Abstract
An outbreak of multidrug-resistant Pseudomonas aeruginosa (MDRPA) infections in a university hospital is described. Phenotypic and genotypic analysis of 240 isolates revealed that 152 patients, mainly in the intensive care unit (ICU), were colonized or infected with MDRPA, the majority with O11. All metallo-β-lactamase (MBL)-positive isolates carried the blaVIM-2 or blaVIM-1 gene. One or more type III secretion system toxin genes were detected in most isolates. Five dominant pulsed-field gel electrophoresis (PFGE) types were characterized, associated with ST235, ST111, ST253, ST309, and ST639.
TEXT
Pseudomonas aeruginosa is an opportunistic pathogen causing severe invasive disease in critically ill and immunocompromised patients. Because of its ubiquitous nature, ability to survive in moist environments, and innate resistance to many antibiotics and antiseptics, it constitutes a common pathogen in hospitals, particularly in intensive care units (ICUs). Its treatment is a therapeutic challenge because of the intrinsic resistance and the ability to easily acquire resistance determinants (1). Multidrug-resistant P. aeruginosa (MDRPA) infections occur mainly in ICU patients (2). The prevalence and epidemiology of MDRPA have become the focus of numerous single- and multicenter surveillance studies (3). The large number of secreted and cell-associated virulence factors is implicated in the pathogenesis of severe infections. The type III secretion system (TTSS), a complex of three proteins, is associated with lung injury, sepsis, and a 6-fold-greater risk of mortality, constituting an important virulence determinant (4, 5). Results on emergence and spread of MDRPA isolates in the University Hospital of Patras (UHP) and their phenotypic and genotypic characteristics are presented in this study.
During a 2-year period, a total of 952 P. aeruginosa isolates were recovered from 430 patients hospitalized in our tertiary-care hospital, located in southwestern Greece, with 700 acute-care beds and about 100,000 admissions annually. Two hundred and forty, the first 10 from every month with no replicate isolates (one isolate per patient), from different wards and a variety of clinical specimens, including true infections and carriage, were selected for further study. Colonizing isolates were recovered from stool and respiratory tract specimens from patients without signs of infection.
P. aeruginosa was identified by standard methods. Colonizing (83) and infection-related (157) isolates were compared for their phenotypes and genotypes. For further analyses, isolates were divided into two groups: those recovered from ICU patients (92) and those from non-ICU patients (148).
Antibiotic susceptibility testing was performed by the agar disk diffusion method against antipseudomonal agents according to CLSI guidelines (6). All isolates resistant to at least three classes of antibiotics were defined as MDRPA (7). The MIC of colistin was determined by the Etest (AB Biodisk, Solna, Sweden). All imipenem-nonsusceptible (IMP-NS) isolates (MICs > 1 mg/liter) were examined for metallo-β-lactamase (MBL) production using the Etest MBL assay (AB Biodisk).
Serotyping was performed by 16 monovalent antisera (Bio-Rad, Marne's-la-Coquette, France) as previously described (8).
Among the IMP-NS isolates, the blaVIM gene was detected using the multiplex PCR-enzyme-linked immunosorbent assay (ELISA) system (hyplex MBL ID PCR module Hyb-module test system; BAG Health Care, Lish, Germany) (9). Types of blaVIM genes were identified by sequencing analysis among selected blaVIM-positive strains after comparison with a data bank (http://blast.ncbi.nlm.nih.gov/Blast.cgi). In all isolates, TTSS genes (exoS, exoT, exoU, exoY) were investigated by PCR (10).
Clones were defined by pulsed-field gel electrophoresis (PFGE) of speI (Roche, Penzberg, Germany) DNA digests. Banding patterns were compared by Fingerprinting II Informatics Software (Bio-Rad, Berkeley, California), and clones were defined according to already established criteria (11). A dendrogram comparing molecular weights of strains' DNA fragments to those from a previous collection by using FPQuest software (Bio-Rad; catalog number 1709300) was computed. Clustering was based on ≥75% similarity. Selected strains of the main PFGE types were characterized by multilocus sequence typing (MLST) (http://pubmlst.org/paeruginosa).
Pearson's chi-square test was used to evaluate the differences in the frequencies of variables in ICU and non-ICU wards, conducted by PASW Statistics 18, release version 18.0.0 (SPSS, Inc., Chicago, IL; www.spss.com). Results were considered significant at a P value of ≤0.05.
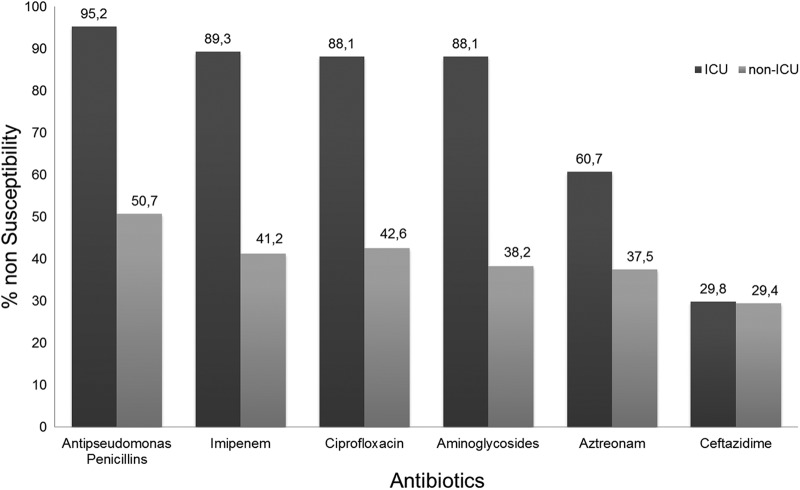
A comparison of nonsusceptibility to antibiotics between ICU and non-ICU P. aeruginosa isolates is presented in Fig. 1. All isolates were susceptible to colistin (MICs ≤ 2 mg/liter). During the study period, a high frequency of MDRPA was detected (152/240 isolates; 63.33%), mainly in ICU isolates (Table 1). There were no statistically significant differences in antibiotic resistance patterns between infecting and colonizing isolates. A blaVIM gene was detected in all 49 MBL-positive isolates. No statistically significant difference was observed between ICU and non-ICU isolates (17.4% versus 22.3%, respectively) (Table 1). The majority of the isolates carried blaVIM-2, while only three strains carried the blaVIM-1 gene.
Fig 1.
Percentages of P. aeruginosa isolates nonsusceptible to various classes of antibiotics among ICU and non-ICU patients. Antipseudomonas penicillins include azlocillin, carbenicillin, piperacillin, and ticarcillin-clavulanic acid. Aminoglycosides include amikacin, netilmicin, and tobramycin.
Table 1.
Characteristics of ICU and non-ICU P. aeruginosa isolatesa
| Characteristic | No. (%) of isolates with each characteristic |
P value | |
|---|---|---|---|
| ICU isolates (n = 92) | Non-ICU isolates (n = 148) | ||
| MDRPA | 84 (91.3) | 68 (46.0) | <0.001 |
| MBL positive | 16 (17.4) | 33 (22.3) | 0.359 |
| Serotype O11 | 68 (73.9) | 50 (33.8) | <0.001 |
| blaVIM | 16 (17.4) | 33 (22.3) | 0.359 |
| exoS | 25 (27.2) | 86 (58.1) | <0.001 |
| exoT | 76 (82.6) | 116 (78.4) | 0.427 |
| exoU | 83 (90.2) | 118 (79.7) | 0.032 |
| exoY | 81 (88.0) | 125 (84.5) | 0.438 |
| Clone a (ST235) | 59 (64.1) | 22 (14.9) | <0.001 |
| Clone d (ST235) | 15 (16.3) | 18 (12.2) | 0.365 |
| Clone b (ST111 or ST235) | 3 (3.3) | 7 (4.3) | 0.745 |
| Clone c (ST253) | 1 (1.0) | 5 (3.4) | 0.410 |
| Clone s (ST309 or ST639) | 0 (0.0) | 6 (4.0) | 0.084 |
| All other clones | 14 (15.2) | 90 (60.8) | <0.001 |
ICU, intensive care unit; MDRPA, multidrug-resistant P. aeruginosa; MBL, metallo-β-lactamase.
Serogroup O11 predominated in the ICU compared to all other wards (Table 1) as well as among MDRPA isolates (75% [63/84] in the ICU and 57.4% [39/68] in non-ICU wards [P = 0.021]).
The majority of isolates (227/240; 94.6%) carried one or more toxin genes, while only 79 (33%) carried all four. Ninety-one isolates (91/240; 37.9%) carried both exoU and exoS genes. exoU was detected mainly among ICU patients, while exoS from non-ICU patients was found with a statistically higher frequency (Table 1). Also, the exoS gene was statistically significant among infection isolates compared to carriage (53.50% versus 31.32%; P = 0.001).
PFGE exhibited five predominant pulsotypes (Table 1; Fig. 2). PFGE type a strains were identified as ST235; type b strains carrying the blaVIM-1 gene belonged to ST111, while those carrying blaVIM-2 were identified as ST235; type c strains were ST253 and type d strains were ST235, while those of type s were classified as ST309 and ST639. The remaining isolates were classified into 78 PFGE types including 1 to 3 strains each. Polyclonality was observed mainly among non-ICU strains (Table 1). Common clones among infecting and colonizing isolates were identified. The MDRPA strains, including MBL-positive strains, belonged mainly to pulsotypes a and d, both characterized as ST235.
Fig 2.

Dendrogram of P. aeruginosa isolates after digestion of DNA with SpeI and PFGE. Comparison of clonal types identified in the present study with previous ones, recovered from patients in the same hospital. No relationship was detected between the recent and the older clones. Lines 8, 9, 6, 7, 2, 3, 10, 11, 4, and 5 are from the present study (clones d, d, a, a, b, b, s, s, c, and c, respectively). Lines 17, 18, 15, 16, 19, 20, 21, 13, and 14 are from a previous study (clones C, C, B, B, D, D, E, A, and A, respectively).
Surveillance studies have documented increases in the frequency of outbreaks, especially in ICU infections caused by strains resistant to multiple classes of antibiotics (12, 13). P. aeruginosa was related to 8% of total infections and carriage during the study period, while among ICU patients, an outbreak occurred due to the spread of one main clone (ST235) of MDRPA. In our hospital setting, MDRPA accounted for 49.5% of P. aeruginosa infections before the present study, reached 63.3% during the studied period, and dropped afterwards to 38.5%.
A higher prevalence of MBL production was observed among IMP-NS P. aeruginosa isolates (33%) than in other studies from Greece and other European countries (14, 15). All MBL-positive isolates in the present study carried the blaVIM gene and were spread in all hospital wards, especially among non-ICU patients (Table 1). VIM-type MBLs are predominant in Europe, particularly in the Mediterranean region, and have been associated with large outbreaks of MDRPA (3, 16, 17). More specifically, blaVIM-2 is the most frequent type in southern European countries (3), while in Greece, blaVIM-17, a variant of blaVIM-2, was identified in another outbreak (16).
Serotype O11 is common in hospital outbreaks and associated with multidrug resistance (2, 12), as is also shown in the present study.
In our collection, 94.6% of P. aeruginosa isolates carried one or more TTSS genes, as reported elsewhere (18). exoS was more frequent in isolates from urinary tract and wound infections from non-ICU patients, a finding that is in accordance with those by other investigators (18). exoU was associated with serotype O11, as reported also by Faure et al. (4), and detected mainly among ICU isolates (P = 0.032). Expression of exoU correlates with acute cytotoxicity and accelerated lung injury playing a role in the development of septic shock in ICU high-risk patients (18, 19).
MDRPA strains belonged mainly to PFGE types a and d of serotype O11 and ST235. A comparison of the recently identified clones with previous ones revealed no relationship (Fig. 2) (20). Studies have reported clonally related nosocomial outbreaks of MDRPA producing IMP-13 MBL (13) and panantibiotic-resistant P. aeruginosa in ICUs (12). The observation that the majority of pulsotype d strains (ST235) carry the blaVIM-2 gene reinforces the theory that clonal spread may have played a role in the outbreak of IMP-NS P. aeruginosa. blaVIM gene spread was identified among clonally related strains in the last decade (1, 17, 21).
The observation that most carriage isolates belonged to the two predominant PFGE types a (38/83) and d (11/83) and to the same clone, ST235, indicates that colonization during ICU hospitalization contributes to infection and spread to other wards. Clinical isolates of ST235 (serotype O11) harboring acquired β-lactamases have been reported worldwide (22). P. aeruginosa ST235 (serotype O11) strains from bloodstream infections were among the three predominant epidemic clones in the Czech Republic (22).
This study describes a clonal outbreak during a 2-year period of MDRPA serotype O11 of the ST235 clone in a university hospital which occurred mainly in the ICU. P. aeruginosa clearly represents one of the most challenging pathogenic bacteria, since MDRPA isolates spread clonally quite frequently. The monitoring of MBL- and exotoxin gene-carrying isolates has epidemiological significance in the identification of drug-resistant and virulent P. aeruginosa isolates, especially in high-risk patients. Our work shows the need for clonal identification, since MDRPA outbreaks require targeted infection control measures.
ACKNOWLEDGMENTS
We are grateful to T. Ajayi for kindly offering the reference P. aeruginosa strain used in the PCRs for exotoxin gene detection.
This research was supported by funds of the Department of Microbiology, University of Patras.
The authors have no conflict of interest to declare.
Footnotes
Published ahead of print 12 December 2012
REFERENCES
- 1. Cornaglia G, Mazzariol A, Lauretti L, Rossolini GM, Fontana R. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-beta-lactamase. Clin. Infect. Dis. 31:1119–1125 [DOI] [PubMed] [Google Scholar]
- 2. Tassios PT, Gennimata V, Spaliara-Kalogeropoulou L, Kairis D, Koutsia C, Vatopoulos AC, Legakis NJ. 1997. Multiresistant Pseudomonas aeruginosa serogroup O:11 outbreak in an intensive care unit. Clin. Microbiol. Infect. 3:621–628 [DOI] [PubMed] [Google Scholar]
- 3. Pena C, Suarez C, Tubau F, Gutierrez O, Domínguez A, Oliver A, Pujol M, Gudiol F, Ariza J. 2007. Nosocomial spread of Pseudomonas aeruginosa producing the metallo-beta-lactamase VIM-2 in a Spanish hospital: clinical and epidemiological implications. Clin. Microbiol. Infect. 13:1026–1029 [DOI] [PubMed] [Google Scholar]
- 4. Faure K, Shimabukuro D, Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. 2003. O-antigen serotypes and type III secretory toxins in clinical isolates of Pseudomonas aeruginosa. J. Clin. Microbiol. 41:2158–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kronish JP. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767–1774 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. Approved standard M7-A6. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Falagas M, Koletsi P, Bliziotis I. 2006. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J. Med. Microbiol. 55:1619–1629 [DOI] [PubMed] [Google Scholar]
- 8. Liu PV, Wang S. 1990. Three new major somatic antigens of Pseudomonas aeruginosa. J. Clin. Microbiol. 28:922–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. 2003. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J. Clin. Microbiol. 41:3526–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deplano A, Denis O, Poirel L, Hocquet D, Nonhoff C, Byl B, Nordmann P, Vincent J, Struelens M. 2005. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 43:1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pagani L, Colinon C, Migliavacca R, Labonia M, Docquier JD, Nucleo E, Spalla M, Bergoli M, Rossolini GM. 2005. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-β-lactamase. J. Clin. Microbiol. 43:3824–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sardelic S, Bedenic B, Colinon-Dupuich C, Orhanovic S, Bosnjak Z, Plecko V, Cournoyer B, Rossolini GM. 2012. Infrequent finding of metallo-β-lactamase VIM-2 in carbapenem-resistant Pseudomonas aeruginosa strains from Croatia. Antimicrob. Agents Chemother. 56:2746–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsakris A, Tassios P, Polydorou F, Papa A, Malaka E, Antoniadis A, Legakis NJ. 2003. Infrequent detection of acquired metallo-beta-lactamases among carbapenem-resistant Pseudomonas isolates in a Greek hospital. Clin. Microbiol. Infect. 9:846–851 [DOI] [PubMed] [Google Scholar]
- 16. Siarkou VI, Vitti D, Protonotariou E, Ikonomidis A, Sofianou D. 2009. Molecular epidemiology of outbreak-related Pseudomonas aeruginosa strains carrying the novel variant blaVIM-17 metallo-β-lactamase gene. Antimicrob. Agents Chemother. 53:1325–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsakris A, Pournaras S, Woodford N, Palepou M, Babini G, Douboyas J, Livermore D. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamood AN, Griswold JA, Duhan CM. 1996. Production of extracellular virulence factors by Pseudomonas aeruginosa isolates obtained from tracheal, urinary tract, and wound infections. J. Surg. Res. 61:425–432 [DOI] [PubMed] [Google Scholar]
- 19. Allewelt M, Coleman FT, Grout M, Priebe GP, Pier GB. 2000. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 68:3998–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drougka E, Panagea T, Chini V, Foka A, Christofidou M, Spiliopoulou I. 2007. Clonal types and serotypes of multidrug-resistant Pseudomonas aeruginosa isolates spread in a university hospital in Greece. Clin. Microbiol. Infect. 13(Suppl 1):37717359321 [Google Scholar]
- 21. Sardelic S, Pallecchi L, Punda-Polic V, Rossolini GM. 2003. Carbapenem-resistant Pseudomonas aeruginosa carrying VIM-2 metallo-β-lactamase determinants, Croatia. Emerg. Infect. Dis. 9:1022–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nemec A, Krizova L, Maixnerova M, Musilek M. 2010. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res. Microbiol. 161:234–242 [DOI] [PubMed] [Google Scholar]