Abstract
Mycoplasma agalactiae isolates from Spain were genetically characterized to investigate their genomic diversity and to better understand their relationship to isolates from other countries. Molecular typing revealed a high genomic homogeneity in Spanish M. agalactiae isolates, which clearly shows the circulation of one endemic clonal population.
TEXT
Mycoplasma agalactiae is the main etiologic agent of contagious agalactia (CA), a serious syndrome affecting small ruminants; the World Organization for Animal Health must be notified of its occurrence because of its high economic significance worldwide. The first genomic studies showed little genomic diversity within M. agalactiae species, apart from that provided by antigenic variation (1, 2). Recently, the development of new sequence-based typing systems has revealed more genetic heterogeneity than previously thought (3–5). To investigate the genomic diversity of Spanish M. agalactiae isolates and to elucidate their relationship to those from other geographic areas, we analyzed isolates from Spain using pulsed-field gel electrophoresis (PFGE), which has been demonstrated to be robust and discriminative for typing different species of mycoplasmas (6–9), including M. agalactiae (3, 10); we also used the most recently developed sequence-based typing techniques, such as multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) (3) and multilocus sequence typing (MLST) (4). Typing systems were selected to obtain a comprehensive approach to the genomic diversity of Spanish M. agalactiae isolates and also to generate suitable data for evolutionary and population studies. Knowledge of the diversity and distribution of M. agalactiae clones will facilitate tracing the source of new international outbreaks as well as contribute to a better understanding of M. agalactiae population genetics and evolution.
Four hundred ten M. agalactiae isolates collected from 171 Spanish sheep flocks from 2008 through 2010, type strain PG2 (Institut Pasteur, Paris, France), and strain Teramo (Mycoplasma Experience, Ltd., Reigate, United Kingdom) were subjected to extensive genomic characterization (see Table S1 in the supplemental material). All the information regarding the sampling and the isolation procedure is detailed in the work of Ariza-Miguel et al. (11). The species designation of the isolates was confirmed by real-time PCR targeting the p40 gene (12, 13). All isolates were analyzed by PFGE with the restriction enzyme SmaI and by MLVA at 4 highly variable VNTR loci (i.e., MagaI VNTR 5, MagaI VNTR 14, MagaI VNTR 17, and MagaI VNTR 19) as previously described (3). MLST analyses (4) were carried out on a subset of 48 field isolates which showed different genomic profiles in the previous analyses, as well as on isolates from different geographic origins and times of isolation selected to yield the highest genetic variability (Table 1). A neighbor-joining dendrogram showing relatedness among isolates on the basis of their MLST allelic profiles was constructed by using BioNumerics v.6.6 software, and BURST analysis was performed with eBURST v3 (http://eburst.mlst.net/). Information about the isolates analyzed, as well as a new allelic profile, was submitted to the PubMLST database (http://pubmlst.org/magalactiae/).
Table 1.
MLST results at 5 housekeeping loci obtained with a subset of 48 Spanish Mycoplasma agalactiae field isolates and strains PG2 and Teramo from 2008 to 2010a
| Isolate | Province of origin | Yr of isolation | PFGE profile | MLST allelic profile for: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| dnaA | gltX | gyrB | metS | tufA | ST | ||||
| 24a | Salamanca | 2008 | II | 1 | 1 | 2 | 2 | 2 | 5 |
| 26a | Zamora | 2008 | II | 1 | 1 | 2 | 2 | 2 | 5 |
| 262a | León | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 276a | Valladolid | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 283a | Palencia | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 286a | Cantabria | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 286b | Cantabria | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 286c | Cantabria | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 287c | Burgos | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 423d | Zamora | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 472d | Salamanca | 2008 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 513a | Salamanca | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 651a | Zamora | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 653a1 | Zamora | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 653a2 | Zamora | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 657c | Valladolid | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 787a | Valladolid | 2009 | IV | 1 | 1 | 2 | 2 | 2 | 5 |
| 787b | Valladolid | 2009 | IV | 1 | 1 | 2 | 2 | 2 | 5 |
| 787c | Valladolid | 2009 | IV | 1 | 1 | 2 | 2 | 2 | 5 |
| 793a | Valladolid | 2009 | V | 1 | 1 | 2 | 2 | 2 | 5 |
| 793b | Valladolid | 2009 | V | 1 | 1 | 2 | 2 | 2 | 5 |
| 799a | Valladolid | 2009 | V | 1 | 1 | 2 | 2 | 2 | 5 |
| 799b | Valladolid | 2009 | V | 1 | 1 | 2 | 2 | 2 | 5 |
| 799c | Valladolid | 2009 | V | 1 | 1 | 2 | 2 | 2 | 5 |
| 1021a | Palencia | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1026b | Zamora | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1032b | León | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1033a | León | 2009 | III | 1 | 1 | 2 | 2 | 2 | 5 |
| 1033b | León | 2009 | III | 1 | 1 | 2 | 2 | 2 | 5 |
| 1033c | León | 2009 | III | 1 | 1 | 2 | 2 | 2 | 5 |
| 1033d | León | 2009 | III | 1 | 1 | 2 | 2 | 2 | 5 |
| 1033e | León | 2009 | III | 1 | 1 | 2 | 2 | 2 | 5 |
| 1043a | Valladolid | 2009 | I | 1 | 6 | 2 | 2 | 2 | 18 |
| 1058b | Valladolid | 2009 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1086a | León | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1114a | Segovia | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1132a | Segovia | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1160a | Palencia | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1423a | Valladolid | 2010 | VI | 1 | 6 | 2 | 2 | 2 | 18 |
| 1423c | Valladolid | 2010 | VI | 1 | 6 | 2 | 2 | 2 | 18 |
| 1506a | Valladolid | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1668a | Zamora | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| 1680a | Burgos | 2010 | I | 1 | 6 | 2 | 2 | 2 | 18 |
| 1700a | León | 2010 | II | 1 | 1 | 2 | 2 | 2 | 5 |
| 1700b | León | 2010 | II | 1 | 1 | 2 | 2 | 2 | 5 |
| 1700c | León | 2010 | II | 1 | 1 | 2 | 2 | 2 | 5 |
| 1703a | Valladolid | 2010 | II | 1 | 1 | 2 | 2 | 2 | 5 |
| 1704b | Salamanca | 2010 | I | 1 | 1 | 2 | 2 | 2 | 5 |
| Teramo | Italy | Unknown | IV | 1 | 1 | 1 | 1 | 1 | 1 |
| PG2 | Spain | 1959 | IV | 1 | 1 | 1 | 1 | 1 | 1 |
PFGE, pulsed-field gel electrophoresis; MLST, multilocus sequence typing; ST, sequence type.
We detected a high genomic homogeneity in M. agalactiae isolates from Spain using three different genotyping tools (i.e., PFGE, MLVA, and MLST). PFGE provided the highest discriminative power and was capable of distinguishing between some isolates which were largely indistinguishable by MLVA or MLST (see Table S1 in the supplemental material). Genomic characterization by PFGE identified 6 different pulsotypes which were closely related and showed very similar fingerprint patterns, with only small size differences in one band among pulsotypes (Table 2). Ninety-five percent of the isolates belonged to the same genomic profile, named pulsotype I, resulting in a Simpson's index of diversity of 0.104. Pulsotype I was found widely distributed in all the provinces sampled; it was the pulsotype of from 87% to 99% of the isolates analyzed per province. The rest of the genomic profiles were found disseminated in 3 neighboring provinces (Table 3). PG2 and Teramo strains belonged to pulsotype IV and clustered with 3 field isolates. Surprisingly, genetic profiles obtained by MLVA were largely indistinguishable, with all field isolates showing the same genetic profile at the 4 highly variable VNTR loci. Moreover, only MLVA at the MagaI VNTR 17 locus was capable of distinguishing between field isolates showing a band at 285 bp and between PG2 and Teramo strains showing a band at 169 bp. The Simpson's index of diversity was determined to be 0.005. Finally, MLST analyses of the 48 field isolates revealed 2 different sequence types (STs). Forty-four out of 48 field isolates (92%) belonged to ST 5 (allelic profile 11222). The other ST was not described at that moment and, after its submission to the PubMLST database, was designated ST 18 (allelic profile 16222) (Table 1).
Table 2.
DNA restriction fragments of 410 Mycoplasma agalactiae isolates generated by PFGE with restriction enzyme SmaI in Spain from 2008 to 2010a
| DNA fragment | Size (kbp) for pulsotype: |
|||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |
| A | 427 | 427 | 454 | 427 | 427 | 427 |
| B | 159 | 159 | 159 | 159 | 159 | 197 |
| C | 124 | 124 | 124 | 124 | 124 | 124 |
| D | 83 | 92 | 83 | 83 | 108 | 83 |
| E | 56 | 56 | 56 | 67 | 56 | 56 |
| F | 9 | 9 | 9 | 9 | 9 | 9 |
| G | 5 | 5 | 5 | 5 | 5 | 5 |
| Genome size | 863 | 872 | 890 | 874 | 888 | 901 |
All the values in the table are expressed in kbp. Boldface indicates restriction fragments showing size differences from those obtained for pulsotype I, which was the most frequently isolated (95% of isolates).
Table 3.
Spatial distribution of PFGE genomic profiles obtained for 410 Spanish Mycoplasma agalactiae field isolates from 2008 to 2010
| Province of origin | PFGE pattern | No. of isolates | Total no. of isolates/province | % of isolates with PFGE pattern/province |
|---|---|---|---|---|
| Burgos | I | 23 | 23 | 100 |
| Cantabria | I | 3 | 3 | 100 |
| León | I | 55 | 63 | 87.3 |
| II | 3 | 4.8 | ||
| III | 5 | 7.9 | ||
| Palencia | I | 56 | 56 | 100 |
| Salamanca | I | 28 | 29 | 96.6 |
| II | 1 | 3.4 | ||
| Segovia | I | 2 | 2 | 100 |
| Valladolid | I | 115 | 127 | 90.6 |
| II | 1 | 0.8 | ||
| IV | 3 | 2.4 | ||
| V | 5 | 3.9 | ||
| VI | 3 | 2.4 | ||
| Zamora | I | 106 | 107 | 99.1 |
| II | 1 | 0.9 |
Overall, molecular typing revealed a high genomic homogeneity in Spanish M. agalactiae isolates, which clearly show the circulation of one endemic clonal population. A similar finding was recently observed in the French western Pyrenees region by Nouvel et al. (5), who reported that the endemic CA repeatedly observed over the past 30 years in that region has been caused by a unique subtype of M. agalactiae. MLVA placed all the French isolates in the same genotype, designated ST 10. Interestingly, all 410 Spanish M. agalactiae isolates analyzed in this study were placed in the same MLVA type, suggesting that the same highly successfully adapted strain has been circulating in Spain and France during the last 2 years. To obtain further information about the endemic clone, a representative isolate, namely, 1668a, was fully sequenced, and future studies will help to clarify the molecular mechanisms involved in the evolutionary success of this clone as well as to provide new insights into the genomic diversity and evolution of the species. In contrast, several studies have reported an unexpectedly high diversity in M. agalactiae Spanish isolates recovered from goats (3, 4, 14). Further studies are necessary to test whether this is because various CA-causing mycoplasmas have been detected in Spanish goat herds (15–17), while M. agalactiae has been the only species detected in sheep, limiting the possibility of genetic exchange (11).
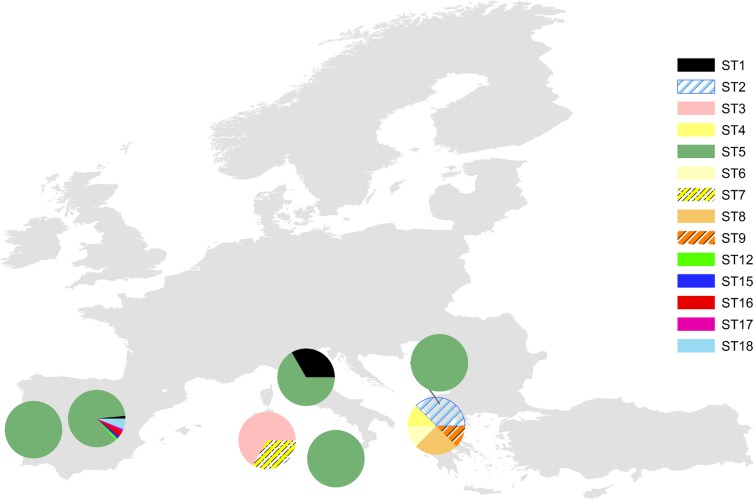
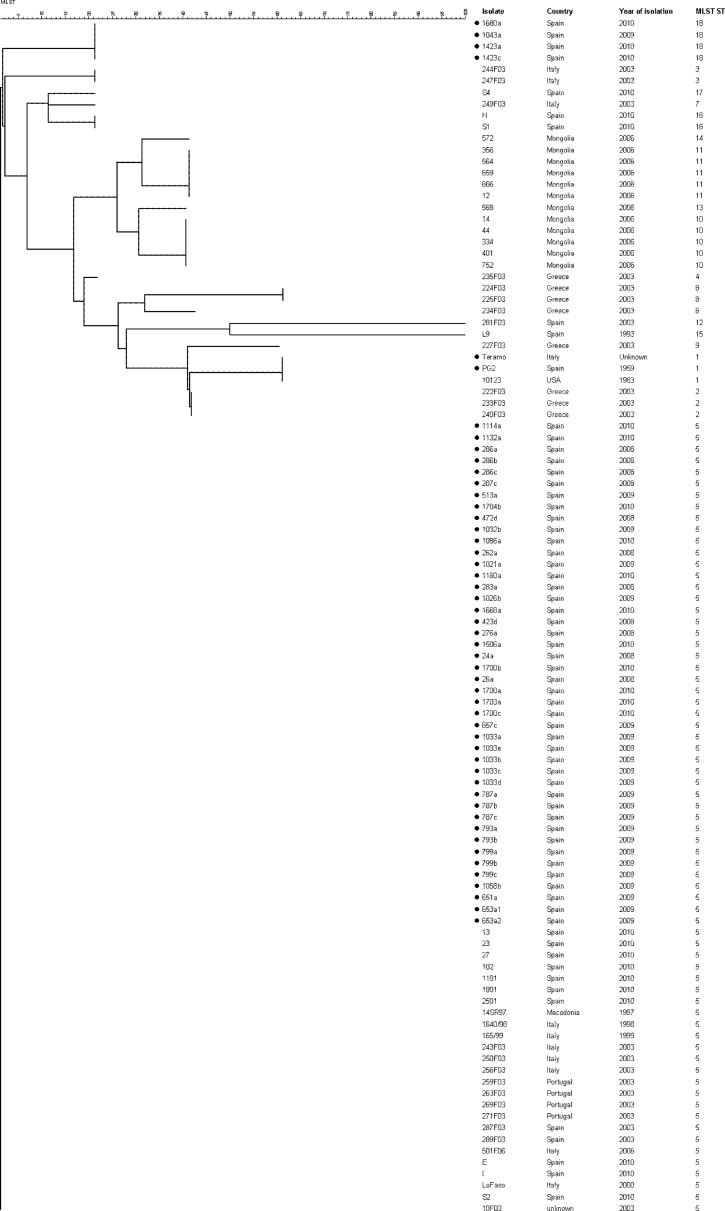
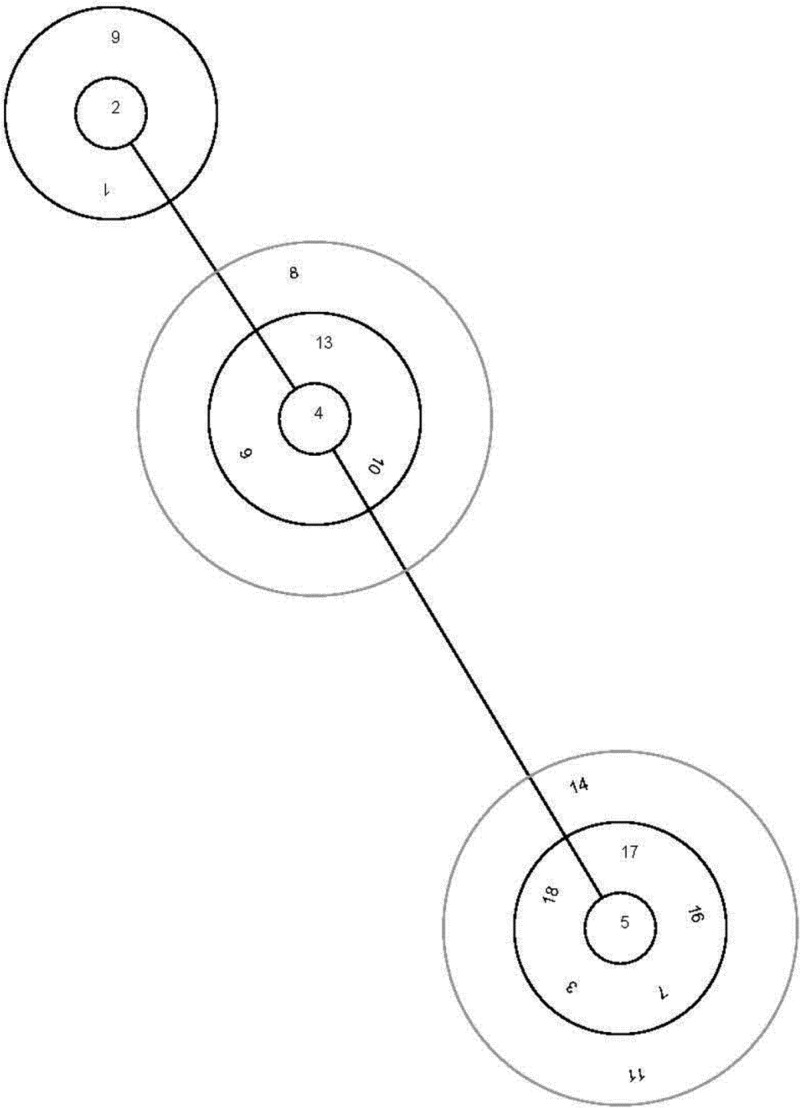
The global relationship of M. agalactiae clones on the basis of available MLST allelic profiles also showed a high genetic homogeneity, with isolates belonging to ST 5 being widely distributed throughout many southern European countries analyzed so far (Fig. 1) (4). The 44 Spanish field isolates belonging to ST 5 examined in this study clustered with previously analyzed isolates from Spain and other southern European countries: Portugal, Italy (including Sicily and Sardinia), and Macedonia. The other 4 field isolates belonging to novel ST 18 clustered closely with the previous one, forming a new branch (Fig. 2). Interestingly, strain Teramo (Italy) and type strain PG2 (Spain) clustered with strain 10123 from the United States, suggesting an evolutionary relationship. We hypothesize that a highly adaptive genotype may have increased rapidly in frequency to produce an epidemic clone in southern Europe. Then, that clone diversified though recombination or mutation to produce minor clonal variants (18). BURST analysis supports this hypothesis, since it defined ST 5 as the adaptive ancestral genotype from which the minor clonal variants have arisen (Fig. 3). Further investigations are necessary to test that hypothesis, and the inclusion of new isolates from other geographic areas and times of isolation will help to clarify the evolution of this pathogen and its current population structure.
Fig 1.
Geographic distribution of 103 European Mycoplasma agalactiae isolates based upon their multilocus sequence typing allelic profiles. Isolates previously subjected to MLST have been included (2). Locations of pie charts indicate the geographic origins of the isolates, and their color reflects the different sequence types (STs).
Fig 2.
Genetic relationships among 104 worldwide Mycoplasma agalactiae isolates based upon allelic differences at 5 housekeeping loci. Name of isolates, years of isolation, countries of origin, and sequence types (MLST) are specified to the right of the branches. Isolates previously analyzed were added to the study (2). Black dots indicate the isolates analyzed in this study. The dendrogram was produced by using the neighbor-joining method of the BioNumerics v.6.6 software.
Fig 3.
BURST analysis of 104 worldwide Mycoplasma agalactiae isolates based upon their multilocus sequence typing allelic profiles. Clonal complexes were defined as groups of multilocus genotypes in which every genetic profile shared at least 3 out of 5 loci with at least one other member of the group. Sequence type 5 was found to be the ancestral genotype. European isolates previously analyzed were added to the study (2).
In conclusion, this study provides a genomic characterization of M. agalactiae in Spain and contributes to the better understanding of the global distribution of clones. Molecular typing revealed a high genomic homogeneity in Spanish M. agalactiae isolates, which clearly show the circulation of one endemic clonal population, and will facilitate the design of prophylactic measures.
ACKNOWLEDGMENTS
This work was supported by project RTA 2008-073 of the Spanish Ministry of Education and Science, Government of Spain.
We thank Nigel Cook (FERA, United Kingdom) for critical revision of the manuscript.
Footnotes
Published ahead of print 5 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02835-12.
REFERENCES
- 1. Solsona M, Lambert M, Poumarat F. 1996. Genomic, protein homogeneity and antigenic variability of Mycoplasma agalactiae. Vet. Microbiol. 50:45–58 [DOI] [PubMed] [Google Scholar]
- 2. Tola S, Idini G, Manunta D, Casciano I, Rocchigiani AM, Angioi A, Leori G. 1996. Comparison of Mycoplasma agalactiae isolates by pulsed field gel electrophoresis, SDS-PAGE and immunoblotting. FEMS Microbiol. Lett. 143:259–265 [DOI] [PubMed] [Google Scholar]
- 3. McAuliffe L, Churchward CP, Lawes JR, Loria G, Ayling RD, Nicholas RAJ. 2008. VNTR analysis reveals unexpected genetic diversity within Mycoplasma agalactiae, the main causative agent of contagious agalactia. BMC Microbiol. 8:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McAuliffe L, Gosney F, Hlusek M, de Garnica ML, Spergser J, Kargl M, Rosengarten R, Ayling RD. 2011. Multilocus sequence typing of Mycoplasma agalactiae. J. Med. Microbiol. 60:803–811 [DOI] [PubMed] [Google Scholar]
- 5. Nouvel LX, Marenda MS, Glew MD, Sagné E, Giammarinaro P, Tardy F, Poumarat F, Rosengarten R, Citti C. 2012. Molecular typing of Mycoplasma agalactiae: tracing European-wide genetic diversity and an endemic clonal population. Comp. Immunol. Microbiol. Infect. Dis. 35:487–496 [DOI] [PubMed] [Google Scholar]
- 6. Arcangioli MA, Aslan H, Tardy F, Poumarat F, Le Grand D. 2012. The use of pulsed-field gel electrophoresis to investigate the epidemiology of Mycoplasma bovis in French calf feedlots. Vet. J. 192:96–100 [DOI] [PubMed] [Google Scholar]
- 7. Marois C, Dufour-Gesbert F, Kempf I. 2001. Comparison of pulsed-field gel electrophoresis with random amplified polymorphic DNA for typing of Mycoplasma synoviae. Vet. Microbiol. 79:1–9 [DOI] [PubMed] [Google Scholar]
- 8. McAuliffe L, Kokotovic B, Ayling RD, Nicholas RAJ. 2004. Molecular epidemiological analysis of Mycoplasma bovis isolates from the United Kingdom shows two genetically distinct clusters. J. Clin. Microbiol. 42:4556–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tardy F, Mercier P, Solsona M, Saras E, Poumarat F. 2007. Mycoplasma mycoides subsp. mycoides biotype large colony isolates from healthy and diseased goats: prevalence and typing. Vet. Microbiol. 121:268–277 [DOI] [PubMed] [Google Scholar]
- 10. Tola S, Idini G, Rocchigiani AM, Manunta D, Angioi PP, Rocca S, Cocco M, Leori G. 1999. Comparison of restriction pattern polymorphism of Mycoplasma agalactiae and Mycoplasma bovis by pulsed field gel electrophoresis J. Vet. Med. B Infect. Dis. Vet. Public Health 46:199–206 [DOI] [PubMed] [Google Scholar]
- 11. Ariza-Miguel J, Rodríguez-Lázaro D, Hernández M. 2012. A survey of Mycoplasma agalactiae in dairy sheep farms in Spain. BMC Vet. Res. 8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleury B, Bergonier D, Berthelot X, Peterhans E, Frey J, Vilei EM. 2002. Characterization of P40, a cytadhesin of Mycoplasma agalactiae. Infect. Immun. 70:5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oravcová K, López-Enríquez L, Rodríguez-Lázaro D, Hernández M. 2009. Mycoplasma agalactiae p40 gene, a novel marker for diagnosis of contagious agalactia in sheep by real-time PCR: assessment of analytical performance and in-house validation using naturally contaminated milk samples. J. Clin. Microbiol. 47:445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De la Fe C, Amores J, Tardy F, Sagne E, Nouvel LX, Citti C. 2012. Unexpected genetic diversity of Mycoplasma agalactiae caprine isolates from an endemic geographically restricted area of Spain. BMC Vet. Res. 8:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corrales JC, Esnal A, De la Fe C, Sánchez A, Assunçao P, Poveda JB, Contreras A. 2007. Contagious agalactia in small ruminants. Small Rumin. Res. 68:154–166 [Google Scholar]
- 16. De la Fe C, Gutiérrez A, Poveda JB, Assunção P, Ramírez AS, Fabelo F. 2007. First isolation of Mycoplasma capricolum subsp. capricolum, one of the causal agents of caprine contagious agalactia, on the island of Lanzarote (Spain). Vet. J. 173:440–442 [DOI] [PubMed] [Google Scholar]
- 17. Gil MC, Peña FJ, Hermoso de Mendoza J, Gomez L. 2003. Genital lesions in an outbreak of caprine contagious agalactia caused by Mycoplasma agalactiae and Mycoplasma putrefaciens. J. Vet. Med. B Infect. Dis. Vet. Public Health 50:484–487 [DOI] [PubMed] [Google Scholar]
- 18. Smith JM, Smith NH, O'Rourke M, Spratt BG. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]