Abstract
Long-term typhoid carriers can simultaneously excrete Salmonella enterica serovar Typhi variants with considerable genetic differences, a situation that complicates the interpretation of the subtyping data used in outbreak investigations and disease surveillance.
TEXT
Salmonella enterica serovar Typhi is a human-restricted pathogen that causes typhoid fever, a disease transmitted primarily through the fecal-oral route. Typhoid fever is most prevalent in south-central and Southeast Asia and was responsible for an estimated 22 million cases and 0.22 million deaths in 2000 (1). Asymptomatic carriers may play an essential role in the evolution and global transmission of S. Typhi (2). Pulsed-field gel electrophoresis (PFGE) and multilocus variable-number tandem repeat (VNTR) analysis (MLVA) are two highly discriminatory subtyping methods for bacteria. An MLVA typing scheme based on 8 highly variable VNTR loci (Sty20, Sty25, Sty37, Sty40, Sty41, Sty42, Sty44, and Sty45) has been demonstrated to be more powerful than PFGE in distinguishing closely related S. Typhi strains and has been recommended as a routine subtyping tool for investigating S. Typhi infection (3). Outbreak investigation and disease surveillance are usually based on a presumptive epidemiological relationship among cases contracted from a common genotype.
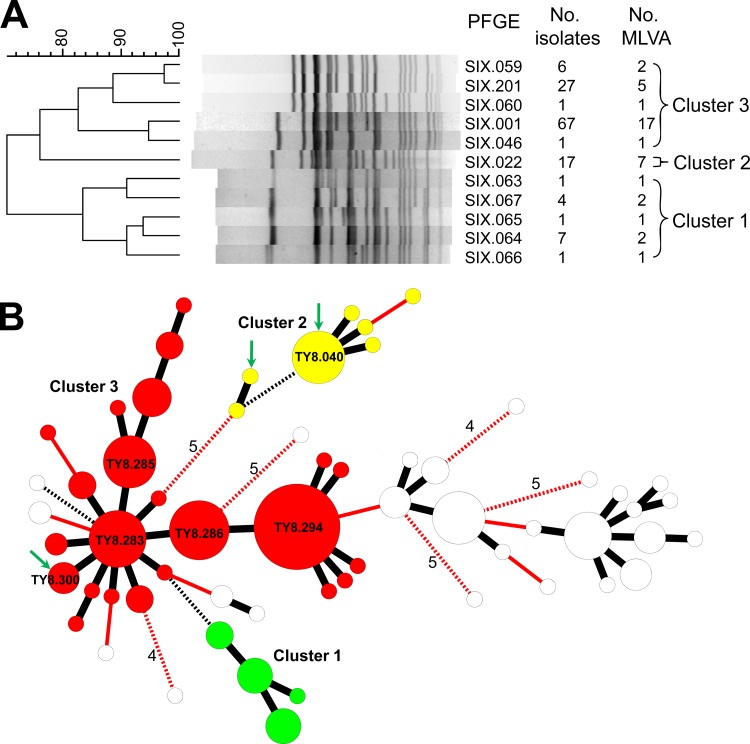
Typhoid fever is rare in Taiwan. In total, 785 cases were confirmed during 1996 to 2011, with a mean annual incidence rate of 0.21 case per 10,000 persons. At least 34% of the cases were imported. In 2005, a cluster (cluster 1) of 14 cases emerged in a 1-month period (from 12 June to 20 July 2005) in Taoyuan County, Taiwan, and it was rather unusual to have so many typhoid cases emerging in such a short period in a single county. Although these infections were very likely to have a common source, this was questioned because the isolates were identified as having 5 XbaI-digested PFGE (PFGE-XbaI) patterns, 5 BlnI-digested PFGE (PFGE-BlnI) patterns, and 4 MLVA profiles (Table 1). The largest differences were up to 8 DNA fragments among the 5 PFGE-XbaI patterns (Fig. 1A). 9 DNA fragments among the 5 PFGE-BlnI patterns, and 2 VNTR loci among the 4 MLVA profiles (Fig. 1B). Variations among the isolates occurred at 3 VNTR loci: Sty25, Sty44, and Sty45 (Table 1). Based on the Tenover criteria for interpreting PFGE patterns, bacterial isolates differing in 7 or more DNA fragments from the outbreak strain should not be part of the same outbreak (4). In interpreting MLVA profiles, several studies have shown that isolates from the same outbreak could allow their MLVA profiles to differ by one VNTR locus (5, 6). Despite the large differences in the genotypic patterns, the isolates from cluster 1 shared a higher PFGE pattern similarity than the other isolates in the Salmonella Typhi fingerprint database built by the Centers for Disease Control of Taiwan. To date, the database contained PFGE and MLVA data for 630 S. Typhi isolates collected since 1996.
Table 1.
PFGE and MLVA types and profiles of 8 VNTR loci from Salmonella enterica serovar Typhi isolates recovered from patients and chronic carriers
| Infection event (no. of isolates) | PFGE-XbaI type | PFGE-BlnI type | MLVA type | VNTR profilea |
No. of isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sty20 | Sty25 | Sty37 | Sty39 | Sty40 | Sty41 | Sty44 | Sty45 | |||||
| Cluster 1 | ||||||||||||
| Patients (14) | SIX.063 | SIN.010 | TY8.131 | 3 | 15 | 10 | 5 | 10 | 6 | 7 | 18 | 1 |
| SIX.064 | SIN.002 | TY8.004 | 3 | 14 | 10 | 5 | 10 | 6 | 7 | 18 | 4 | |
| TY8.005 | 3 | 14 | 10 | 5 | 10 | 6 | 6 | 18 | 3 | |||
| SIX.065 | SIN.034 | TY8.005 | 3 | 14 | 10 | 5 | 10 | 6 | 6 | 18 | 1 | |
| SIX.066 | SIN.033 | TY8.005 | 3 | 14 | 10 | 5 | 10 | 6 | 6 | 18 | 1 | |
| SIX.067 | SIN.003 | TY8.004 | 3 | 14 | 10 | 5 | 10 | 6 | 7 | 18 | 1 | |
| TY8.038 | 3 | 14 | 10 | 5 | 10 | 6 | 7 | 19 | 3 | |||
| Cluster 2 | ||||||||||||
| Patients (15) | SIX.022 | SIN.019 | TY8.040 | 1 | 8 | 25 | 5 | 16 | 12 | 13 | 13 | 9 |
| TY8.041 | 1 | 8 | 24 | 5 | 16 | 12 | 13 | 13 | 1 | |||
| TY8.042 | 1 | 8 | 27 | 5 | 16 | 12 | 11 | 15 | 1 | |||
| TY8.152 | 1 | 8 | 25 | 5 | 16 | 12 | 13 | 14 | 1 | |||
| TY8.279 | 1 | 9 | 25 | 5 | 16 | 12 | 13 | 13 | 1 | |||
| TY8.280 | 1 | 8 | 21 | 5 | 16 | 12 | 12 | 14 | 1 | |||
| SIX.022 | SIN.078 | TY8.040 | 1 | 8 | 25 | 5 | 16 | 12 | 13 | 13 | 1 | |
| Carriers (2) | SIX.022 | SIN.019 | TY8.281 | 1 | 8 | 27 | 5 | 16 | 12 | 11 | 16 | 1 |
| TY8.040 | 1 | 8 | 25 | 5 | 16 | 12 | 13 | 13 | 1 | |||
| Cluster 3 | ||||||||||||
| Patient (1) | SIX.001 | TY8.300 | 3 | 8 | 10 | 5 | 10 | 13 | 7 | 25 | 1 | |
| Carriers (101) | SIX.001 | TY8.085 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 27 | 2 | |
| TY8.283 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 25 | 13 | |||
| TY8.284 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 22 | 3 | |||
| TY8.285 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 24 | 11 | |||
| TY8.286 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 23 | 15 | |||
| TY8.287 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 15 | 1 | |||
| TY8.288 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 19 | 1 | |||
| TY8.289 | 3 | 8 | 11 | 5 | 10 | 13 | 7 | 26 | 1 | |||
| TY8.290 | 3 | 8 | 11 | 5 | 10 | 14 | 7 | 25 | 1 | |||
| TY8.291 | 3 | 8 | 11 | 5 | 10 | 12 | 7 | 24 | 1 | |||
| TY8.295 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 25 | 3 | |||
| TY8.296 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 24 | 5 | |||
| TY8.299 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 27 | 1 | |||
| TY8.300 | 3 | 8 | 10 | 5 | 10 | 13 | 7 | 25 | 3 | |||
| TY8.301 | 3 | 8 | 10 | 5 | 10 | 13 | 7 | 26 | 1 | |||
| TY8.302 | 3 | 8 | 12 | 5 | 12 | 13 | 7 | 24 | 3 | |||
| TY8.303 | 3 | 8 | 12 | 5 | 12 | 14 | 7 | 25 | 1 | |||
| SIX.046 | TY8.305 | 3b | 8 | 12 | 5 | 12 | 13 | 7 | 24 | 1 | ||
| SIX.059 | TY8.294 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 23 | 5 | ||
| TY8.296 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 24 | 1 | |||
| SIX.060 | TY8.294 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 23 | 1 | ||
| SIX.201 | TY8.292 | 3 | 8 | 12 | 5 | 10 | 14 | 7 | 23 | 1 | ||
| TY8.293 | 3 | 8 | 12 | 5 | 10 | 15 | 7 | 23 | 1 | |||
| TY8.294 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 23 | 23 | |||
| TY8.297 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 22 | 1 | |||
| TY8.298 | 3 | 8 | 12 | 5 | 10 | 13 | 7 | 20 | 1 | |||
VNTR loci that showed variations among the isolates from the cluster are highlighted in boldface.
This allele harbored 3 repeat units with a 9-bp insertion in a flanking region.
Fig 1.
Genetic relatedness of Salmonella enterica serovar Typhi strains. (A) Dendrogram constructed using the PFGE-XbaI patterns of the strains from 3 clusters of infection. (B) Minimum spanning tree constructed using the MLVA profiles of 8 VNTR loci for the isolates from 3 clusters of infection and 57 isolates with the SIX.001 PFGE type recovered from other patients and asymptomatic carriers between 1998 and 2011. Each circle represents an MLVA type, and the size of the circle is proportional to the number of isolates belonging to the MLVA type. A distance of one locus between two MLVA types is indicated by a thick black line, a distance of two loci by a thin red line, a distance of three loci by a broken black line, and a distance of four or more loci by a numbered broken red line. The MLVA types for the 2 isolates from the carrier in cluster 2 and for the isolate from the patient in cluster 3 are indicated by green arrows. The MLVA types for the isolates from clusters 1, 2, and 3 are marked in green, yellow, and red, respectively, while those for other isolates with the SIX.001 type are marked in white.
From 12 January to 23 May 2010, a cluster (cluster 2) of 4 typhoid cases was identified that had an epidemiological link to a high-tech electronics factory in Hsinchu County. Four isolates from the 4 patients had an indistinguishable PFGE-XbaI pattern (SIX.022), two PFGE-BlnI patterns (SIN.019 and SIN.078), and an identical VNTR profile (TY8.040). SIN.019 and SIN.078 differed by one fragment. Although the source of infection was not found, the genotype (SIX.022:SIN.019:TY8.040) had been previously identified in 2 isolates that emerged in 2007 and 2008 in the same county. In late April to early May 2011, a cluster of 4 cases emerged that were linked to the same electronics factory with the typhoid outbreak in 2010. The 4 isolates from the 4 cases displayed the same PFGE patterns (SIX.022 and SIN.019) but different MLVA patterns: 3 had a TY8.040 type and 1 had a TY8.279 type, which differed from TY8.040 at 1 VNTR locus (Table 1). The epidemiologists were aware that these victims had eaten noodles ordered from a nearby Indonesian-style food booth. S. Typhi was detected in two stool specimens obtained in early May on 2 consecutive days from the cook of the booth, a female Indonesian immigrant of approximately 60 years of age who was asymptomatic. An isolate from the first stool specimen displayed SIX.022 and SIN.019 PFGE patterns and an MLVA type (TY8.281) that differed from TY8.040 at 3 VNTR loci; however, an isolate from the second stool specimen belonged to the major SIX.022:SIN.019:TY8.040 genotype. Two additional cases emerged in mid-May 2011, with one having an MLVA type (TY8.280) that differed from TY8.040 at 3 VNTR loci. In total, 15 of the cases emerging in the area since 2007 could have an infection source from this carrier. Variations in the isolates from the 15 cases and the carrier occurred at 4 VNTR loci, Sty25, Sty37, Sty44, and Sty45. The multiple MLVA types detected in the isolates from the carrier and the cases in the outbreak of 2011 suggested that variants with considerable genetic differences could be excreted simultaneously from a long-term carrier. However, the evidence is not sufficiently strong to support this speculation.
In mid-April 2012, our laboratory received an S. Typhi isolate recovered from a blood specimen from a 92-year-old female in New Taipei City. Stool specimens were collected from close contacts of the patient, and S. Typhi was detected in a stool specimen from a female Indonesian caregiver of the patient. However, the PFGE-XbaI type (SIX.001) for the isolate from the patient differed from the type (SIX.201) for an isolate from the caregiver in 4 DNA fragments (Fig. 1A). Furthermore, the MLVA type (TY8.300) for the isolate from the patient differed from the type (TY8.294) for the isolate from the caregiver at 2 VNTR loci. The stool sample from the caregiver was streaked onto Salmonella-Shigella agar plates and Hektoen enteric agar plates to recover additional S. Typhi isolates. Surprisingly, the colonies grown on the two types of media all appeared to have the same type of morphology. One hundred colonies were selected from the primary culture plates, and all of them were subsequently identified as S. Typhi. All of the isolates were subjected to PFGE and MLVA genotyping. In total, 5 PFGE-XbaI and 23 MLVA types were detected among the 102 isolates from the patient and the carrier. The largest difference among the 5 PFGE-XbaI patterns was up to 8 DNA fragments. SIX.001 and SIX.201 were the major PFGE-XbaI types. The largest difference among the 23 MLVA profiles was 5 VNTR loci: variations occurred at Sty20 (2 alleles), Sty37 (3 alleles), Sty40 (2 alleles), Sty41 (4 alleles), and Sty45 (9 alleles). TY8.283, TY8.285, TY8.286, and TY8.294 were the major MLVA types. In general, isolates with different PFGE-XbaI patterns had different MLVA profiles. Only 2 MLVA types (TY8.294 and TY8.296) were found in the isolates with different PFGE-XbaI types: TY8.294 was detected in the isolates with PFGE genotypes SIX.059, SIX.060, SIX.201, and TY8.296 in the isolates with PFGE types SIX.001 and SIX.059 (Table 1). The SIX.001:TY8.300 genotype for the isolate from the patient was found in only 3 of the 101 isolates from the carrier.
SIX.001 was a major genotype in the Salmonella Typhi fingerprint database of the Taiwan Centers for Disease Control. The white circles in Fig. 1B represent the MLVA types for 57 isolates with the SIX.001 genotype, of which 45 were recovered from Indonesian migrant workers in 2008 and 2009 and 12 from Taiwanese travelers who acquired infections in Indonesia between 1998 and 2011. Some of these isolates had MLVA profiles close to those recovered from cluster 3 by a distance of only 2 VNTR loci (Fig. 1B). In contrast, the isolates that were simultaneously excreted from the carrier of cluster 3 could be more distantly related than those epidemiologically unrelated isolates. The largest distance among the isolates from cluster 3 was up to 5 VNTR loci, as seen in TY8.290 and TY8.305 (Table 1). The comparison indicates that in such a circumstance, it is very unlikely MLVA data can be used alone to identify an outbreak without sufficient epidemiological data.
PFGE and MLVA are two highly discriminatory genotyping methods for discerning bacterial strains. PFGE has been adopted as the standard subtyping tool in the PulseNet food-borne disease surveillance network (7, 8). The MLVA typing scheme based on rapidly evolving VNTR loci is more discriminatory than PFGE in discerning very closely related strains of S. Typhi and other bacterial organisms (3, 5, 6, 9). However, both methods seem to be too discriminatory for S. Typhi in these 3 outbreaks; the use of PFGE and MLVA data alone would exclude the epidemiological association of the cases from the corresponding outbreak. Single nucleotide polymorphism (SNP) typing and ribotyping could be alternative methods to balance the discriminatory power of PFGE and MLVA (10, 11). The markers of SNP and ribotyping evolve much slowly than those of PFGE and MLVA do. The isolates from each of the 3 outbreaks could be typed as the same SNP type and ribotype. Although SNP typing and ribotyping could be useful to reduce the complexity of PFGE and MLVA data for the isolates from chronic typhoid carriers, they are not sufficiently discriminatory in discerning the monomorphic S. Typhi isolates for the purposes of short-term epidemiological studies, i.e., outbreak investigation and disease surveillance.
The genotypic data for the isolates from the carrier of cluster 3 revealed that S. Typhi variants with considerable genetic variations could be excreted simultaneously from a long-term carrier. Chromosomal rearrangements due to the recombination between the rrn operons of host-adapted S. Typhi frequently occur over time within the human host (12), which could result in large differences in the PFGE patterns for variants, as seen in clusters 1 and 3. The present study indicated that the genetic variations of S. Typhi could occur frequently in the VNTR loci, generating variants with differences at more than 2 VNTR loci. Because outbreak investigations and disease surveillance are usually based on a presumptive epidemiological relationship among cases contracted from a common genotype, the simultaneous excretion of variants with large difference in their PFGE and MLVA patterns would increase the complexity of the interpretation of the PFGE and MLVA data.
ACKNOWLEDGMENT
Funding for this work was provided by the Department of Health, Taiwan, grant no. DOH101-DC-2201.
Footnotes
Published ahead of print 12 December 2012
REFERENCES
- 1. Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull. World Health Organ. 82:346–353 [PMC free article] [PubMed] [Google Scholar]
- 2. Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, Achtman M. 2006. Evolutionary history of Salmonella typhi. Science 314:1301–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tien YY, Ushijima H, Mizuguchi M, Liang SY, Chiou CS. 2012. Use of multilocus variable-number tandem repeat analysis in molecular subtyping of Salmonella enterica serovar Typhi isolates. J. Med. Microbiol. 61:223–232 [DOI] [PubMed] [Google Scholar]
- 4. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liang SY, Watanabe H, Terajima J, Li CC, Liao JC, Tung SK, Chiou CS. 2007. Multilocus variable-number tandem repeat analysis for molecular typing of Shigella sonnei. J. Clin. Microbiol. 45:3574–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Noller AC, McEllistrem MC, Pacheco AG, Boxrud DJ, Harrison LH. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 8. Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiou CS, Hung CS, Torpdahl M, Watanabe H, Tung SK, Terajima J, Liang SY, Wang YW. 2010. Development and evaluation of multilocus variable number tandem repeat analysis for fine typing and phylogenetic analysis of Salmonella enterica serovar Typhimurium. Int. J. Food Microbiol. 142:67–73 [DOI] [PubMed] [Google Scholar]
- 10. Le TA, Fabre L, Roumagnac P, Grimont PA, Scavizzi MR, Weill FX. 2007. Clonal expansion and microevolution of quinolone-resistant Salmonella enterica serotype Typhi in Vietnam from 1996 to 2004. J. Clin. Microbiol. 45:3485–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Octavia S, Lan R. 2009. Multiple locus variable number of tandem repeat analysis of Salmonella enterica serovar Typhi. J. Clin. Microbiol. 47:2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matthews TD, Rabsch W, Maloy S. 2011. Chromosomal rearrangements in Salmonella enterica serovar Typhi strains isolated from asymptomatic human carriers. mBio 2(3):e00060–11 doi:10.1128/mBio.00060-11 [DOI] [PMC free article] [PubMed] [Google Scholar]