Abstract
We report a simple, single step reaction that transforms riboflavin, which is insoluble in benzene, to tetraphenylacetyl riboflavin (TPARF), which is soluble in this solvent to over 250 mM. Electrochemical analysis of TPARF both alone and in complexes with two benzene-soluble flavin receptors: dibenzylamidopyridine (DBAP) and monobenzylamidopyridine (MBAP), prove that this model system behaves similarly to other previously studied flavin model systems which were soluble only in more polar solvents such as dichloromethane. Binding titrations in both benzene and dichloromethane show that the association constants of TPARF with its ligands are over an order of magnitude higher in benzene than dichloromethane, a consequence of the fact that benzene does not compete for H-bonds, but dichloromethane does. The alteration induced in the visible spectrum of TPARF in benzene upon the addition of DBAP is very similar to the difference produced by transfer to dichloromethane, further indicating that the flavin head group engages in a similar degree of hydrogen bonding with dichloromethane as with its ligands. This work underlines the importance of using a truly nonpolar solvent for the analysis of the effects of hydrogen bonding on the energetics of any biomimetic host-guest model system.
Introduction
Protein interiors are perhaps best envisioned as inhomogeneous matrices of low dielectric constants irregularly interrupted by dipoles which can serve as hydrogen bond donors and acceptors and, less commonly, of cationic and anionic charged groups. Moreover since these groups are not free to reorient, they do not greatly increase the overall dielectric constant. Indeed, the exact placement of these dipoles and charges is an important component of both biological recognition and protein modulation of the energetics of binding partners, for example the pKas or reduction potentials of bound protein cofactors. Consequently, systems intended to mimic the effects of a protein environment must both be nonpolar overall and exert a high degree of control of the identity, position and orientation of those functionalities that are intended to represent polar interactions originating from the protein.
The host-guest interaction between flavin cofactors (Scheme 1) and their protein partners is one example of a biologically important interaction pair for which many mimetic complexes have been constructed.1-8 Flavin cofactors are present in over 20% of proteins which have an electron transfer as some part of their reaction mechanism.9 These cofactors are capable of one- and two-electron reduction chemistry, coupled to the binding of one or two protons to the central diazabutadiene moiety of the isoalloxazine headgroup (see Fig. 1). Thus, they exhibit a remarkably diverse range of catalytic abilities as found in signaling and transcription activation, light activated DNA repair, respiratory electron chains, and dehydrogenation, dehalogenation, hydroxylation and oxygenation reactions.10
Scheme 1.
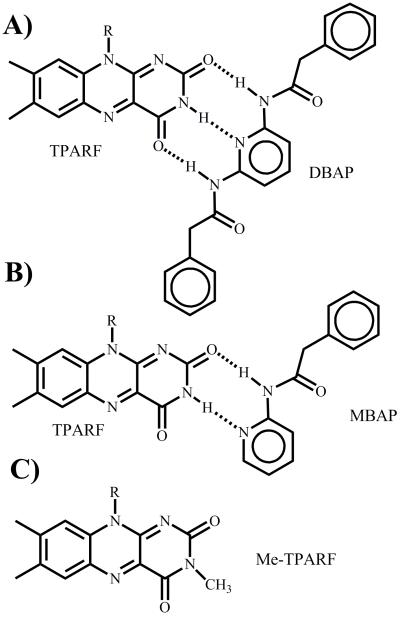
Fig. 1.
The benzene-soluble flavin analogues and host receptors. TPARF-DBAP complex A). TPARF-MBAP complex B) and the N(3)-methylated TPARF (Me-TPARF) C).
The differing catalytic capabilities of the flavin in different enzymes result from modulation of the highly conjugated flavin isoalloxazine electronic structure by the protein environment.4, 10-12 As most, if not all, flavin-catalyzed reactions involve electron transfer as part of their catalytic mechanism, modulation of the reduction potentials of the bound cofactor plays a central role in the selective promotion of a particular catalytic activity in each flavoenzyme. This is accomplished via specific interactions, electrostatic and otherwise, between the cofactor and its protein host. Nonetheless, several decades of research on flavoenzymes has underlined the difficulty in quantifying such effects due to the complexity of the protein environment, as well as the aqueous medium, which makes it almost impossible to isolate specific physiologically relevant flavin-protein interactions.
One approach to this problem consists of constructing minimalist synthetic models that are soluble in simplifying aprotic solvents. These permit the examination of the energetics of interactions between flavins and small molecule mimics of specific interactions found in proteins. Several groups have constructed chloroform-soluble model flavin compounds and studied the effects of their interactions with small molecules intended to simulate hydrogen bonding, π-stacking, and conformational modification by proteins.1-3, 7, 8 In one chloroform-soluble compound in particular, a three-point hydrogen-bonding interaction between N(10)-isobutyl isoalloxazine and 2,6-diethylamidopyridine, modified the flavin one-electron reduction potential by 100 mV or over.2
These model flavin systems are attractive spectroscopic targets, promising insight into the manner in which these isolated intermolecular interactions modulate the electronic properties of protein-bound flavins, thus specifying their catalytic function. However, chloroform is a poor choice of solvent due to its own property to donate a hydrogen bond.13-15 Quantification of the energetics of mimetic complexes dissolved in chloroform is therefore subject to uncertainties caused by extensive hydrogen bonding between the ‘free’ flavin and the solvent; thus, the cost of displacing solvent significantly detracts from the apparent favorability of the new interactions formed.16 Moreover, spectra collected in the absence of the designed H-bonding partner do not probe interaction-free flavins.
We have previously reported the construction of a new flavin analogue, N(10)-2,2-dibenzylethylisoalloxazine, which is soluble in less polar solvents.5 This molecule was soluble in dichloromethane to 20 mM and even soluble in benzene to a concentration of over 1 mM. The synthetic route for the construction of this molecule was intentionally designed for the inexpensive incorporation of 15N isotopic labels in the isoalloxazine ring. This, coupled with the solubility of this compound in volatile hydrophobic solvents enabled the first observation of the complete chemical shift tensors for three of the four nitrogen atoms and every carbon atom in the isoalloxazine moiety using solid state NMR in the oxidized and reduced state.17 These experiments confirmed earlier ab initio calculations that predicted that several of the principal components of the chemical shift tensor, in particular the σ11 components of the N(1) and N(5) tensors, strongly reflect the H-bond accepting lone pairs of the flavin.18
In order to perform solid state NMR experiments in frozen solution, a molecule must be soluble to at minimum 100 mM concentration in a solvent that freezes above -70° C. In order for a solvent to dissolve a model flavin it must be able to interact favorably with some part of it. An additional parameter of importance is the ease and expense of constructing the molecule with isotopic labels at the appropriate positions. We report here the development of a simple process which can transform riboflavin, which can readily be extracted in isotopically labeled form from bacteria grown in labeled media, into a compound designed to be soluble in benzene to greater than 250 mM. We have chosen this flavin-solvent pair because of the ability of benzene to dissolve the flavin through π–stacking interactions. Because we wish to study this flavin alone and in complexes with binding partners which modify the cofactor’s reduction potential, we examined its interaction with some benzene-soluble analogues of the 2,6-diethylamidopyridine ligand developed by Breinlinger et al.2 We found large differences between the binding affinities of these ligands depending on whether the solvent used was benzene or dichloromethane. We report spectroscopic evidence that this difference can be attributed to the weak hydrogen bonding ability of dichloromethane. This highlights the importance of using truly noninteractive solvents both in spectroscopically examining biomimetic molecules alone and in making conclusions about the energetics of their interactions with ligands.
Results
TPARF synthesis
Biochemists have employed tetraacetylriboflavin (TARF)† for over three decades to study flavins in polar organic solvents such as methanol and acetone.19 Constructed by the reaction of riboflavin (Scheme 1) with acetic anhydride, TARF has the advantage that the parent riboflavin molecule can be prepared in isotopically labeled form by isolating it from microorganisms grown on labeled minimal media. We surmised that addition of a larger, more hydrophobic sidechain to each ribose hydroxyl group should increase the resulting derivative’s solubility in polar solvents.
An initial attempt to attach benzyl groups to the riboflavin ribose side chain using benzoic anhydride generated a single benzyl ester instead of four (data not shown). Reaction with benzoyl chloride resulted in pentabenzylated riboflavin, with the fifth addition taking place at the N(3) position. Reactions with benzoic acid in the presence of catalytic 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, or by the creation of an asymmetric anhydride using POCl3, were also unsuccessful.
Addition of a less sterically hindered hydrophobic molecule, phenylacetic acid, via the formation of an asymmetric anhydride using POCl3 was successful. The resultant TPARF molecule proved to be soluble to over 250 mM in benzene, which is more than sufficient for solid state NMR studies. Similarly, the ligand molecule 2,6-diethylamidopyridine was also insufficiently soluble in benzene, was replaced by a more soluble analog with benzylamido groups replacing the two ethylamido groups (2,6-dbenzylamidopyridine, DBAP, see Fig. 1).
The TPARF-DBAP complex in benzene and dichloromethane
Spectroscopic analyses of the interactions between the oxidized TPARF and DBAP (Fig. 1) in benzene and dichloromethane are presented in Fig. 2. Addition of DBAP to TPARF in both solvents results in an increase in the extinction coefficient (ε) of the flavin in concert with a 2-nm red shift from 445 to 447 nm of the main absorption band (Fig. 2A), a result typical of hydrogen bonding to the flavin isoalloxazine moiety.20, 21 No change is observed for Me-TPARF upon the addition of either ligand (data not shown), suggesting that TPARF-DBAP interactions are mediated at least in part by the N(3) of the flavin. Fig. 2B displays the progressive difference spectra of TPARF in benzene upon the addition of DBAP. These reveal the absorption change (Δε) in the shoulder-peak-shoulder absorptions of TPARF, with maximal changes at 428, 455 and 487 nm respectively for the TPARF-DBAP complex vs unliganded TPARF in benzene. The maximum change occurs at 487 nm. The inset in Fig. 2B shows a plot of the absorbance change at 487 nm versus DBAP concentration in benzene solvent. Fitting these data with equation 2 yields a dissociation constant Kd of 420 μM for the oxidized TPARF-DBAP complex in benzene.
Fig. 2.
TPARF interactions with DBAP in dichloromethane and in benzene. The absolute spectra of TPARF are shown in A): TPARF in benzene (black), TPARF in dichloromethane (grey), TPARF with DBAP in benzene (black dashed line) and TPARF with DBAP in dichloromethane (grey dashed line). The difference spectra of TPARF with DBAP relative to TPARF in benzene solvent, is shown in B). The inset displays the curve fit to a plot of the difference absorbance at 487 nm vs added DBAP in benzene. The same set of spectra and curve fit in dichloromethane is illustrated in C).
The difference spectrum of TPARF in dichloromethane upon the addition of DBAP is shown in Fig. 2C. Qualitatively similar behavior to that seen in benzene is observed in dichloromethane. However, the magnitude of the absorbance change is 30% less than that produced by TPARF-DBAP complex formation in benzene. Furthermore, the plot of the change in absorbance at 487 nM vs. DBAP concentration for TPARF in dichloromethane (inset) shows that the dissociation constant Kd of the oxidized TPARF is 4,120 μM, an order of magnitude weaker than in benzene.
TPARF-MBAP complex in benzene and dichloromethane
DBAP is proposed to bind TPARF in aprotic solvents using a tridentate hydrogen bonding interaction to both carbonyls and the N(3) hydrogen, based on the well-studied analogous complex formed between N(10)-isobutyl isoalloxazine and diethylamidopyridine.2 In order to separate the relative energetic consequence of hydrogen bonding to carbonyls from hydrogen bonding to the N(3), measurements were carried out by adding monobenzylamidopyridine (MBAP) to TPARF in benzene and dichloromethane. This ligand, which contains a single amide functional group, can only donate a hydrogen bond to one of the two carbonyls of TPARF. Therefore there are two possible orientations of the flavin-ligand complex – one as shown and one in which the MBAP ligand rotates 180° around its pyridine N(1)-C(4) axis, thereby forming a bidentate complex which hydrogen bonds to the flavin N(3) hydrogen and the C(4) carbonyl instead of the C(2) carbonyl.
Addition of MBAP to TPARF in benzene does not cause a shift in the UV/Vis spectrum maximum (Fig. 3A) and the change in spectral amplitude upon the addition of MBAP to TPARF was less than one third the magnitude of that observed for the TPARF:DBAP complex in the same solvent (Fig 2B). Furthermore, the difference spectrum of the TPARF-MBAP complex (Fig. 3B) is much broader than that of TPARF-DBAP, likely due to the presence of two superimposed difference spectra, one for each MBAP orientation.
Fig. 3.
TPARF interactions with MBAP in dichloromethane and in benzene. The absolute spectra of TPARF are shown in A): TPARF in benzene (black), TPARF in dichloromethane (grey), TPARF with MBAP in benzene (black dashed line) and TPARF with MBAP in dichloromethane (grey dashed line). The difference spectra of TPARF with MBAP relative to TPARF in benzene solvent, is shown in B). The inset displays the curve fit to a plot of the difference absorbance at 464 nm vs added MBAP in benzene. The same set of spectra and curve fit in dichloromethane is illustrated in C).
The attenuated spectral differences are accompanied by a change in binding affinity: the dissociation constant of MBAP from TPARF in benzene is 1,600 μM (Fig. 3B), four times weaker than that of the DBAP:TPARF complex. This is a result of the loss of one of the amide-carbonyl hydrogen bonds of the TPARF-DBAP complex. The addition of MBAP to TPARF in dichloromethane results in a difference spectrum (Fig. 4C) similar to that of the same observed in benzene but 30% weaker, as was the case for the TPARF:DBAP complex. Similarly, to either the dissociation constant of the MBAP:TPARF complex in dichloromethane is 14,890 μM (Fig. 3C), four times greater than that of the TPARF:DBAP complex in dichloromethane (Table 1). The weaker interaction between TPARF and MBAP is consistent with it being stabilized by fewer H-bonds than the TPARF:DPAP complex. Our results indicate that TPARF behaves substantially differently in benzene than in dichloromethane although both solvents are considered to be nonpolar aprotic media.
Fig. 4.

Solvent interactions with TPARF. The absolute spectra of TPARF in benzene (bottom), TPARF in dichloromethane (middle) and TPARF in chloroform (top). The inset shows the difference spectra of TPARF+DBAP minus TPARF only in benzene (a), TPARF only in chloroform minus TPARF in benzene (b, dashed line), and TPARF only in dichloromethane minus TPARF in benzene (c).
Table 1.
Electrochemical and binding properties of TPARF/Me-TPARF.
| DBAP/MBAP binding | ||||
|---|---|---|---|---|
| E1/2 (mv vs ferrocene) |
ΔE (mV) [E1/2(bound) - E1/2(unbound)] DBAP/MBAP |
Kd (ox)† (mM) DBAP/MBAP |
Kd (red)§ (mM) DBAP/MBAP |
|
| benzene | ||||
| TPARF | −1252 | 100/<10* | 420 ±20/1600 ±360 | 8.5/1100 |
| Me-TPARF | −1290 | - | - | - |
| dichloromethane | ||||
| TPARF | −1180 | 100/<10* | 4,120 ±180/14,890 ±940 | 84/10,100 |
| Me-TPARF | −1243 | - | - | - |
Values obtained from UV/Vis absorption measurements
Although the change in E1/2 values is small and comparable to the expected error, in our repeated experiments there was always a change, but it never exceeded 10 mV.
Calculated by using eqn. 1.
Solvent interactions with TPARF
The large difference in ligand affinity led us to examine the interactions between unliganded TPARF and its solvent. The electronic spectra of TPARF in chloroform, dichloromethane and benzene are detailed in Fig. 4.‡ (TPARF does not appreciably interact with itself as the flavin spectrum is unchanged over a range of TPARF concentrations from 150 nM to 1.5 mM in benzene or dichloromethane, data not shown.)
The changes in the absorption spectrum of TPARF appear to be due principally to hydrogen bonding interactions with the solvent, since they are almost identical to the changes in the spectrum generated by hydrogen bonding to DBAP in benzene (inset, Fig. 4, spectrum a). Furthermore, there is no appreciable difference between the spectra of the TPARF:DBAP complexes in benzene vs. dichloromethane. This indicates that the spectral differences observed for unliganded TPARF in different solvents are primarily the result of hydrogen bonding and not due to changes in the dielectric constant of the solvent. This interpretation is further supported by the observation of differences between the spectra of TPARF:MBAP complexes in benzene and dichloromethane (Figs. 3B and 3C). This complex only binds to one of the two carbonyls of the isoalloxazine ring, leaving one available to hydrogen bond to the solvent.
Transfer of TPARF from benzene to dichloromethane increases the extinction coefficient at 487 nm 30% of the increase observed upon addition of DBAP in benzene (the maximum increase observed). Moreover, the spectral effects of replacing benzene with dichloromethane, and the addition of DBAP are additive, suggesting that either 30% of the TPARF molecules in dichloromethane participate in hydrogen bonding interactions, all of the TPARF molecules in that solvent participate in hydrogen bonds which are 70% weaker than those of DBAP, or some combination of the two.
These hydrogen bonding interactions between TPARF and dichloromethane clearly play a role in the order of magnitude difference between the binding constants of the ligand complexes in dichloromethane vs. benzene. Although we have chosen not to examine in depth the behavior of TPARF in chloroform, we note that the electronic spectra indicate that the interactions between this solvent and TPARF are even greater than in dichloromethane (spectrum B in the inset of Fig. 4).
Cyclic voltammetry of TPARF complexes in benzene and dichloromethane
If the differences between benzene and dichloromethane are indeed due to increasing H-bonding, then they should produce different flavin reduction potentials.2 TPARF and Me-TPARF were each examined electrochemically in dichloromethane and in benzene both alone and in the presence of DBAP or MBAP, with Me-TPARF providing a negative control for flavin complex formation. The results of these studies are presented in Table 1. Cyclic voltammograms of TPARF and Me-TPARF in benzene containing 0.5 M tetrahexylammonium perchlorate (Hx4NClO4) performed using a 1.6-mm diameter platinum working electrode are depicted in Fig. 5A. The necessary inclusion of a supporting electrolyte introduces another species capable of hydrogen bonding. We cannot determine the change in the binding affinities between the ligands and the reduced flavin caused by the presence of the electrolyte because the electrochemical experiments require a minimum of 0.1 M Hx4NClO4. The experiments performed in this study show that Hx4NClO4 has minimum or no effects on the TPARF:DBAP association constants in comparison to the changes caused by solvent substitution, which is the focus of this paper.
Fig. 5.

Cyclic voltammetry of TPARF and Me-TPARF in benzene with 0.5 M Hx4NClO4. The voltammograms were measured at 22 oC vs ferrocene at a scan rate of 50 mV s−1. Cyclic voltammograms measured with a 1.6-mm diameter platinum electrode of 1 mM TPARF (line) and 1 mM Me-TPARF (dashed line) are shown in A) (negative peaks denote oxidation). The low-current cyclic voltammograms shown in B) were measured with a 10-μm diameter platinum electrode. When DBAP is added to 40 μM TPARF as shown in C), the voltammograms shift to positive potential and no change is observed after the addition of 1 mM of DBAP (inset).
These voltammograms are similar in appearance to those reported by Rotello’s group in the studies of N(10)-isobutyl isoalloxazine in dichloromethane.2, 4 The three wave pattern in the voltammogram has been elegantly explained by Niemz and coworkers:4, 22 the single reduction peak corresponds to two one-electron processes that follow one another in rapid succession with acquisition of one proton in between, in an ECE (electrochemical, chemical, electrochemical) cascade. Thus, upon reduction, the resulting semiquinone radical anion (Fl•−) draws a proton from excess flavin forming the neutral radical (Fl•H) which undergoes essentially instantaneous reduction to the flavin hydroquinone anion (FlH−). The two negative peaks in the voltammogram of TPARF (Fig. 5A) correspond to the oxidation of Fl-H− to Fl and Fl•− to Fl, respectively, with the latter couple having a lower reduction potential (E1/2). Since the source of the proton in the ECE scheme is from the N(3) position of the flavin unit,4 the voltammogram is reduced to two waves, centered at -1290 mV (vs. ferrocene) for the reduction (to Fl•−) and reoxidation of Me-TPARF in benzene (Fig. 5A).
Although the electrochemical behavior of TPARF and Me-TPARF in benzene is similar to that of these flavins in dichloromethane (not shown), the E1/2 values are likely to be affected by the high ohmic resistivity of benzene. Since the ohmic distortion (iR drop) in benzene is minimized with the use of very small-diameter electrodes,23 a 10-μm diameter platinum microelectrode was employed in order to generate low-current cyclic voltammograms as shown in Figs. 5B and 5C. This method has the added advantage that scan rates can be increased until they out compete proton transfer, resulting in replacement of the ECE pattern with a simple one-electron oxidation peak. As shown in Fig. 5B, E1/2 = -1252 mV (vs. ferrocene) for TPARF in benzene with 0.5 M Hx4NClO4, while the E1/2 of Me-TPARF under the same conditions is - 1290 mV. The E1/2 values of TPARF and Me-TPARF in dichloromethane containing 0.5 M Hx4NClO4 are -1180 and -1243 mV respectively (Table 1, data not shown).
DBAP addition to TPARF
The interaction between TPARF and DBAP in benzene increases the flavin E1/2 to less negative potentials, as shown in Fig. 5C. A limiting E1/2 value is reached when the DBAP concentration is greater than 1mM (inset, Fig. 5C) and the maximal change in E1/2 (ΔE = E1/2,bound – E1/2,unbound) is +100 mV. The use of a thermodynamic cycle, consisting of a pair of ligand binding equilibria and oxidation/reduction allows the derivation of eqn. 1 (see Scheme 2):22
| (1) |
This allows calculation of the dissociation constant for the reduced TPARF:DBAP complex: 8.4 μM. Thus, the binding affinity between TPARF and DBAP increases by a factor of 50 upon reduction of the flavin in benzene.
Scheme 2.

Although TPARF exhibits different basal E1/2 values for one-electron reduction in benzene and in dichloromethane, the limiting change in reduction potential at >20 mM DBAP is again +100 mV when DBAP is added in dichloromethane (Table 1). This ΔE value in dichloromethane again corresponds to a 50-fold decrease in the dissociation constant of the reduced TPARF:DBAP complex (84 μM) over the oxidized flavin (4,120 μM). In the case of benzene and dichloromethane, while both the flavin E1/2 values and ligand affinities for both oxidation states vary significantly, the energies coupling ligand binding and reduction in the two solvents are equal.
MBAP addition to TPARF
Formation of the MBAP:TPARF complex has little effect on the flavin E1/2. In our measurements, ΔE does not exceed +10 mV in either dichloromethane or benzene. The small magnitude of ΔE means that binding of MBAP to TPARF is not significantly enhanced upon the one-electron reduction of the flavin analogue (Kd(red)/Kd(ox) ~ 1.5). As in the case of DBAP, the change in medium causes large differences in binding affinity and reduction potentials while not affecting the coupling energy between complex formation and oxidation/reduction. Thus, the removal of one of the two hydrogen bonds to the flavin uracil ring carbonyl groups has the effect of greatly decreasing the cooperativity between the two processes.
The removal of one of the amide hydrogen bonding moieties from DBAP results in a 4-fold decrease in binding affinity to oxidized TPARF in either solvent. Furthermore, while hydrogen bonding to N(3) and a single carbonyl does not significantly modulate the electron affinity of the oxidized flavin, hydrogen bonding to both carbonyls extensively modulates the electronic structure of the flavin as evinced by both the coupling energies and the large change in the electronic spectra of TPARF upon complex formation. This decrease in ligand affinity corresponds to a 0.80 kcal/mol increase in the binding energy. Because of the ability of MBAP to bind in two orientations, however, it is not possible to isolate this energy loss to one particular hydrogen bonding interaction. FTIR and NMR investigations to quantify any orientational preference in MBAP binding to oxidized and reduced TPARF are currently underway.
Use of the thermodynamic box to analyze energetic coupling between two processes has the benefit that it isolates the two processes from their environment. Thus the coupling between binding and reduction potentials, ΔΔG, has the same value in both solvents despite the fact that each of the individual interactions varies significantly with solvent. There is a dramatic increase in the magnitude of the coupling energy, from +10 mV to +100 mV, when the complex switches from a two-point hydrogen bonding to a three-point hydrogen bonding system. This change, equivalent to 2.3 kcal/mol, is almost threefold larger than the change in binding energy to the oxidized state. This is a result of an even larger decrease in the binding affinity of the ligand to the reduced state upon removal of one amide-carbonyl hydrogen bond. This suggests that flavin reduction evinces a significant increase in the electron density at each carbonyl group, and that the increased ligand affinity of anionic flavin semiquinone radical is principally mediated by hydrogen bonding contacts of these carbonyls, as previously noted.22
Conclusions
The linkage between binding energy and the reduction potential of redox-active cofactors, as well as the biological importance of this coupling mechanism was described by Wyman over 50 years ago.24 Binding interactions coupled to the E1/2 effect redox tuning. For a complex cofactor such as the flavin, which has many possible sites of interaction with a protein, the mechanism of coupling is perhaps best investigated using simplified biomimetic complexes which can isolate an individual or a limited set of interactions in noninteractive solvents. Our original goal was to create and characterize an improved flavin analogue which is highly soluble in an apolar solvent with a high freezing point. The extreme solubility of this analogue in nonpolar, noninteractive solvents allowed us to compare its behavior both in one of the more commonly utilized apolar solvents, dichloromethane, with that in benzene, a less interactive solvent which has been impractical in the past with less soluble flavin models.
The large differences, both in ligand binding affinity and the electronic spectra of TPARF suggests that even dichloromethane donates a significant amount of hydrogen bonding to its solutes. Thus, ligand affinities measured in dichloromethane and chloroform conceal much of the true energetics of any host guest interaction. Similarly, these solvents do not provide good baselines for spectroscopic studies such as NMR, IR or Raman spectroscopies of unliganded model compounds. This is of some concern, as many ligand binding studies of other flavin analogues are performed in chloroform.2, 25 While it is possible that π- stacking interactions may alter TPARF’s interaction energetics,26 it is clear that benzene is a better choice than dichloromethane for studying the effects of hydrogen bonding on the energetics of a biomimetic host-guest model system.
Materials and methods
Chemicals and solvents
Ferrocene (>98%), anhydrous benzene, dichloromethane and chloroform were purchased from Sigma Chemical Company (St. Louis, MO). Electrochemical tetrahexylammonium perchlorate (GFS Chemicals Inc., Powell, Ohio) was purified by recrystallizing 3 times from ethanol.
Synthesis
All reactions were carried out under an inert atmosphere. Commercial reagents were used without further purification unless otherwise noted. For analytical thin-layer chromatography, precoated glass silica gel plates (Analtech HLF 250 μm) were used. All products were purified using silica gel (Fisher 40-63 μm) and/or by recrystallization. Melting points are uncorrected and were determined using a Fisher-Johns melting point apparatus. NMR spectra were recorded in CDCl3 with TMS as an internal standard at room temperature on a Varian Unity 500 operating at 500 MHz for 1H and 125 MHz for 13C. When necessary, two-dimensional homonuclear correlation (COSY) spectra were run in order to confirm assignments.
Tetraphenylacetylriboflavin (1)
Phosphorous oxychloride (75 μL, 0.798 mmol) was added dropwise to a solution of phenylacetic acid (688 mg, 5.05 mmol) in anhydrous pyridine (3 mL). Riboflavin (100 mg, 0.266 mmol) was added to the resulting solution, and the reaction was stirred at 73 °C for 4.5 hours. The reaction was cooled to room temperature and poured into saturated NaHCO3. This mixture was extracted twice with chloroform. The combined organics were washed with brine, dried (MgSO4) and concentrated in vacuo. The resulting crude product was purified by column chromatography on silica with a single step gradient from 1:3 ethyl acetate-hexanes to 3:1 ethyl acetate-hexanes to afford 1 (184.5 mg, 82%) as an orange glass. Rf 0.52 [3:1 ethyl acetate:hexanes +1% triethylamine]; δH 8.26 [1H, s, C(6)-H], 7.97 [1H, s, C(9)-H], 7.10-7.40 [20H, m, PhH], 6.78 [2H, d, J= 6 Hz, ribityl-N-CH2], 5.63 [1H, br d, J = 8.0 Hz, ribityl-CH], 5.37 [1H, m, ribityl-CH], 5.31 [1H, m, ribityl-CH], 4.38 [1H, d, J = 12.5, ribityl-CH], 4.05 [1H, dd, J = 12.5 and 6.0, ribityl-CH], 3.83 [1H, d, J = 16, PhCH], 3.75 [1H, d, J = 15.5, PhCH], 3.70 [2H, s, PhCH2], 3.54 [2H, s, PhCH2], 3.31 [1H, d, J = 15, PhCH], 3.20 [1H, d, J = 14.5, PhCH], 2.41 [3H, s, C(8)-CH3], 2.40 [3H, s, C(7)-CH3]; δC 171.2, 171.0, 170.7, 170.6 [BnC=O], 159.4 [C(4)], 154.3 [C(2)], 150.7 [C(10a)], 148.3 [C(8)], 137.0 [C(7)], 136.1 [C(4a)], 134.7 [C(5a)], 133.9, 133.8, 133.4 [i-Ph], 133.1 [C(6)], 132.7 [C(9a)], 129.8, 129.6, 129.5, 129.0, 128.9, 128.8, 128.7 [o/m-Ph], 127.7, 127.6, 127.4, 127.3 [p-Ph], 115.6 [C(9)], 70.8, 63.8, 69.5, 62.0 [ribityl-C], 44.5 [C(10α)], 41.2, 40.8 [PhCH2], 21.6 [C(8) CH3], 19.7 [C(7)CH3]
N(3)-Methyl-tetraphenylacetylriboflavin (2)
TPARF (0.500 g, 0.590 mmol) and Cs2CO3 (0.254 g, 0.831 mmol) were dissolved in 50 mL anhydrous dimethyl formamide containing 3 g dry molecular sieves (4Ǻ). Methyl iodide (0.797 g, 5.62 mmol) was added via cannula and the reaction stirred at room temperature for 12 hours.6 The reaction was extracted twice with saturated aqueous Na2CO3, dried (MgSO4), filtered and concentrated in vacuo. Flash chromatography of the residue (silica gel, 4:1 MeCl2:ethyl acetate) resulted in 0.40 g (79%) 2 as a yellow-orange powder. Rf 0.72 [3:1 ethyl acetate:hexanes +1% triethylamine]; δH 7.90 [1H, s, C(9)-H], 7.36-7.06 [20H, m, PhH], 6.75 [2H, d, J = 6.8 Hz, ribityl-N-CH2], 5.64 [1H, br d, J = 7.0 Hz, ribityl-CH], 5.38 [1H, br s, ribityl-CH], 5.32 [1H, br s, ribityl-CH], 4.38 [1H, d, J = 12.0 Hz, ribityl-CH], 4.06 [1H, q, J = 6.0 Hz, ribityl-CH], 3.80 [1H, q, J = 11.0 Hz, PhCH], 3.71 [2H, s, PhCH2], 3.54 [2H, s, PhCH2], 3.49 [3H, s, NCH3], 3.30 [1H, d, J = 16.0 Hz, PhCH], 3.20 [1H, br d, J = 14.0 Hz, PhCH], 2.41 [3H, s, C(8)-CH3], 2.39 [3H, s, C(7)-CH3]; δC 170.9, 170.8, 170.4, 170.3 [BnCO], 159.9 [C(4)], 155.23 [C(2)], 148.8 [C(10a)], 147.4 [C(8)], 136.3 [C(7)], 135.5 [C(4a)], 134.5 [C(5a)], 133.6 [i-Ph], 133.2 [C(6)], 132.5 [C(9a)], 129.5 [br, m-Ph], 128.5 [br, o-Ph], 127.1 [br, p-Ph], 115.2 [C(9)], 70.5, 69.5, 69.4, 69.3, 61.7 [ribityl-C], 43.8 [C(10a)], 41.0 [br, BnCH3], 28.7 [N(3)CH3], 21.3 [C(8) CH3], 19.4 [C(7)CH3]
2,5-dibenzylamidopyridine (3)
2,6-diaminopyridine (4.38 g, 40 mmol) was dissolved in 30 mL anhydrous pyridine. Phenylacetyl chloride (15.46 g, 100 mmol) was added dropwise with stirring over 30 minutes 27. After the addition was complete, the solution was refluxed for 16 h. After cooling to room temperature, pyridine was removed by rotary evaporation and the resultant black oil was dissolved in 600 mL ethyl acetate then subsequently washed with water, 5% tartaric acid and saturated aqueous NaHCO3. The organic layer was dried (MgSO4), filtered and concentrated in vacuo. The crude residue was purified using flash chromatography (3:1 hexanes/ethyl acetate with 1% triethylamine) and recrystallized from ethyl acetate resulting in 4.6 g (37%) of pale yellow crystalline 10: mp 143 °C; Rf 0.29 [3:1 hexanes:ethyl acetate +1% triethylamine]; δH 7.88 [2H, d, J = 7.8 Hz, m-PyrH], 7.66 [1H, t, J = 8.1 Hz, p-PyrH], 7.45 [2H, br s, NH], 7.38 [4H, t, J = 7.3 Hz, o-PheH], 7.33 [2H, d, J = 7.8 Hz, p-PhH], 7.29 [4H, d, J = 7.4 Hz, m-PhH], 3.68 [4H, s, CH2Ph]; δC 169.2 [C=O], 149.1 [o-Pyr], 140.8 [p-Pyr], 133.9 [i-Ph], 129.5, 129.4, 129.2 [o/m-Ph], 127.8, 127.6 [p-Ph], 109.6 [m-Pyr], 44.9 [CH2]
UV/Vis absorption spectroscopy
Spectra were obtained using an Agilent 8453 UV/Vis diode array spectrometer equipped with an optical filter to cut off light below 400 nm in order to avoid photoreduction of the flavin during the measurements. The initial flavin in either benzene or dichloromethane (ca. 70 μM) was titrated with addition of substoichiometric amount of ligand in solvent containing flavin at the same concentration as in the cuvette. In this manner, optical changes due to flavin dilution were minimized. Data were fit with:
| (2) |
where ΔA is the change in absorbance at the wavelength of maximum perturbation, ΔAT is the change in absorbance at this wavelength at saturating ligand concentrations, Kd is the dissociation constant for the TPARF-ligand complex, LT is the total ligand concentration, and FT is the total concentration of TPARF.28
Cyclic voltammetry
Cyclic voltammograms were collected on a BAS 100B workstation (Bioanalytical Systems, Inc., W. Lafayette, IN) equipped with a low current module. A Pt electrode (either a 1.6-mm or 10-μM diameter disk, from Bioanalytical Systems, Inc.) was used as the working, while a Pt wire and Ag wire were used as counter and pseudo reference electrodes, respectively. The three electrodes were fitted into a Claisen Adapter (Kontes Glass Company, Vineland, NJ) with working and reference electrodes arranged as close as possible to minimize ohmic resistance and contained in a Faraday cage under the dry anoxic (O2 < 1 ppm) N2 atmosphere of a Lab Master 130 glove box (Mbraun Inc, Stratham, NH). 0.5 M Hx4NClO4 was used as the electrolyte in all experiments. Binding experiments performed in solvents containing this electrolyte gave the same results, within error, of binding experiments performed in its absence. Reduction potentials in both solvents did not change when the electrolyte concentration was varied between 0.1 and 0.5 M
Supplementary Material
Acknowledgments
This work was supported by DOE (DE-FG02-05ER46223) MRSEC NSF (award #DMR05-20020) and NIH (41048).
Footnotes
Abbreviations: cyclic voltammetry (cv), 2,6-dbenzylamidopyridine (DBAP), Fourier transform infrared spectroscopy (FTIR), monobenzylamidopyridine (MBAP), N(3)-methylated TPARF (Me-TPARF), tetraacetylriboflavin (TARF), tetrahexylammonium perchlorate (Hx4NClO4), tetraphenylacetyl riboflavin (TPARF).
Notes and References
- 1.Cuello AO, McIntosh CM, Rotello VM. J. Am. Chem. Soc. 2000;122:3517–3521. [Google Scholar]
- 2.Breinlinger E, Niemz A, Rotello VM. J. Am. Chem. Soc. 1995;117:5379–5380. [Google Scholar]
- 3.Breinlinger EC, Keenan CJ, Rotello VM. J. Am. Chem. Soc. 1998;120:8606–8609. [Google Scholar]
- 4.Niemz A, Imbriglio J, Rotello VM. J. Am. Chem. Soc. 1997;119:887–892. [Google Scholar]
- 5.Koder RL, Lichtenstein BR, Cerda JF, Miller AF, Dutton PL. Tetrahedron Lett. 2007;48:5517–5520. doi: 10.1016/j.tetlet.2007.05.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epple R, Wallenborn EU, Carell T. J. Am. Chem. Soc. 1997;119:7440–7451. [Google Scholar]
- 7.Akiyama T, Simeno F, Murakami M, Yoneda F. J. Am. Chem. Soc. 1992;114:6613–6620. [Google Scholar]
- 8.Hasford JJ, Kemnitzer W, Rizzo CJ. J. Org. Chem. 1997;62:5244–5245. [Google Scholar]
- 9.Massey V. Biochem. Soc. Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 10.Hemmerich P, Massey V, editors. The Role of Apoprotein in Directing Pathways of Flavin Catalysis. Pergamon Presss; Oxford: 1982. [Google Scholar]
- 11.Schopfer LM, Ludwig ML, Massey V. A working proposal for the role of the apoprotein in determining the redox potential of the flavin in flavoproteins: correlations between potentials and flavin pKs. Walter de Gruyter; New York: 1990. [Google Scholar]
- 12.Koder RL, Haynes CA, Rodgers ME, Rodgers DW, Miller AF. Biochemistry. 2002;41:14197–14205. doi: 10.1021/bi025805t. [DOI] [PubMed] [Google Scholar]
- 13.Gordy W. J. Chem. Phys. 1939;7:163–166. [Google Scholar]
- 14.Kamlet MJ, Taft RW. J. Chem. Soc., Perkin Trans. 1. 1979:349–356. [Google Scholar]
- 15.Kamlet MJ, Dickinson C, Taft RW. J. Chem. Soc., Perkin Trans. 2. 1981:353–355. [Google Scholar]
- 16.Walsh JD. Ph.D. The Johns Hopkins University; 2000. [Google Scholar]
- 17.Koder RL, Walsh JD, Pometum MS, Dutton PL, Wittebort RJ, Miller AF. J. Am. Chem. Soc. 2006;128:15200–15208. doi: 10.1021/ja0648817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh JD, Miller AF. J. Phys. Chem. B. 2003;107:854–863. [Google Scholar]
- 19.Muller F. In: Chemistry and Biochemistry of Flavoenzymes. Muller F, editor. vol. I. CRC Press; Boca Raton, FL: 1991. pp. 2–72. Editon edn. [Google Scholar]
- 20.Nishimoto K, Watanabe Y, Yagi K. Biochim. Biophys. Acta. 1978;526:34–41. doi: 10.1016/0005-2744(78)90287-5. [DOI] [PubMed] [Google Scholar]
- 21.Yagi K, Ohishi N, Nishimoto K, Choi JD, Song PS. Biochemistry. 1980;19:1553–1557. doi: 10.1021/bi00549a003. [DOI] [PubMed] [Google Scholar]
- 22.Niemz A, Rotello VM. Acc. Chem. Res. 1999;32:44–52. [Google Scholar]
- 23.Bond AM, Mann TF. Electrochim. Acta. 1987;32:863–870. [Google Scholar]
- 24.Wyman J. Adv. Protein Chem. 1948;4:407–531. doi: 10.1016/s0065-3233(08)60011-x. [DOI] [PubMed] [Google Scholar]
- 25.Legrand YM, Gray M, Cooke G, Rotello VM. J. Am. Chem. Soc. 2003;125:15789–15795. doi: 10.1021/ja036940b. [DOI] [PubMed] [Google Scholar]
- 26.Breinlinger EC, Rotello VM. J. Am. Chem. Soc. 1997;119:1165–1166. [Google Scholar]
- 27.Feibush B, Figueroa A, Charles R, Onan KD, Feibush P, Karger BL. J. Am. Chem. Soc. 1986;108:3310–3318. [Google Scholar]
- 28.Koder RL, Haynes CA, Rodgers DW, Miller AF. Flavins and Flavoproteins 2002. Rudolph Weber; Berlin: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.