LETTER
Beginning in 2007, we began noticing a high level (∼80%) of resistance to ampicillin and co-trimoxazole among Gram-negative urinary tract isolates from inpatients at the Komfo Anokye Teaching Hospital (KATH) in Kumasi, Ghana (1). Also, we noticed resistance to expanded-spectrum cephalosporins among urinary tract isolates from outpatients in the community. To investigate whether specific pathogenic genotypes were associated with resistance, we characterized 156 Escherichia coli isolates from blood, urine, sputum, and wound swab specimens as well as infected body site aspirates (collected February to April 2008 and March to July 2009). This study was approved by the Joint Committee on Human Research Publications and Ethics of the School of Medical Sciences at the Kwame Nkrumah University of Science and Technology, only patients with a diagnosed infection were considered, and only a single isolate per patient was obtained.
Over half of all isolates were resistant to amoxicillin-clavulanic acid (67%), ampicillin (92%), cefpodoxime (65%), cefuroxime (58%), gentamicin (56%), nalidixic acid (62%), co-trimoxazole (90%), and chloramphenicol (76%), and roughly half (49%) carried blaTEM, blaSHV, blaCTX-M, or some combination of these genes. Extended-spectrum β-lactamase (ESBL) production was found in 77 of the 156 (49.4%) isolates and was significantly associated (chi-square test; P = 0.009) with nosocomial cases (53 of 91 isolates; 58.2%) compared to outpatient cases (24 of 65 isolates; 36.9%) but was not associated with patient age, gender, or the source of the clinical sample (P = 0.101).
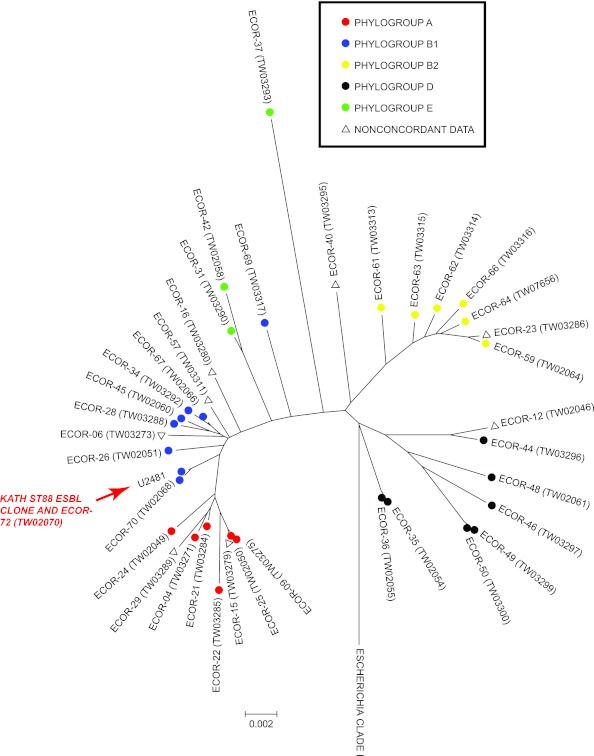
Due to the widespread dissemination of the ESBL-producing extraintestinal pathogenic E. coli (ExPEC) clone, identified by multilocus sequence typing (MLST) as sequence type 131 (ST131) (2, 3), we selected 29 ESBL-producing E. coli isolates at random for MLST. All isolates were identical and belonged to a previously identified sequence type, ST88, according to the STEC Center database (http://www.shigatox.net). ST88 belongs to the E. coli B1 phylogroup (Fig. 1) and includes a pyelonephritis strain, E. coli reference (ECOR) strain 72 (ECOR-72). Screening for the presence of 37 virulence genes (4–6) confirmed the presence of fimbrial genes (c1936, fimA, ppdD, and yehA) previously shown to be associated with attachment and virulence in the urinary tract (4, 6).
Fig 1.
Phylogenetic relationships among isolates of the ECOR collection and the ESBL-producing ST88 clone from KATH (red arrow). Isolates are labeled according to ECOR numbers and STEC Center identification numbers (“TW”). Colors are used to indicate E. coli phylogroup designations, and only unique STs are shown (i.e., other ECOR representatives of the same ST are not shown). Inconsistencies between phylogroup designations between studies (11, 12) are labeled for reference with open triangles. A representative of the closest common ancestor to E. coli (Escherichia clade I) was used as an outgroup in the dendrogram (13).
ECOR-72 has been circulating in humans for some time (7, 8) and was shown to be sensitive to 14 antibiotics (9). Susceptibility testing (Vitek 2; bioMérieux Inc., Durham, NC) of 22 randomly selected ST88 isolates from this study, however, indicated that all were resistant to ampicillin and ampicillin-clavulanic acid as well as narrow-spectrum and expanded-spectrum cephalosporins (cefazolin and ceftriaxone), 2 of the 3 aminoglycosides (gentamicin and tobramycin), and, with a single exception, fluoroquinolones (ciprofloxacin and moxifloxacin).
There is growing concern over the emergence of non-ST131, fluoroquinolone-resistant (FQr) ExPEC (10). Given that all ST88 KATH isolates tested but one were resistant to both ciprofloxacin and moxifloxacin, it seems they may be an important reservoir of fluoroquinolone resistance genes. More data are needed to determine the directionality of these events and whether resistant ST88 ExPEC strains are common in other regions. Regardless, the widespread availability of antibiotics without prescription in the Kumasi area may have contributed to the fact that ∼37% of all community-acquired and ∼60% of all nosocomial, clinical ExPEC isolates at our center are now ESBL-producing ST88.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (United States) grant 1K01AI09728101A1 (to S.T.W.), by National Research Service Awards grant HL07749-15 (to S.T.W.), and by the University of Michigan (to C.X.).
In 2010 to 2011, P. Feglo was a visiting scholar with the University of Michigan African Presidential Scholars Program (UMAPS). The UMAPS program is administered by the African Studies Center and funded through the Office of the President and the Department of Afroamerican and African Studies. The African Studies Center is part of The International Institute.
Footnotes
Published ahead of print 12 December 2012
REFERENCES
- 1. Feglo P. 2007. Antimicrobial sensitivity of urine isolates at Komfo Anokye Teaching Hospital (KATH) Kumasi 2000-2005. Ghana J. Allied Health Sci. 1:43–49 [Google Scholar]
- 2. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 3. Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 4. Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HL. 2011. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect. Immun. 79:4753–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tobias J, Vutukuru SR. 2012. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol. Res. 167:564–570 [DOI] [PubMed] [Google Scholar]
- 6. Vigil PD, Stapleton AE, Johnson JR, Hooton TM, Hodges AP, He Y, Mobley HL. 2011. Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio. 2(3):e00066–11 doi:10.1128/mBio.00066-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caugant DA, Levin BR, Lidin-Janson G, Whittam TS, Svanborg Eden C, Selander RK. 1983. Genetic diversity and relationships among strains of Escherichia coli in the intestine and those causing urinary tract infections. Prog. Allergy 33:203–227 [DOI] [PubMed] [Google Scholar]
- 8. Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazel D, Dychinco B, Webb VA, Davies J. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Platell JL, Trott DJ, Johnson JR, Heisig P, Heisig A, Clabots CR, Johnston B, Cobbold RN. 2012. Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob. Agents Chemother. 56:3898–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herzer PJ, Inouye S, Inouye M, Whittam TS. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75:6534–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]