway genes in both the lung and spleen of infected mice (Fig. 3), thus validating the transcriptional data obtained by microarray and further substantiating that FtfabI encodes a clinically relevant drug target.
Fig 3.
F. tularensis requires de novo fatty acid biosynthesis during infection of the lung and spleen at 96 h postinfection. Total RNA was isolated from the lungs and spleen of mice at 96 h postinfection with F. tularensis Schu4. Gene-specific primers were run against RNA from infected and uninfected mouse tissue and compared to generate ΔCT values. Gene expression patterns in both the lung (A) and spleen (B) indicate the necessity of the F. tularensis de novo biosynthesis pathway during infection.
DISCUSSION
The development of new drugs has numerous hurdles, including verification that the target of interest is necessary under conditions associated with disease and is therefore a clinically relevant drug target. Clinically relevant targets for drug discovery can be experimentally defined as those proteins that are essential, whose functions are not bypassed by available exogenous nutrients, and that are active during, and at sites of, the infection. Recently, there has been a debate about the essentiality of the bacterial FAS-II system based on the ability of some bacteria to incorporate exogenous lipids, thereby bypassing de novo fatty acid synthesis and rendering the pathway conditionally nonessential and, thus, a poor target for the development of novel therapeutics (7). It has been shown, however, that this phenomenon is a result of the ability of individual species of bacteria to take up and utilize available nutrients (8, 9). Notably, there have been numerous publications that have substantiated enoyl-ACP reductases as valid drug targets (15, 25–31).
Because we have a drug discovery effort targeting enoyl-ACP reductases, there is a need to demonstrate that FabI is a clinically relevant drug target in F. tularensis, despite the fact that we have previously shown the efficacy of novel FabI inhibitors both in vitro and in vivo (4, 28, 32). To further substantiate FabI as a relevant drug target, we investigated the expression and essentiality of FtfabI in the presence of exogenously supplied fatty acids, examined whether exogenous fatty acids affect the potency of known FabI inhibitors, and determined if the F. tularensis-encoded FAS-II system is transcriptionally active during infection.
Transcriptional profiling of F. tularensis grown in the presence of exogenous lipids and recovered from the spleen revealed that the expression of fatty acid biosynthetic genes is unaffected by availability of lipids and that these genes are transcriptionally active at the site of infection. This response indicates that, despite the availability of lipids supplied exogenously or as part of the host environment, de novo fatty acid biosynthesis is still necessary to synthesize the full complement of fatty acids required for the F. tularensis cell wall. In combination, these transcriptional studies demonstrate that de novo fatty acid synthesis is not bypassed by the availability of lipids and potential short-chain fatty acid intermediates. Consistent with the lack of transcriptional response to the availability of lipids, an auxotrophic mutant could not be created. This further substantiates that exogenous lipids cannot be readily used to circumvent fatty acid biosynthesis.
Although an auxotrophic mutant could not be created, an FtfabI knockout strain was pursued in an FtfabI-merodiploid strain. Even though a second copy of FtfabI was present, generation of a stable FtfabI knockout strain of F. tularensis proved unobtainable. Further investigation via gene dosage expression studies revealed that artificial expression of FtfabI and production of FtFabI were not well tolerated. These studies demonstrated that only low-level expression was stable. This observation is consistent with observations in that the M. tuberculosis fabI, inhA, is not induced equivalently to other fatty acid biosynthetic genes (23). These studies demonstrate that FabI activity under native control is essential, which is consistent with transposon mutant studies that did not identify FabI as a gene that could tolerate transposon insertion (33–39).
We are currently testing novel FabI inhibitors against F. tularensis as well as priority pathogens Y. pestis and B. pseudomallei and the medically important pathogen M. tuberculosis in an effort to develop broad-spectrum antibiotics that target the FAS-II system of bacterial pathogens. Therefore, we determined if the availability of exogenous lipids altered the minimal inhibitory activity of a known FabI inhibitor and a novel FabI inhibitor developed in our drug discovery program (4, 6, 28, 32). It was shown that there was no difference in the MICs of FabI inhibitors compared to testing without exogenous long-chain lipids. In addition, the inhibitory activity of the positive-control fatty acid inhibitors cerulenin and thiolactomycin that target the keto-acyl-ACP synthase were also unaffected by the availability of lipids. The MIC studies in the presence of exogenous lipids further substantiate that FabI in F. tularensis is a clinically relevant drug target.
Importantly, we showed FtfabI transcriptional activity both in the presence of exogenously supplied long-chain lipids and in vivo during infection, signifying that FtfabI and de novo fatty acid biosynthesis have an essential metabolic role during in the host and during infection. These data demonstrate the importance of establishing the essentiality of a drug target as well as clinical relevance based on the target being actively expressed during the infection and therefore readily targetable for therapeutic intervention. As the development of broad-spectrum chemotherapeutics targeting FAS-II advances, work with priority pathogens can be translated to medically important infectious diseases that affect a much larger number of patients. Together, this information can be used to substantiate fatty acid biosynthesis as well as FtFabI as a clinically relevant drug target in F. tularensis.
ACKNOWLEDGMENTS
This work was supported by RP006 of the Rocky Mountain Regional Center of Excellence funded by NIH NIAID grant AI065357 (R.A.S. and P.J.T.).
Footnotes
Published ahead of print 9 November 2012
REFERENCES
- 1. Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285: 2763– 2773 [DOI] [PubMed] [Google Scholar]
- 2. Miller SD, Snyder MB, Kleerekoper M, Grossman CH. 1989. Ulceroglandular tularemia: a typical case of relapse. Henry Ford Hosp. Med. J. 37: 73– 75 [PubMed] [Google Scholar]
- 3. Penn RL, Kinasewitz GT. 1987. Factors associated with a poor outcome in tularemia. Arch. Intern. Med. 147: 265– 268 [PubMed] [Google Scholar]
- 4. England K, am Ende C, Lu H, Sullivan TJ, Marlenee NL, Bowen RA, Knudson SE, Knudson DL, Tonge PJ, Slayden RA. 2009. Substituted diphenyl ethers as a broad-spectrum platform for the development of chemotherapeutics for the treatment of tularaemia. J. Antimicrob. Chemother. 64: 1052– 1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu N, Cummings JE, England K, Slayden RA, Tonge PJ. 2011. Mechanism and inhibition of the FabI enoyl-ACP reductase from Burkholderia pseudomallei. J. Antimicrob. Chemother. 66: 564– 573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu H, England K, am Ende C, Truglio JJ, Luckner S, Reddy BG, Marlenee NL, Knudson SE, Knudson DL, Bowen RA, Kisker C, Slayden RA, Tonge PJ. 2009. Slow-onset inhibition of the FabI enoyl reductase from Francisella tularensis: residence time and in vivo activity. ACS Chem. Biol. 4: 221– 231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458: 83– 86 [DOI] [PubMed] [Google Scholar]
- 8. Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. 2010. Essentiality of FASII pathway for Staphylococcus aureus. Nature 463: E3 doi:10.1038/nature08667 [DOI] [PubMed] [Google Scholar]
- 9. Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. 2011. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U. S. A. 108:15378– 15383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schiebel J, Chang A, Lu H, Baxter MV, Tonge PJ, Kisker C. 2012. Staphylococcus aureus FabI: inhibition, substrate recognition, and potential implications for in vivo essentiality. Structure 20: 802– 813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nichols PD, Mayberry WR, Antworth CP, White DC. 1985. Determination of monounsaturated double-bond position and geometry in the cellular fatty acids of the pathogenic bacterium Francisella tularensis. J. Clin. Microbiol. 21: 738– 740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. 2004. Novel modification of lipid A of Francisella tularensis. Infect. Immun. 72: 5340– 5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LoVullo ED, Molins-Schneekloth CR, Schweizer HP, Pavelka MS., Jr 2009. Single-copy chromosomal integration systems for Francisella tularensis. Microbiology 155: 1152– 1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bergler H, Hogenauer G, Turnowsky F. 1992. Sequences of the envM gene and of two mutated alleles in Escherichia coli. J. Gen. Microbiol. 138: 2093– 2100 [DOI] [PubMed] [Google Scholar]
- 15. Mdluli K, Slayden RA, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane DD, Musser JM, Barry CE., III 1998. Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science 280: 1607– 1610 [DOI] [PubMed] [Google Scholar]
- 16. Miller WH, Seefeld MA, Newlander KA, Uzinskas IN, Burgess WJ, Heerding DA, Yuan CC, Head MS, Payne DJ, Rittenhouse SF, Moore TD, Pearson SC, Berry V, DeWolf WE, Jr, Keller PM, Polizzi BJ, Qiu X, Janson CA, Huffman WF. 2002. Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI). J. Med. Chem. 45: 3246– 3256 [DOI] [PubMed] [Google Scholar]
- 17. Park HS, Yoon YM, Jung SJ, Kim CM, Kim JM, Kwak JH. 2007. Antistaphylococcal activities of CG400549, a new bacterial enoyl-acyl carrier protein reductase (FabI) inhibitor. J. Antimicrob. Chemother. 60: 568– 574 [DOI] [PubMed] [Google Scholar]
- 18. Ramnauth J, Surman MD, Sampson PB, Forrest B, Wilson J, Freeman E, Manning DD, Martin F, Toro A, Domagala M, Awrey DE, Bardouniotis E, Kaplan N, Berman J, Pauls HW. 2009. 2,3,4,5-Tetrahydro-1H-pyrido[2,3-b and e][1,4]diazepines as inhibitors of the bacterial enoyl ACP reductase, FabI. Bioorg. Med. Chem. Lett. 19: 5359– 5362 [DOI] [PubMed] [Google Scholar]
- 19. Seefeld MA, Miller WH, Newlander KA, Burgess WJ, DeWolf WE, Jr, Elkins PA, Head MS, Jakas DR, Janson CA, Keller PM, Manley PJ, Moore TD, Payne DJ, Pearson S, Polizzi BJ, Qiu X, Rittenhouse SF, Uzinskas IN, Wallis NG, Huffman WF. 2003. Indole naphthyridinones as inhibitors of bacterial enoyl-ACP reductases FabI and FabK. J. Med. Chem. 46: 1627– 1635 [DOI] [PubMed] [Google Scholar]
- 20. Luckner SR, Liu N, am Ende CW, Tonge PJ, Kisker C. 2010. A slow, tight binding inhibitor of InhA, the enoyl-acyl carrier protein reductase from Mycobacterium tuberculosis. J. Biol. Chem. 285: 14330– 14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sullivan TJ, Truglio JJ, Boyne ME, Novichenok P, Zhang X, Stratton CF, Li H-J, Kaur T, Amin A, Johnson F, Slayden RA, Kisker C, Tonge PJ. 2006. High affinity InhA inhibitors with activity against drug-resistant strains of Mycobacterium tuberculosis. ACS Chem. Biol. 1: 43– 53 [DOI] [PubMed] [Google Scholar]
- 22. Xu H, Sullivan TJ, Sekiguchi J, Kirikae T, Ojima I, Mao W, Rock FL, Alley MRK, Johnson F, Walker SG, Tonge PJ. 2008. Mechanism and inhibition of saFabI, the enoyl reductase from Staphylococcus aureus. Biochemistry 47: 4228– 4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slayden RA, Lee RE, Barry CE., III 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38: 514– 525 [DOI] [PubMed] [Google Scholar]
- 24. Kingry LC, Troyer RM, Marlenee NL, Bielefeldt-Ohmann H, Bowen RA, Schenkel AR, Dow SW, Slayden RA. 2011. Genetic identification of unique immunological responses in mice infected with virulent and attenuated Francisella tularensis. Microbes Infect. 13: 261– 275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 274: 11110– 11114 [DOI] [PubMed] [Google Scholar]
- 26. Heath RJ, White SW, Rock CO. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 58: 695– 703 [DOI] [PubMed] [Google Scholar]
- 27. Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273: 30316– 30320 [DOI] [PubMed] [Google Scholar]
- 28. Lu H, Tonge PJ. 2008. Inhibitors of FabI, an enzyme drug target in the bacterial fatty acid biosynthesis pathway. Acc. Chem. Res. 41: 11– 20 [DOI] [PubMed] [Google Scholar]
- 29. McMurry LM, McDermott PF, Levy SB. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob. Agents Chemother. 43: 711– 713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stewart MJ, Parikh S, Xiao G, Tonge PJ, Kisker C. 1999. Structural basis and mechanism of enoyl reductase inhibition by triclosan. J. Mol. Biol. 290: 859– 865 [DOI] [PubMed] [Google Scholar]
- 31. Ward WH, Holdgate GA, Rowsell S, McLean EG, Pauptit RA, Clayton E, Nichols WW, Colls JG, Minshull CA, Jude DA, Mistry A, Timms D, Camble R, Hales NJ, Britton CJ, Taylor IW. 1999. Kinetic and structural characteristics of the inhibition of enoyl (acyl carrier protein) reductase by triclosan. Biochemistry 38: 12514– 12525 [DOI] [PubMed] [Google Scholar]
- 32. Boyne ME, Sullivan TJ, am Ende CW, Lu H, Gruppo V, Heaslip D, Amin AG, Chatterjee D, Lenaerts A, Tonge PJ, Slayden RA. 2007. Targeting fatty acid biosynthesis for the development of novel chemotherapeutics against Mycobacterium tuberculosis: evaluation of A-ring-modified diphenyl ethers as high-affinity InhA inhibitors. Antimicrob. Agents Chemother. 51: 3562– 3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U. S. A. 104: 1009– 1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawula TH, Hall JD, Fuller JR, Craven RR. 2004. Use of transposon-transposase complexes to create stable insertion mutant strains of Francisella tularensis LVS. Appl. Environ. Microbiol. 70: 6901– 6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maier TM, Casey MS, Becker RH, Dorsey CW, Glass EM, Maltsev N, Zahrt TC, Frank DW. 2007. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect. Immun. 75: 5376– 5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maier TM, Pechous R, Casey M, Zahrt TC, Frank DW. 2006. In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl. Environ. Microbiol. 72: 1878– 1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin A, Mann BJ. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6: 69 doi:10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, Allen LA. 2009. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77:1324– 1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tempel R, Lai XH, Crosa L, Kozlowicz B, Heffron F. 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 74: 5095– 5105 [DOI] [PMC free article] [PubMed] [Google Scholar]