Abstract
In this study, we investigated the prevalence of human immunodeficiency virus type 1 (HIV-1) drug resistance mutations and genetic variability among Senegalese patients undergoing highly active antiretroviral therapy (ART) in the public health system. We conducted a cross-sectional study of 72 patients with suspected therapeutic failure. HIV-1 genotyping was performed with Viroseq HIV-1 Genotyping System v2.0 or the procedure developed by the ANRS AC11 resistance study group, and a phylogenetic analysis was performed. The median follow-up visit was at 40 (range, 12 to 123) months, and the median viral load was 4.67 (range, 3.13 to 6.94) log10 copies/ml. The first-line therapeutic regimen was nucleoside reverse transcriptase inhibitors (NRTIs) plus efavirenz (EFV) or NRTIs plus nevirapine (NVP) (54/72 patients; 75%), and the second-line therapy was NRTIs plus a protease inhibitor (PI/r) (18/72; 25%). Fifty-five patients (55/72; 76.39%) had at least one drug resistance mutation. The drug resistance rates were 72.22 and 88.89% for the first-line and second-line ARTs, respectively. In NRTI mutations, thymidine analog mutations (TAMs) were found in 50.79% and the M184V mutation was found in 34.92% of the samples. For non-NRTI resistance, we noted a predominance of the K103N mutation (46.27%). For PI/r, several cases of mutations were found with a predominance of M46I and L76V/F at 24% each. The phylogenetic analysis revealed CRF02_AG as the predominant circulating recombinant form (43/72; 59.72%). We found a high prevalence of resistance mutations and a high rate of TAMs among Senegalese patients in the public health system. These findings emphasize the need to improve virological monitoring in resource-limited settings.
INTRODUCTION
Phylogenetic analyses of many HIV-1 strains isolated from diverse geographic origins have revealed four distinct groups of viruses, namely, groups M, N, O, and P. Group M, which is responsible for the global pandemic, can be further subdivided into subtypes (A to D, F to H, J, and K), subsubtypes, and numerous circulating recombinant forms (CRF01 to CRF54; http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html). In Senegal, circulating HIV-1 strains have been characterized and show a high level of genetic diversity, with the predominance of CRF02_AG among members of the general population (1, 2) and female sex workers (3). However, a high prevalence of subtype C was noted among men who have sex with men (4).
In developing countries where non-B subtypes are predominant, antiretroviral therapy (ART) has become increasingly available, resulting in the improved health and survival of patients infected with HIV-1. The emergence of HIV resistance, due to mutations in the viral protease and reverse transcriptase (RT) genes, is the major cause of ART failure. The development of drug resistance requires the occurrence of both antiretroviral drug exposure and ongoing viral replication (5). One of the first ART initiatives sponsored by an African government was launched in Senegal in 1998. Patients were monitored clinically and biologically by plasma HIV-1 RNA load determination, CD4 cell counting at the baseline and at 6-month time intervals, and monthly medical examinations (6). According to the 2010 World Health Organization (WHO) guidelines, the first-line drug regimen for adults and adolescents in resource-limited settings should contain two nucleoside RT inhibitors (NRTIs) and one non-NRTI (NNRTI). After first-line ART failure, a boosted protease inhibitor with two NRTIs has been recommended for second-line ART (7). In a prospective observational cohort study, it was shown that the implementation of highly active ART (HAART), in combination with clinical, biological, and logistical monitoring, reduced the emergence of resistant strains in Africa (8). The WHO guidelines also suggest that the first-line ART regimen should be switched after immunological or clinical failure when plasma HIV-1 RNA load tests are not available (7). However, previous studies reported the development of resistance mutations when the switch was performed solely on the basis of immunological or clinical monitoring of patients (9, 10). A similar study of patients receiving first-line ART in Africa revealed a high rate of thymidine analog mutation (TAM) accumulation in patients with immunological or clinical failure when following the WHO guidelines (11). Most published data on ART treatment in Senegal have resulted from structural prospective studies, including a biological follow-up study according to the best standards of care (6, 12, 13). However, no data on HIV drug resistance (HIVDR) was available from patients undergoing HAART in the public health system. The aim of this study was to investigate the prevalence of HIVDR mutations and the genetic diversity in a routinely treated general population group.
MATERIALS AND METHODS
Specimen collection and viral load and resistance testing.
We obtained blood samples by venipuncture from patients at three different clinical sites in Dakar, the capital of Senegal, and collected them in EDTA tubes. The patients were enrolled in the Senegalese Antiretroviral Drug Access Initiative (ISAARV) from 2001 to 2012. We conducted a cross-sectional study of patients with known virological failure among patients suspected to have experienced therapeutic failure. The samples were selected on the basis of a viral load of >3 log10 copies/ml by the Abbott RealTime HIV-1 m2000rt quantitative assay (Abbott Laboratories, Chicago, IL). Resistance testing was performed on the plasma samples using Celera Diagnostics ViroSeq HIV-1 Genotyping System version 2.0 according to the manufacturer's instructions (Celera Diagnostics, San Francisco, CA) or the procedure developed by the ANRS AC11 resistance study group (http://www.hivfrenchresistance.org/). For ViroSeq, RNA was amplified by reverse transcription, followed by a one-step PCR to generate a fragment of 1,800 bp. The PCR products were then purified and used for direct sequencing on an ABI 3100-Avant system using the BigDye Terminator v3.1 kit (Applied Biosystems, Courtaboeuf, France). Sequencing was performed to cover the entire protease gene and RT amino acids 1 to 320. Sequences were analyzed using the ViroSeq HIV-1 Genotyping System software v2.8 (Celera Diagnostics, San Francisco, CA), which generated a drug resistance report. For the ANRS AC11 resistance method, RNA extraction from plasma was performed using the QIAamp Viral RNA kit (Qiagen, Valencia, Spain) according to the manufacturer's instructions. The PCR products were purified (Qiagen, Valencia, Spain) and directly sequenced on ABI3100-Avant using BigDye Terminator v3.1 (Applied Biosystems, Courtaboeuf, France). The sequences generated were edited using the SeqMan II software program from the DNAStar package v.5.08 (Lasergene, Madison, WI). To determine the resistance mutations, the entire protease gene and at least the first 240 amino acids encoded by the RT gene were analyzed with the Stanford HIVDR database tools (http://hivdb.stanford.edu/).
Phylogenetic analysis.
HIV-1 subtype and CRF designations were determined by phylogenetic tree analysis. The nucleotide sequences generated were aligned, and a neighbor-joining tree was drawn with 100 bootstrap replicates, as implemented in the Seaview software (14). All pure subtypes and CRFs available in the Los Alamos database and circulating in West Africa were included in the analysis. To determine whether the viruses were recombinant or not, similarity analysis and bootscanning were performed with the Simplot version 3.5.1 software. The final tree was drawn with the minimal number of references, i.e., excluding those that were not represented in the data set. Phylogenies over trimmed alignment were inferred by using PhyML version 3 under the GTR+I+G model of nucleotide substitution. An approximate likelihood ratio test was used to assess confidence in topology.
Nucleotide sequence accession numbers.
The sequences of the HIV-1 isolates used in this study have been deposited in GenBank under accession numbers JN673567, JN673569, JN673574, JN673576, JN673579, JN673583, JN673584, JN673595 to JN673598, JN673603 to JN673618, JN673621, JN673628, JN673637, JN673647, JN673677, JN673678, JN673683 to JN673686, JN673688, JN673691 to JN673693, JN673697, JN673698, JN673703, JN673708, JQ855853 to JQ855855, JX187611 to JX187622, JX227940, JX187624 to JX187630, and KC176534 to KC176537.
RESULTS
Patient group characteristics.
Table 1 summarizes the virological, drug resistance, and therapeutic patient group data. The sex ratio was 0.60, including 45 (60.81%) women and 27 (37.50%) men. The 72 participants' ages ranged from 20 to 57 years in the selected group. The median follow-up visit was at 40 (range, 12 to 123) months, and the median viral load was 4.73 (range, 3.13 to 6.94) log10 copies/ml. The first-line therapeutic regimen was NRTIs plus efavirenz (EFV)/nevirapine (NVP) (NRTIs-EFV/NVP; 54/72 patients; 75%) (where a slash [/] indicates that either EFV or NVP was used), and the second-line therapy was NRTIs plus a protease inhibitor (NRTIs-PI/r; 19/73; 25%).
Table 1.
Virological, drug resistance, and therapeutic patient group data
| Parametera | No. (%) of patients |
|
|---|---|---|
| First-line ART (n = 54) | Second-line ART (n = 18) | |
| Viral loads (log10) | ||
| 3–3.67 | 12 (22.22) | 2 (11.11) |
| 3.67–4 | 8 (14.81) | 1 (5.56) |
| 4–5 | 22 (40.74) | 7 (38.89) |
| >5 | 12 (22.22) | 8 (44.44) |
| Treatments | ||
| AZT + 3TC + NVP/EFV | 38 (70.37) | |
| d4T + 3TC + NVP/EFV | 8 (14.81) | |
| DDI + 3TC + EFV | 4 (7.41) | |
| TDF + 3TC + NVP/EFV | 3 (5.56) | |
| 3TC + DDI + NVP | 1 (1.85) | |
| TDF + ABC/DDI/3TC/FTC + LPV/r | 12 (66.67) | |
| AZT + 3TC/ABC + LPV/r | 4 (22.22) | |
| ABC + DDI + DRV/r | 1 (5.56) | |
| DDI + 3TC + IDV/r | 1 (5.56) | |
| ART times (mo) | ||
| 12 | 11 (20.37) | |
| 12–24 | 10 (18.52) | 3 (16.67) |
| >24 | 33 (61.11) | 15 (83.33) |
| Drug resistance | ||
| NRTIs | 1 (1.85) | 1 (5.56) |
| NNRTIs | 5 (9.26) | 2 (11.11) |
| NRTIs + PI/r | 4 (22.22) | |
| NNRTIs + NRTIs | 31 (57.41) | 7 (38.89) |
| All drugs | 2 (3.70) | 2 (11.11) |
| Any drug | 15 (27.78) | 2 (11.11) |
VL, viral load; AZT, zidovudine; 3TC, lamivudine; FTC, emtricitabine; d4T, stavudine; NVP, nevirapine; EFV, efavirenz; DDI, didanosine; TDF, tenofovir; ABC, abacavir; LPV/r, lopinavir/ritonavir; DRV/r, darunavir/ritonavir; IDV/r, indinavir/ritonavir.
Phylogenetic analyses.
As indicated in Materials and Methods, sequences were aligned with HIV-1 references and also with sequences from ART-naïve patients who were included in the ISAARV cohort and collected since 1997 or who were diagnosed as newly infected since 2003 (not shown). These sequences were used to report on resistance mutations in naïve patients from Senegal by Diop-Ndiaye et al. (1). Also, some of the earliest samples from the ISAARV cohort were used to document, for one of the first times, the minor mutations in non-B subtypes (15). Our goal was to check any duplicate patient samples between the ART-naïve patient samples and the 72 samples from the present study to avoid any sequence overlap in database accessions. We found five patients who were previously included in the above studies under accession numbers FN599718, FN599740, FN599686, FN599697, and AJ286986. However, 7 to 10 years later, the patients harbored several drug resistance mutations and consequently, we submitted the corresponding sequences.
The overall subtype distribution was as follows: CRF02_AG (n = 43; 59.72%), C (n = 5; 6.94%), A3 (n = 3; 4.17%), B (n = 2; 2.78%), D (n = 2; 2.78%), CRF06_cpx (n = 4; 5.56%), and one each of the CRF09_cpx and CRF01_AE strains (1.39%). In addition, 11 unique recombinant forms (URFs) were also found, including CRF02_AG/A3 (n = 3; 4.17%), CRF02_AG/A (n = 2; 2.78%), and one (1.39%) each of CRF02_AG/CRF06_cpx, CRF02_AG/U/CRF02_AG, CRF09_cpx/CRF02_AG, G/CRF02_AG/A, CRF02_AG/C/A, and CRF11_cpx/CRF02_AG. Unique recombinants with the same profile, i.e., CRF02_AG/A3 and CRF02_AG/A, did not cluster together in the phylogenetic tree. Figure 1 shows the phylogenetic relationships between the sequences in the present study. We found one very probable transmission chain between two strains originating from one man and one woman.
Fig 1.
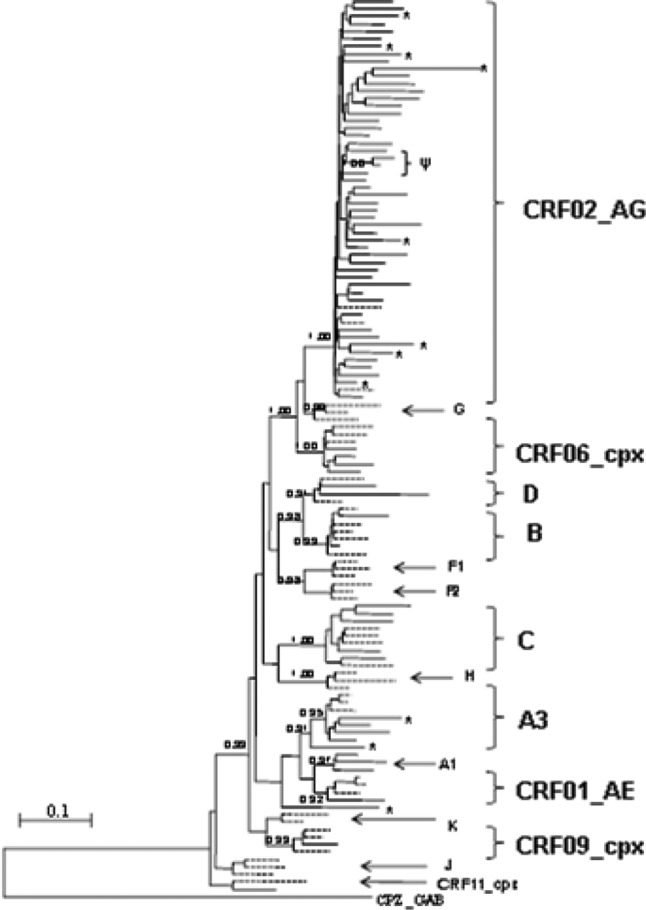
Maximum-likelihood tree depicting the phylogenetic relationships between the sequences in this study. The tree was constructed as indicated in Materials and Methods. aLRT statistics were used to support the tree topology and are indicated on the main branches. The letter Ψ indicates a very probable transmission chain between two sequences collected from one man and one woman. Asterisks indicate unique recombinant strains. HIV-1 reference sequences from the Los Alamos database are indicated by dashed lines, and sequences from the present study are indicated by solid lines.
Drug resistance analysis.
Table 1 summarizes the findings on drug resistance mutations among the 72 HIV-1-infected patients known to have experienced virological failure (≥3 log10 copies/ml) during first-line (n = 54) and second-line (n = 18) ART. The first-line therapeutic regimen was NRTIs-EFV/NVP (54/72; 75%), and the second-line therapy was NRTIs-PI/r (18/72; 25%). Fifty-five patients (55/72; 76.39%) had at least one drug resistance mutation. The drug resistance rates were 72.22 and 88.89% for the first- and second-line ARTs, respectively. For patients on the first-line ART, the high rate of resistance mutations was associated with NRTI-NNRTI combinations (n = 31; 57.41%). However, the rates of resistance mutation with second-line ART were 38.89 and 22.22% with the NRTI-NNRTI and NRTI-PI/r combinations, respectively.
For NRTI mutations (Table 2), the CRF02_AG strain contained all of the mutations associated with resistance to NRTIs, except the K65R mutation. However, the subtypes B, CRF09_cpx, CRF02_AG/CRF06_cpx, and CRF09_cpx/CRF02_AG displayed no mutations associated with resistance to NRTIs. Among the mutations associated with resistance to NRTIs (126 mutations), we observed a predominance of TAMs (n = 64) which was equivalent to 50.79% of the set. We also observed a high rate of mutations at the M184 position, which represents 44 (34.92%) of the total of 126 mutations. The CRF02_AG form also displayed mutational combinations, including several cases of TAMs-M184V, two cases of TAMs and an insertion at position 69, three cases of TAMs-E44D, and five cases of M184V-L74V. The M184V mutation was also found in only two variants.
Table 2.
Mutations in the RT gene at key positions associated with NRTI resistancea
| Subtype | No. of strains | M41 | D67 | K70 | L210 | T215 | K219 | K65 | L74 | Y115 | M184 | E44 | V118 | 69 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A3 | 3 | V1 | ||||||||||||
| B | 2 | |||||||||||||
| C | 5 | V4 | ||||||||||||
| CRF01_AE | 1 | A1, F1 | E1 | V1 | N1 | |||||||||
| CRF02_AG | 43 | L7 | N8, G1 | R5, K/N1, G/R1, G1 | W4 | Y8, F6, I1, S/Y1, I/V1 | E4, Q3, N1 | V3 | F1 | V26, I1 | D3 | I3 | N1, P/T1 | |
| CRF02_AG/A | 2 | L1 | V2 | |||||||||||
| CRF02_AG/A3 | 3 | V1 | V3 | |||||||||||
| CRF02AG/CRF06_cpx | 1 | |||||||||||||
| CRF02_AG/U/CRF02_AG | 1 | N1 | Q1 | V1 | ||||||||||
| CRF06_cpx | 4 | V2 | ||||||||||||
| CRF09_cpx | 1 | _ | ||||||||||||
| CRF09_cpx/CRF02_AG | 1 | |||||||||||||
| D | 2 | N1 | R1 | Q1 | V1 | |||||||||
| G/CRF02_AG/A | 1 | L1 | W1 | I1 | V1 | I1 | ||||||||
| CRF02_AG/C/A | 1 | R1 | F1 | V1 | ||||||||||
| CRF11_cpx/CRF02_AG | 1 | S/Y1 | V1 | |||||||||||
| Total | 72 | 9 | 11 | 9 | 5 | 19 | 11 | 1 | 5 | 2 | 44 | 3 | 4 | 3 |
For example, in V2, the letter represents the amino acid substitution and the number indicates the number of strains with that mutation.
Table 3 displays all of the mutations in our study population that are associated with resistance to NNRTIs. The CRF02_AG strains showed all of the mutations associated with resistance to NNRTIs, except mutations at positions K101P/E and V106I. The subtype D, URFs (CRF09_cpx/CRF02_AG, CRF11_xpx/CRF02_AG, and CRF02_AG/CRF06_cpx) and CRF09_cpx contained any mutations that were associated with resistance to NNRTIs. In addition, we observed a predominance of mutations at position K103N (31/67; 46.27%), followed by additional mutations at positions V108, G190, and Y181. Several cases of K103N combined with other mutations (e.g., L100I and P225H) were noted. The mutation K103N was also found once in each of 10 patients.
Table 3.
Mutations in the RT gene at key positions associated with NNRTI resistancea
| Subtype | No. of strains | L100 | K101 | K103 | V106 | V108 | Y181 | Y188 | G190 | P225 |
|---|---|---|---|---|---|---|---|---|---|---|
| A3 | 3 | P1, E1 | N1 | I1 | A1 | |||||
| B | 2 | N1 | ||||||||
| C | 5 | N2 | A2 | |||||||
| CRF01_AE | 1 | C1 | C1 | |||||||
| CRF02_AG | 43 | I2 | N21 | I7 | C1, C/Y1 | L2 | A3, A/G1 | H3 | ||
| CRF02_AG/A | 2 | N2 | C1 | H2 | ||||||
| CRF02_AG/A3 | 3 | N1 | ||||||||
| CRF02_AG/CRF06_cpx | 1 | |||||||||
| CRF02_AG/U/CRF02_AG | 1 | N1 | ||||||||
| CRF06_cpx | 4 | N1 | C1 | |||||||
| CRF09_cpx | 1 | |||||||||
| CRF09_cpx/CRF02_AG | 1 | |||||||||
| D | 2 | |||||||||
| G/CRF02_AG/A | 1 | N1 | I1 | |||||||
| CRF02_AG/C/A | 1 | C1 | L1 | |||||||
| CRF11_cpx/CRF02_AG | 1 | |||||||||
| Total | 72 | 2 | 2 | 31 | 10 | 6 | 3 | 7 | 5 |
For example, in N1, the letter represents the amino acid substitution and the number indicates the number of strains with that mutation.
In contrast to the high rate of mutations associated with resistance to NRTIs and NNRTIs, there were few mutations associated with PI/r resistance. We documented 25 positions of mutations among 57 HIV-1-infected patients who developed this type of resistance. Several cases of mutations were found with a predominance of the M46I and L76V/E mutation types, and each represented 24% of the total. The CRF01_AE and CRF02_AG strains harbored most of the mutational positions. One subtype D and one URF (G/CRF02_AG/A) developed two mutations, at positions I50 and L76 and positions M46 and I84, respectively. Subtypes A3, B, C, and D, CRF06_cpx, CRF09_cpx, and the URFs (CRF02_AG/A, CRF02_AG/A3, CRF02_AG/CRF06_cpx, CRF02_AG/U/CRF02_AG, CRF09_cpx/CRF02_AG, CRF02_AG/C/A, and CRF11_cpx/CRF02_AG) contained any mutations associated with PI/r resistance.
DISCUSSION
In this study, we investigated the prevalence of HIV-1 drug resistance mutations and the genetic variability among Senegalese patients undergoing HAART in the public health system. This work is in contrast to structural prospective studies, as we conducted a cross-sectional study of 72 patients with known virological failure (≥3 log10 copies/ml) while undergoing treatment. This study identified a predominance of the CRF02_AG strains (43/72, 59.72%), the circulation of eight subtypes/CRFs, and 11 URFs, representative of a very high level of genetic diversity, in accordance with previously published studies (16). However, the subtype/CRF distribution can change over time in some high-risk groups, such as female sex workers (3). A very high level of genetic diversity with a substantial number of URFs and an increased prevalence of subtype C strains was previously described in untreated Senegalese patients from 1998 to 2007 (1).
In a structural cohort in Senegal, an increase in AIDS-defining illnesses was noted in patients with immunological and virological failure or with only virological failure (17). Studies in Senegal have demonstrated a low rate of drug resistance mutations in patients in a structural cohort (6, 12, 13). In the present study, we examined patients from the public health system and found a high rate of drug resistance mutations: 76.39% of the samples tested (55/72) had at least one drug resistance mutation after a median follow-up period of 40 months. This rate of drug resistance mutation is similar to that recently reported in studies in decentralized areas of Senegal (18) and in other countries such as the Central African Republic (19) and the Republic of South Africa using the same first-line regimens (20). In contrast, a relatively low prevalence of resistant viruses has been reported in Cameroon after 12 and 24 months, probably because of the shorter median follow-up period of 17 months (21). The drug resistance rates among patients with virological failure were 72.22 and 88.89% for first- and second-line ART, respectively. For patients on first-line ART, the highest rate of resistance mutations was associated with a combination of NRTIs-NNRTIs (n = 31; 57.41%). This finding confirms the need to increase the number of patients on second-line ART, but the financial cost is higher than that of the first-line drugs (22). At sites with viral load monitoring, the patients are switched earlier than at sites without viral load monitoring (23). While expensive, viral load monitoring has the potential to save costs on expensive second-line drugs by confirming that they are needed (7). However, the rates of resistance mutations for second-line ART were 36.84 and 21.05% with combinations of NRTIs-NNRTIs and NRTIs-PI/r, respectively. This high level of patients who harbored resistance to NRTIs-NNRTIs was due to the re-emergence of archived mutations as a result of failure of first-line ART with NRTI- or NNRTI-containing regimens. These results showed a high rate of HIV-1 drug resistance among patients undergoing first- and second-line therapeutic regimens, while third-line regimens are more expensive and not yet available in our countries. The main solution for those patients resistant to both first- and second-line regimens is to use another boosted PI/r and etravirine, a second-generation NNRTI (13). In the public health system, as ART availability continues to increase throughout Africa, the monitoring of HIV drug resistance mutations should be a priority (24). In Senegal, a total of 18,352 patients were undergoing HAART from 1998 to 2011 (http://www.unaids.org/en/dataanalysis/knowyourresponse/countryprogressreports/2012countries/ce_SN_Narrative_Report[1].pdf) and the few data available showed a high rate of HIV-1 drug resistance among treated patients (18). HIV infection cohorts should have an excellent and important platform to preserve current first-line drugs for the long term. There is also a need for some form of cheap and practical viral load monitoring and drug resistance testing for urban and rural environments in resource-limited settings (24, 25).
As mentioned in Tables 2 and 3, the CRF02_AG strains harbored all of the mutations associated with resistance to NRTIs, except the K65R mutation, and all of the mutations associated with resistance to NNRTIs, except those at positions K101P/E and V106I. Most of the mutated positions associated with PI/r resistance (Table 4) were harbored by the CRF01_AE and CRF02_AG viruses. These findings indicate that the rate of antiretroviral drug resistance mutation selection appears to be influenced by viral genetic diversity, as previously observed (26), despite the fact that the prevalence of patients carrying the CRF02_AG strain was higher than that of patients in the study set carrying other subtypes, CRFs, and URFs. Mutations associated with NRTI, NNRTI, and PI/r resistance have already been reported in CRF02_AG strains from Burkina Faso (26).
Table 4.
Mutations in the protease gene at key positions associated with resistance to PI/ra
| Subtype | No. of strains | V32 | D30 | M46 | I47 | G48 | I50 | I54 | Q58 | T74 | L76 | V82 | I84 | N88 | L90 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A3 | 3 | ||||||||||||||
| B | 2 | ||||||||||||||
| C | 5 | ||||||||||||||
| CRF01_AE | 1 | I1 | V1 | V1 | A1 | V1 | |||||||||
| CRF02_AG | 43 | I4 | V2 | V3 | V2 | V4 | M1 | ||||||||
| CRF02_AG/A | 2 | ||||||||||||||
| CRF02_AG/A3 | 3 | ||||||||||||||
| CRF02_AG/CRF06_cpx | 1 | ||||||||||||||
| CRF02_AG/U/CRF02_AG | 1 | ||||||||||||||
| CRF06_cpx | 4 | ||||||||||||||
| CRF09_cpx | 1 | ||||||||||||||
| CRF09_cpx/CRF02_AG | 1 | ||||||||||||||
| D | 2 | A1 | F1 | ||||||||||||
| G/CRF02_AG/A | 1 | I1 | V1 | ||||||||||||
| CRF02_AG/C/A | 1 | ||||||||||||||
| CRF11_cpx/CRF02_AG | 1 | ||||||||||||||
| Total | 72 | 6 | 3 | 2 | 3 | 3 | 6 | 1 | 1 |
For example, in I3, the letter represents the amino acid substitution and the number indicates the number of strains with that mutation.
Among the mutations associated with resistance to NRTIs, we observed a predominance of TAMs (n = 64; 50.79%), followed by the M184V/I mutation, which represents 44 (34.92%) of the total of 126. The higher rate of TAMs and the M184V/I mutation observed in this study has previously been described in Malawi and could be related to the duration of treatment failure before genotyping in patients being treated in a public health system (27). A high rate of TAM accumulation in patients with immunological or clinical failure in response to WHO-recommended treatment has also been described in South Africa (28), and it is higher than that observed in the Central Africa Republic (24%). These treatment guidelines may also be associated with a higher frequency of the M184V mutation, conferring increased sensitivity to zidovudine (AZT) and stavudine (d4T) (29). CRF02_AG strains also revealed several cases of mutational combinations of TAMs and M184V, two cases of TAMs plus an insertion at position 69, three cases of TAMs plus E44D, and five cases of M184V plus L74V. The M184V mutation was also found solely in CRF06_cpx. These mutations have been identified in limited-resource settings, where the first-line regimen drugs contained AZT or d4T (28). The M184V mutation alone does not significantly affect the in vivo virological responses to abacavir (ABC) (30). In adults infected with HIV-1 subtype B, the M184V mutation in the presence of additional TAMs increased the susceptibility to AZT (31). Furthermore, this mutation has been demonstrated to delay the emergence of TAMs, suggesting that it would remain advantageous to maintain emtricitabine (FTC)-containing treatment even for patients with a detectable viral load (32). The presence of amino acid insertions at codon 69 generally occurred in the presence of multiple TAMs, as observed in this study. The combination of TAMs plus an insertion at position 69 is associated with intermediate resistance to lamivudine (3TC) and FTC and high-level resistance to each of the remaining NRTIs (33). The E44D mutation may have an accessory role in increasing resistance to NRTIs in the presence of TAMs (34). The presence of the L74V mutation in combination with the M184V mutation is enough to get high-level resistance to both ABC and didanosine (DDI) (35).
For resistance to NNRTIs, we have reported a predominance of K103N mutations (31/67; 46.27%), followed by mutations at positions V108, Y181, and K101. A higher prevalence of the K103N mutation and resistance to NNRTIs have also been observed after 24 months of first-line ART treatment in adults in Bangui (19). The Y181C mutation confers high-level resistance to nevirapine (36). However, combined with the K103N mutation or K103N/V108I, it may confer significant cross-resistance to all of the currently approved NNRTIs (37). Several cases of combination of the K103N mutation with another mutation, such as L100I or P225H, have been reported and resulted in lower susceptibility and a poorer virologic response than the K103N mutation alone (38).
In contrast to the high rate of mutations associated with resistance to NRTIs and NNRTIs, there are only a few mutations associated with PI/r resistance. The antiretroviral regimens based on boosted PI/r might be a better alternative to NNRTI-based regimens when patient compliance is imperfect and viral load monitoring is infrequent, as in many HIV programs conducted in resource-limited settings (39). We identified mutations at 25 positions among the 57 HIV-1-infected patients who developed resistance to PI/r, with a predominance of the M46I and L76V/E mutations, each representing 24% of the total. These common mutations have been associated with a decrease in susceptibility to lopinavir (LPV)/ritonavir and indinavir (IDV)/ritonavir but an increase in susceptibility to atazanavir and saquinavir (40).
Despite the low number of genotyped samples, this study points out the problem of the high prevalence of HIV-1 drug resistance and the high rate of TAMs when viral load monitoring is not done regularly. These findings emphasize the need to improve the monitoring of ART in resource-limited settings.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Health through the National Division against AIDS and STI, the CHAIN project.
We thank all of the people who directly or indirectly contributed to the successful completion of this study.
We have no competing interests.
Footnotes
Published ahead of print 12 December 2012
REFERENCES
- 1. Diop-Ndiaye H, Toure-Kane C, Leye N, Ngom-Gueye NF, Montavon C, Peeters M, Mboup S. 2010. Antiretroviral drug resistance mutations in antiretroviral-naive patients from Senegal. AIDS Res. Hum. Retroviruses 26:1133–1138 [DOI] [PubMed] [Google Scholar]
- 2. Toure-Kane C, Montavon C, Faye MA, Gueye PM, Sow PS, Ndoye I, Gaye-Diallo A, Delaporte E, Peeters M, Mboup S. 2000. Identification of all HIV type 1 group M subtypes in Senegal, a country with low and stable seroprevalence. AIDS Res. Hum. Retroviruses 16:603–609 [DOI] [PubMed] [Google Scholar]
- 3. Hamel DJ, Sankalé JL, Eisen G, Meloni ST, Mullins C, Gueye-Ndiaye A, Mboup S, Kanki PJ. 2007. Twenty years of prospective molecular epidemiology in Senegal: changes in HIV diversity. AIDS Res. Hum. Retroviruses 23:1189–1196 [DOI] [PubMed] [Google Scholar]
- 4. Ndiaye HD, Toure-Kane C, Vidal N, Niama FR, Niang-Diallo PA, Dièye T, Gaye-Diallo A, Wade AS, Peeters M, Mboup S. 2009. Surprisingly high prevalence of subtype C and specific HIV-1 subtype/CRF distribution in men having sex with men in Senegal. J. Acquir. Immune Defic. Syndr. 52:249–252 [DOI] [PubMed] [Google Scholar]
- 5. Tournoud M, Etard JF, Ecochard R, DeGruttola V. 2010. Adherence to antiretroviral therapy, virological response, and time to resistance in the Dakar cohort. Stat. Med. 29:14–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laurent C, Diakhaté N, Gueye NF, Touré MA, Sow PS, Faye MA, Gueye M, Lanièce I, Touré Kane C, Liégeois F, Vergne L, Mboup S, Badiane S, Ndoye I, Delaporte E. 2002. The Senegalese government's highly active antiretroviral therapy initiative: an 18-month follow-up study. AIDS 16:1363–1370 [DOI] [PubMed] [Google Scholar]
- 7. World Health Organization 2010. Antiretroviral therapy for HIV infection in adults and adolescents. Recommendations for a public health approach: 2010 revision. World Health Organization, Geneva, Switzerland: http://www.who.int/hiv/pub/arv/adult2010/en/ [PubMed] [Google Scholar]
- 8. Vergne L, Kane CT, Laurent C, Diakhaté N, Gueye NF, Gueye PM, Sow PS, Faye MA, Liégeois F, Ndir A, Lanièce I, Peeters M, Ndoye I, Mboup S, Delaporte E. 2003. Low rate of genotypic HIV-1 drug-resistant strains in the Senegalese government initiative of access to antiretroviral therapy. AIDS 17:S31–38 [DOI] [PubMed] [Google Scholar]
- 9. Cozzi-Lepri A, Phillips AN, Martinez-Picado J, Monforte A, Katlama C, Eg Hansen AB, Horban A, Bruun J, Clotet B, Lundgren JD, EuroSIDA Study Group 2009. Rate of accumulation of thymidine analogue mutations in patients continuing to receive virologically failing regimens containing zidovudine or stavudine: implications for antiretroviral therapy programs in resource-limited settings. J. Infect. Dis. 200:687–697 [DOI] [PubMed] [Google Scholar]
- 10. Kantor R, Shafer RW, Follansbee S, Taylor J, Shilane D, Hurley L, Nguyen DP, Katzenstein D, Fessel WJ. 2004. Evolution of resistance to drugs in HIV-1-infected patients failing antiretroviral therapy. AIDS 18:1503–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hosseinipour MC, van Oosterhout JJ, Weigel R, Phiri S, Kamwendo D, Parkin N, Fiscus SA, Nelson JA, Eron JJ, Kumwenda J. 2009. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS 23:1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laurent C, Ngom Gueye NF, Ndour CT, Gueye PM, Diouf M, Diakhaté N, Touré Kane NC, Lanièce I, Ndir A, Vergne L, Ndoye I, Mboup S, Sow PS, Delaporte E, ANRS 1215/1290 Study Group 2005. Long-term benefits of highly active antiretroviral therapy in Senegalese HIV-1-infected adults. J. Acquir. Immune Defic. Syndr. 38:14–17 [DOI] [PubMed] [Google Scholar]
- 13. Wallis C, Mellors J, Venter W, Sanne I, Stevens W. 2009. Protease inhibitor resistance is uncommon in patients on failing second-line lopinavir/r-containing regimen in South Africa. Antivir. Ther. 14(Suppl 1):A183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 15. Vergne L, Peeters M, Mpoudi-Ngole E, Bourgeois A, Liegeois F, Toure-Kane C, Mboup S, Mulanga-Kabeya C, Saman E, Jourdan J, Reynes J, Delaporte E. 2000. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. J. Clin. Microbiol. 38:3919–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peeters M, Aghokeng AF, Delaporte E. 2010. Genetic diversity among human immunodeficiency virus-1 non-B subtypes in viral load and drug resistance assays. Clin. Microbiol. Infect. 16:1525–1531 [DOI] [PubMed] [Google Scholar]
- 17. De Beaudrap P, Etard JF, Diouf A, Ndiaye I, Ndèye GF, Sow PS, Ndèye KC, Ecochard R, Delaporte E, ANRS 1215 Study Group 2010. Incidence and determinants of new AIDS-defining illnesses after HAART initiation in a Senegalese cohort. BMC Infect. Dis. 10:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diouara AAM, Kebe K, Diop-Ndiaye H, Thiam M, Leye N, Diallo S, Thiackpe E, Diallo A, Faye M, Sakine M, Ouattara B, Diouf S, Toure-Kane C, Mboup S. 2011. Apport du papier buvard dans le génotypage du VIH-1 en milieu décentralisé au Senegal, WEAA0504. 16ème Conférence Internationale sur le Sida et les IST en Afrique, Addis Abeba, Ethiopie [Google Scholar]
- 19. Péré H, Charpentier C, Mbelesso P, Dandy M, Matta M, Moussa S, De Dieu Longo J, Grésenguet G, Abraham B, Bélec L. 2012. Virological response and resistance profiles after 24 months of first-line antiretroviral treatment in adults living in Bangui, Central African Republic. AIDS Res. Hum. Retroviruses 28:315–323 [DOI] [PubMed] [Google Scholar]
- 20. El-Khatib Z, Ekström AM, Ledwaba J, Mohapi L, Laher F, Karstaedt K, Charalambous S, Petzold M, Katzenstein K, Lynn M. 2010. Viremia and drug resistance among HIV-1 patients on antiretroviral treatment: a cross-sectional study in Soweto, South Africa. AIDS 24:1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kouanfack C, Montavon C, Laurent C, Aghokeng A, Kenfack A, Bourgeois A, Koulla-Shiro S, Mpoudi-Ngole E, Peeters M, Delaporte E. 2009. Low levels of antiretroviral-resistant HIV infection in a routine clinic in Cameroon that uses the World Health Organization (WHO) public health approach to monitor antiretroviral treatment and adequacy with the WHO recommendation for second-line treatment. Clin. Infect. Dis. 48:1318–1322 [DOI] [PubMed] [Google Scholar]
- 22. Long L, Fox M, Sanne I, Rosen S. 2010. The high cost of second-line antiretroviral therapy for HIV/AIDS in South Africa. AIDS 24:915–919 [DOI] [PubMed] [Google Scholar]
- 23. ART-LIC of leDEA Study Group, Keiser O, Tweya H, Boulle A, Braitstein P, Schecter M, Brinkhof MW, Dabis F, Tuboi S, Sprinz E, Pujades-Rodriguez M, Calmy A, Kumarasamy N, Nash D, Jahn A, MacPhail P, Lüthy R, Wood R, Egger M. 2009. Switching to second-line antiretroviral therapy in resource-limited settings: comparison of programmes with and without viral load monitoring. AIDS 23:1867–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price M, Wallis CL, Lakhi S, Karita E, Kamali A, Anzala O, Sanders EJ, Bekker LG, Twesigye R, Hunter E, Kaleebu P, Kayitenkore K, Allen S, Ruzagira E, Mwangome M, Mutua G, Amornkul PN, Stevens G, Pond SL, Schaefer M, Papathanasopoulos MA, Stevens W, Gilmour J, IAVI Early Infection Cohort Study Group 2011. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res. Hum. Retroviruses 27:5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Phillips A, Pillay D, Miners AH, Bennett DE, Gilks CF, Lundgren JD. 2008. Outcomes from monitoring of patients on antiretroviral therapy in resource-limited settings with viral load, CD4 cell count, or clinical observation alone: a computer simulation model. Lancet 371:1443–1451 [DOI] [PubMed] [Google Scholar]
- 26. Brenner BG. 2007. Resistance and viral subtypes: how important are the differences and why do they occur? Curr. Opin. HIV AIDS 2:94–102 [DOI] [PubMed] [Google Scholar]
- 27. Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. 2010. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. J. Acquir. Immune Defic. Syndr. 53:480–484 [DOI] [PubMed] [Google Scholar]
- 28. Marconi VC, Sunpath H, Lu Z, Gordon M, Koranteng-Apeagyei K, Hampton J, Carpenter S, Giddy J, Ross D, Holst H, Losina E, Walker BD, Kuritzkes DR, South Africa Resistance Cohort Study Team 2008. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin. Infect. Dis. 46:1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2002. The M184V mutation reduces the selective excision of zidovudine 5′-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 76:3248–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Katlama C, Clotet B, Plettenberg A, Jost J, Arasteh K, Bernasconi E, Jeantils V, Cutrell A, Stone C, Ait-Khaled M, Purdon S. 2000. The role of abacavir (ABC, 1592) in antiretroviral therapy-experienced patients: results from a randomized, double-blind, trial. CNA3002 European Study Team. AIDS 14:781–789 [DOI] [PubMed] [Google Scholar]
- 31. Larder BA, Kemp SD, Harrigan PR. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696–699 [DOI] [PubMed] [Google Scholar]
- 32. Ait-Khaled M, Rakik A, Griffin P, Cutrell A, Fischl MA, Clumeck N, Greenberg SB, Rubio R, Peters BS, Pulido F, Gould J, Pearce G, Spreen W, Tisdale M, Lafon S, CNA3003 International Study Team 2002. Mutations in HIV-1 reverse transcriptase during therapy with abacavir, lamivudine and zidovudine in HIV-1-infected adults with no prior antiretroviral therapy. Antivir. Ther. 7:43–51 [PubMed] [Google Scholar]
- 33. Prado JG, Franco S, Matamoros T, Ruiz L, Clotet B, Menéndez-Arias L, Martínez MA, Martinez-Picado J. 2004. Relative replication fitness of multi-nucleoside analogue-resistant HIV-1 strains bearing a dipeptide insertion in the fingers subdomain of the reverse transcriptase and mutations at codons 67 and 215. Virology 326:103–112 [DOI] [PubMed] [Google Scholar]
- 34. Romano L, Venturi G, Bloor S, Harrigan R, Larder BA, Major JC, Zazzi M. 2002. Broad nucleoside-analogue resistance implications for human immunodeficiency virus type 1 reverse-transcriptase mutations at codons 44 and 118. J. Infect. Dis. 185:898–904 [DOI] [PubMed] [Google Scholar]
- 35. Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. 2003. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J. Infect. Dis. 188:992–1000 [DOI] [PubMed] [Google Scholar]
- 36. Casado JL, Hertogs K, Ruiz L, Dronda F, Van Cauwenberge A, Arnó A, Garcia-Arata I, Bloor S, Bonjoch A, Blazquez J, Clotet B, Larder B. 2000. Non-nucleoside reverse transcriptase inhibitor resistance among patients failing a nevirapine plus protease inhibitor-containing regimen. AIDS 14:F1–7 [DOI] [PubMed] [Google Scholar]
- 37. Bacheler L, Jeffrey S, Hanna G, D'Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, Baker D, Gallagher K, Scarnati H, Tritch R, Rizzo C. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scherrer AU, Hasse B, von Wyl V, Yerly S, Böni J, Bürgisser P, Klimkait T, Bucher HC, Ledergerber B, Günthard HF. 2009. Prevalence of etravirine mutations and impact on response to treatment in routine clinical care: the Swiss HIV Cohort Study (SHCS). HIV Med. 10:647–656 [DOI] [PubMed] [Google Scholar]
- 39. Adlington R, Richens J, Shahmanesh M. 2009. First-line antiretroviral therapy in resource-limited settings: time to reconsider? J. Infect. Dis. 199:1407–1408 [DOI] [PubMed] [Google Scholar]
- 40. Young TP, Parkin NT, Stawiski E, Pilot-Matias T, Trinh R, Kempf DJ, Norton M. 2010. Prevalence, mutation patterns, and effects on protease inhibitor susceptibility of the L76V mutation in HIV-1 protease. Antimicrob. Agents Chemother. 54:4903–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]


