Abstract
Long-term infection with high-risk human papillomavirus (HPV) is the leading cause of cervical cancer, while infection with low-risk HPV is the major reason for condylomata acuminata. An accurate, rapid, and convenient assay that is able to simultaneously detect, genotype, and quantify HPV would be of great clinical value yet remains to be achieved. We developed a three-color real-time PCR assay that is able to analyze 30 predominant HPV types in three reactions. The amplification curves indicated the presence of HPV, melting curve analysis identified the HPV genotype, and the quantification cycle value determined the quantity. We applied this assay to 647 cervical swab samples, and the results were compared with those obtained with a commercial genotyping system. The proposed assay had a limit of detection of 5 to 50 copies per reaction and a dynamic range of 5 × 101 to 5 × 106 copies per reaction. A comparison study showed that the overall sample concordance with the comparison method was 91.6% and the type agreement was greater than 98.7%. The quantification study demonstrated that the loads of HPV type 16 in 30 samples with cervical intraepithelial neoplasia grade III (CIN III) lesions were significantly higher than those in samples with CIN I lesions or CIN II lesions, and the results were concordant with those of the comparison method. The increased information content, high throughput, and low cost would facilitate the use of this real-time PCR-based assay in a variety of clinical settings.
INTRODUCTION
Human papillomavirus (HPV) is one of the most important and complex targets in molecular diagnosis (1, 2). The importance lies in its direct association with cervical cancer, and the complexity is caused by its large number of genotypes (3), which should be differentiated in order to clarify the disease risks and to implement vaccine programs (4, 5). Moreover, quantification of HPV is required to determine disease status and monitor the progression of HPV infection (6–9). Preferably, a diagnostic method for HPV infection should enable simultaneous detection, genotyping, and quantification of HPV. Ideally, such a method should be undertaken in a high-throughput, easily automated, and cost-effective way, in particular, when considering its use in large-scale population screening programs (10, 11).
A variety of assays for detection and/or genotyping of HPV have been described. The Hybrid Capture 2 assay (HC2; Qiagen Inc., Gaithersburg, MD), cleared by the U.S. Food and Drug Administration (FDA), is considered the “gold standard” to detect 13 high-risk HPV types from cervical scrapings and has been used in routine screening for years (12). However, this assay does not allow identification of the specific HPV type and cannot discriminate multiple infections. These deficiencies have partially been solved with the introduction of PCR-based assays by using consensus or type-specific primers combined with numerous post-PCR analysis procedures, such as genotype-specific hybridization (13–15), minisequencing (16, 17), and sequencing (18, 19). Most of these assays allow identification of a broad range of HPV types, with the major disadvantage being that they are expensive and difficult to automate due to the complicated post-PCR analysis.
Real-time PCR-based methods, being free of post-PCR analysis, have gained increasing popularity in HPV diagnosis. This trend is reflected in the recent reports of several automated commercial systems. For example, the newly FDA-approved Cobas 4800 HPV test features fully automated sample preparation combined with real-time PCR technology to detect 14 high-risk HPV genotypes (20). Although designed to be a qualitative test utilizing real-time PCR technology, the test also provides the cycle number, which is generally indicative of the amount of viral DNA present in the sample. Similar features are found in the Abbott real-time high-risk HPV assay (21). LG Life Sciences (Seoul, South Korea) has also developed a real-time PCR assay (the AdvanSure HPV screening real-time PCR), which can detect and distinguish the signals for HPV types 16 and 18 (HPV-16/18) from 39 non-HPV-16/18 types as well as an internal control in a single reaction (22). Because current real-time thermocyclers have a limited number of detection channels and are thus unable to distinguish many different fluorescent dyes, the real-time PCR-based systems described above preclude full genotyping of dozens of HPV types in a single reaction. Alternatively, type-specific real-time PCR approaches with high sensitivity and specificity have been developed; however, many separate reactions are required in order to cover all common HPV types (23).
To maintain the simplicity of real-time PCR while overcoming the channel limitation, we describe here a new strategy that allows the simultaneous detection, quantification, and genotyping of 30 HPV types in three reactions on a three-color real-time PCR machine. Our previous work had demonstrated that dually labeled, self-quenched probes could be used for both real-time PCR detection and melting curve analysis under asymmetric PCR conditions (24). Because real-time PCR detection offers quantitative information, while melting curve analysis reveals the sequence information, use of the combination of them would allow the simultaneous quantification and genotyping from one sample in a single assay. By assigning a predefined melting temperature to one HPV type, up to five different HPV types could be differentiated in one detection channel. A multichannel real-time PCR machine would therefore allow a large number of HPV types to be differentiated in one reaction. The established assay was evaluated regarding the sensitivity, accuracy, and dynamic ranges. Both the typing and quantification results were compared with those of commercial and approved assays by analyzing clinical samples.
MATERIALS AND METHODS
Clinical samples and synthetic plasmids.
Cervical swab specimens were obtained from 647 females ages 19 to 68 years (mean, 33 years; median, 32 years); 494 of them were referred to the Molecular Diagnostics Laboratory, Maternal and Child Health Hospital of Xiamen, Xiamen, China, for a routine gynecologic examination, and 153 of them were referred for colposcopy after a suspected diagnosis of HPV infection. Genomic DNA was extracted from each swab using a kit supplied with the HPV GenoArray test kit (Hybribio Ltd., Chaozhou, China). DNA extracts were first used for GenoArray analysis. After storage at −20°C for 3 to 4 months, the same DNA extracts were thawed and used for real-time PCR assay. The Research Ethics Committee of Xiamen University approved the study protocol.
Plasmids containing the GP5+/GP6+ flanked region of the respective 30 HPV types were synthesized by Sangon (Shanghai, China) on the basis of the type-specific sequences provided by the Genome Sequencing Project in GenBank (http://www.ncbi.nlm.nih.gov). The concentration of each plasmid was determined by an ND-1000 spectrometer (Nanodrop Technologies, Rockland, DE). To determine the limit of detection (LOD) of the real-time PCR assay for each HPV type, the plasmids were diluted to a series of concentrations of 106, 105, 104, 103, 102, 101, and 100 copies/μl in the presence of 20 ng of SW-480 cell genomic DNA (HPV negative) for each concentration. These plasmid preparations were also used to study the reproducibility of the melting temperature (Tm) measurement as well as the specificity of HPV genotyping.
Primers and probes.
The design of consensus primers for the 30 HPV types was based on the previously reported GP5+/GP6+ primer pair, which proved to be able to efficiently amplify 23 mucosotropic HPV types (25). We introduced 2 degenerate bases in GP5+ and 2 degenerate bases in GP6+; the resulting GP5+′/GP6+′ primer pair could efficiently amplify all 30 HPV types. One additional primer pair was designed for amplification of the human β-globin gene (HBB), which was used as an internal positive control (IPC). The sequences of the primers are given in Table S1 in the supplemental material.
Thirty dually labeled fluorogenic probes were designed for the 30 respective HPV types. The rule of the probe design was that each probe would hybridize with its specific target at a distinct Tm value and that each probe would have no cross-hybridization with any other known mucosotropic HPV types detectable at above 40°C. Moreover, the Tm decrease induced by the intratype variants within each type would cause no crossover with other HPV types. The Tm value of each probe-target hybrid was controlled by the length and G+C content of the probe. According to our experimental design, three reactions were used to test 30 HPV types. Reaction 1 tests for 10 high-risk HPV types (HPV types 68, 58, 82, 59, and 73 in the Cy5 channel; HPV types 31, 33, 16, 35, and 18 in the carboxy-X-rhodamine [ROX] channel). Reaction 2 tests for 5 high-risk HPV types (HPV types 56, 39, 45, 52, and 51 in the Cy5 channel) and 5 low-risk HPV types (HPV types 66, 53, 26, 11, and 6 in the ROX channel). Reaction 3 tests for 10 low-risk HPV types (HPV types 72, 70, 61, 54, and 44 in the Cy5 channel; HPV types 43, 69, 81, 42, and 40 in the ROX channel). In all three reactions, HBB was detected in the 6-carboxyfluorescein (FAM) channel. Therefore, each reaction mixture contained 10 HPV probes and 1 HBB probe. Five of the HPV probes were labeled with 5′-ROX and 3′-black hole quencher 2 (BHQ2), another five HPV probes were labeled with 5′-Cy5 and 3′-BHQ2, and the HBB probe was labeled with 5′-FAM and 3′-BHQ1. The sequences of the probes are given in Table 1.
Table 1.
Probe sequences, amplification efficiencies, and Tm values for 30 HPV types in the real-time PCR assay
| HPV type or gene | Probe sequence (5′–3′) | Efficiency (%) | Tm value (°C) |
|---|---|---|---|
| 18 | ROX-TCTACACAGTCTCCTGTACCTGGGCAATATGATGCTACCA-BHQ2 | 85.59 | 74.6 |
| 35 | ROX-TGCTGTGTCTTCTAGTGACAGTACATATAAAAATGAC-BHQ2 | 103.17 | 67.0 |
| 16 | ROX-TTATGTGCTGCCATATCTACTTCAGAAA-BHQ2 | 99.52 | 63.4 |
| 33 | ROX-ACAAGTAACTAGTGACAGTACA-BHQ2 | 85.10 | 55.8 |
| 31 | ROX-CAAACAGTGATACTACATT-BHQ2 | 93.41 | 50.8 |
| 73 | Cy5-TGTAGGTACACAGGCTAGTAGCTCTACTACAACGTATGCC-BHQ2 | 103.56 | 73.7 |
| 59 | Cy5-TTCTGTGTGTGCTTCTACTACTTCTTCTATTCCTAA-BHQ2 | 95.94 | 66.9 |
| 82 | Cy5-TTACTCCATCTGTTGCACAAACATTTAC-BHQ2 | 94.88 | 63.3 |
| 58 | Cy5-TGAAGTAACTAAGGAAGGTACA-BHQ2 | 88.69 | 58.3 |
| 68 | Cy5-TTTGTCTACTACTACTGAAT-BHQ2 | 99.16 | 52.0 |
| 6 | ROX-TGCATCCGTAACTACATCTTCCACATACACCAATTCTGAT-BHQ2 | 99.87 | 71.1 |
| 11 | ROX-CATCTGTGTCTAAATCTGCTACATACACTAATTCA-BHQ2 | 97.72 | 66.0 |
| 26 | ROX-ATTAGTACATTATCTGCAGCATCTGCAT-BHQ2 | 84.63 | 62.0 |
| 53 | ROX-CCACACAGTCTATGTCTACATATA-BHQ2 | 100.27 | 57.1 |
| 66 | ROX-AAGCACATTAACTAAATATGA-BHQ2 | 90.57 | 51.9 |
| 51 | Cy5-TAACTATTAGCACTGCCACTGCTGCGGTTTCCCCAAC-BHQ2 | 90.30 | 74.0 |
| 52 | Cy5-CATGACTTTATGTGCTGAGGTTAAAAAGGAAAGC-BHQ2 | 96.93 | 69.3 |
| 45 | Cy5-ACATTATGTGCCTCTACACAAAATCCTGT-BHQ2 | 91.58 | 63.9 |
| 39 | Cy5-ATCTACCTCTATAGAGTCTTCCATA-BHQ2 | 94.07 | 57.8 |
| 56 | Cy5-AACAGTTAAGTAAATATGAT-BHQ2 | 89.08 | 48.0 |
| 40 | ROX-TGCCACACAGTCCCCCACACCAACCCCATATAATAACAGT-BHQ2 | 103.46 | 74.9 |
| 42 | ROX-TTGTGTGCCACTGCAACATCTGGTGATACATATA-BHQ2 | 96.19 | 68.9 |
| 81 | ROX-AATTTTACTATTTGCACAGCTACATCTGCT-BHQ2 | 96.38 | 64.3 |
| 69 | ROX-CACTATTAGTACTGTATCTGCACAAT-BHQ2 | 87.28 | 58.8 |
| 43 | ROX-TCACTATTAGTACTGTATC-BHQ2 | 90.25 | 48.1 |
| 44 | Cy5-TGCTGCCACTACACAGTCCCCTCCGTCTACATATACTAG-BHQ2 | 102.76 | 77.8 |
| 54 | Cy5-AGTACTAACCTAACATTGTGTGCTACAGCATCCAC-BHQ2 | 86.23 | 68.9 |
| 61 | Cy5-ATCTGAATATAAAGCCACAAGCTTTAGGGA-BHQ2 | 103.68 | 63.9 |
| 70 | Cy5-ACACGTAGTACTAATTTTACATTGTCT-BHQ2 | 92.72 | 59.2 |
| 72 | Cy5-TGTAACTATTTGTACTGCC-BHQ2 | 87.75 | 52.0 |
| HBB | FAM-TCTACCCTTGGACCCAGAGGTTCTTT-BHQ1 | 83.13 | 65.1 |
Real-time PCR assay.
The real-time PCR assay was performed in 3 separate reactions. Each reaction mixture (25 μl) contained 75 mmol/liter Tris-HCl (pH 9.0), 20 mmol/liter (NH4)2SO4, 4 mmol/liter MgCl2, 0.01% (vol/vol) Tween 20, 1 unit Taq DNA polymerase, 200 μmol/liter of each deoxynucleoside triphosphate, 100 nmol/liter primer GP5+′, 1 μmol/liter primer GP6+′, 100 nmol/liter primer IPC-F, 1 μmol/liter primer IPC-R, and 250 nmol/liter of each of the 10 HPV and HBB probes. In each reaction mixture, 5 μl template DNA was added. The PCR and dissociation were performed in an Applied Biosystems 7500 real-time PCR system (Foster City, CA). PCR started with denaturation at 95°C for 5 min, followed by 45 cycles of 95°C for 10 s, 40°C for 32 s, and 75°C for 30 s. Melting curve analysis consisted of 95°C for 15 s, 40°C for 60 s, and 85°C for 15 s, with the ramp rate between 40°C and 85°C set to 5%. The fluorescence signal was acquired at the annealing step of each cycle during amplification and throughout the final ramp between 40°C and 85°C in three detection channels: FAM (510 nm), ROX (610 nm), and Cy5 (660 nm). The existence of HPV in the sample was judged by the appearance of the amplification curve, and the HPV genotype was called by the predefined Tm value in the melting curve.
Quantification of HPV in a sample.
The amount of HPV was determined by the quantification cycle (Cq) value of the amplification curve and calculated by its external calibration curve. For a sample infected with a single HPV type, quantification can be directly achieved through the Cq value. For a sample infected with multiple types of HPV that occur in different channels or identical channels but different reactions, each type can be quantified separately as in the infection with a single type. If the coinfecting HPV types occur in identical channels of the same reaction, the Cq value obtained represents the sum of the values for the coinfecting types. To calculate the amount of an individual genotype, the quantity ratio of different genotypes needs to be determined. We explored this situation by using coinfection with HPV-16 and HPV-18 as a model. These two genotypes occur in the ROX channel of reaction 1. First, we hypothesized that the quantity ratio of HPV-16 to HPV-18 has a linear relationship with the height ratio of their respective melting peaks, according to previous reports (26). This hypothesis was confirmed by the observation that in the range of 90:10 to 10:90, the quantity ratio of HPV-16 to HPV-18 was linear to the height ratio of their melting peaks, and this linear relationship (y = −3.58 + 3.99x, where y is the concentration ratio, and x is the peak height ratio; R2 = 0.9905) was kept constant over their concentration range from 5 × 102 copies to 5 × 106 copies per reaction (see Fig. S1 in the supplemental material). These observations allowed us to deduce the quantity ratio of these two genotypes from the melting curves. Second, we reasoned that once the equations of the calibration curves of both HPV-16 and HPV-18 are known, the equation of the calibration curve of their mixture at a certain ratio could be obtained. To do so, five mimic samples containing different percentage ratios of HPV-16 to HPV-18 plasmid DNA (100:0, 75:25, 50:50, 25:75, and 0:100) were prepared, and the respective equation of the calibration curve was obtained by detecting the Cq values at a series of different concentrations (see Fig. S2 in the supplemental material). Five linear equations were obtained from the five ratios in terms of Cq = a + b × lg (viral load), where a is the initial value, and b is the slope. Using a polynomial regression fit, a could be obtained as a function of the percentage of HPV-16 (y) as
| (1) |
and b could be obtained as
| (2) |
Therefore, using equations 1 and 2, an equation for the mixture of HPV-16 and HPV-18 at a certain ratio could be calculated. Consequently, a quantification guideline was formulated to quantify both HPV-16 and HPV-18 of different ratios (see “Quantification rule” in the supplemental material). This quantification is also applicable to other dual, triple, or even higher levels of multiple infections.
Comparison methods.
The HPV GenoArray test is a Chinese FDA-approved assay for HPV genotyping. It is an L1 consensus primer-based PCR assay and is capable of typing 21 HPV genotypes (15). The assay utilizes a flowthrough hybridization technique by actively directing the targeting molecules toward the immobilized probes within the membrane fibers, with the complementary molecules being retained by the formation of duplexes. After a stringent wash, the hybrids were detected by the addition of a streptavidin-horseradish peroxidase conjugate, which binds to the biotinylated PCR products, and a substrate to generate a purple precipitate at the probe dot. The results were interpreted by direct visualization. The Diagnostic kit for HPV (types 16 and 18) is a Chinese FDA-approved real-time PCR assay for quantification of HPV-16 and HPV-18 (Kehua Ltd., Shanghai, China). This is a 5′-nuclease assay that can quantify both HPV-16 and HPV-18. Sequencing with type-specific primers (27) was used to confirm those discrepant samples detected by the new method and the comparison methods. Statistical analysis of the comparison data was carried out by using the statistical software SPSS (version 13.0, SPSS Inc., Chicago, IL).
RESULTS
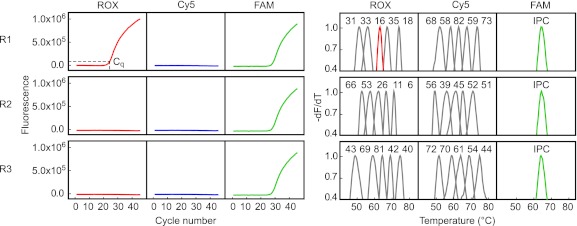
The proposed assay is a 3-tube, 3-color, duplex real-time PCR combined with melting curve analysis. Its temperature program contains two stages, amplification and melting, which can be finished in consecutive procedures within 2.5 h. A typical result from a sample infected with HPV-16 is shown in Fig. 1. The appearance of an amplification signal from the ROX channel of reaction 1 demonstrated that the sample was HPV positive. The Tm value obtained from the melting curve identified HPV-16 according to the predefined Tm values (63.4°C). The quantity of HPV-16 in the sample was determined to be 2.19 × 105 copies/μl by referring the Cq value (21.7) to the external calibration curve of HPV-16.
Fig 1.
Readout of the real-time PCR assay showing a typical result from a sample infected with HPV-16. One positive amplification curve appears only in the ROX channel of reaction 1. Red line, melting peak of HPV-16; gray lines, melting peaks of 29 other HPV types; green lines, melting peaks of IPC. All melting curves were first normalized to between 0 and 1, and the data for values between 0.4 and 1.0 are shown. The Cq value is marked in the amplification curve. R1, R2, and R3, reactions 1, 2, and 3, respectively.
Using plasmid DNA as the amplification template, the LOD of the assay was 5 to 50 copies per reaction. The amplification efficiency was 84.63% to 103.68%. A linear relationship (R2 > 0.99) between the Cq value and the logarithmic DNA concentration was achieved in the range of 5 × 101 to 5 × 106 copies per reaction. All HPV types could be identified by their corresponding Tm values, which mostly had a window of larger than 4°C between neighboring HPV types in the same channel (Table 1).
The Tm values listed in Table 1 were obtained from the plasmid template containing the sequence of the reference HPV genotype. To detect whether the presence of intratype variants could cause Tm overlapping between two neighboring genotypes in one channel, five HPV genotypes were studied using synthetic oligonucleotides as target mimics to hybridize with their respective probes. The results showed that the Tm shifts caused by these variants were smaller than 2°C and none of them could induce crossover with neighboring types.
The new assay was then subjected to evaluation by a blind test of 647 cervical swab samples, and the results were compared with those of the GenoArray method. The overall agreement between the two methods was 91.6% (593/647). Among the 54 discrepant samples, 12 samples were found to be infected with HPV types not included in the GenoArray technique. The results for all 12 of these samples were concordant with sequencing results. Of the remaining 42 discrepant samples, sequencing analysis revealed that the results for 35 (83.3%) samples were concordant with those of our assay and the results for 7 (16.7%) samples agreed with those of the GenoArray method. We then compared the type-specific concordance between the two assays (Table 2). Except for the 9 HPV types that were not included in the GenoArray, the concordance between the two assays was higher than 98%. The degree of concordance (κ) was more than 0.75 for all the genotypes, with the exception of HPV-44. There were in total 48 discrepant results (from 42 samples) in 2 × 21 × 647 typing results (from 647 samples), yielding an overall disagreement rate of 0.18%. Sequencing results demonstrated that, among these 48 discrepant typing results, results for 41 (85.4%) agreed with those of our method and results for 7 (14.6%) agreed with those of the GenoArray method.
Table 2.
Kappa values and P values by McNemar's test for HPV types detected by two assaysa
| Oncogenic potential | Genotype | No. of genotypes found to be positive by: |
No. of discrepant results | Agreement (%) | κ value (95% CId) | ||
|---|---|---|---|---|---|---|---|
| rtPCRb | GAc | rtPCR and GA | |||||
| High | 16 | 48 | 47 | 44 | 7 | 98.9 | 0.920 (0.890–0.950)f |
| 18 | 14 | 12 | 11 | 4 | 99.4 | 0.843 (0.766–0.920)f | |
| 31 | 8 | 8 | 8 | 0 | 100 | 1.000 (1.000–1.000)e | |
| 33 | 18 | 19 | 17 | 3 | 99.5 | 0.917 (0.869–0.965)f | |
| 35 | 3 | 2 | 2 | 1 | 99.8 | 0.799 (0.602–0.996)f | |
| 39 | 6 | 6 | 5 | 2 | 99.7 | 0.832 (0.715–0.949)f | |
| 45 | 3 | 2 | 2 | 1 | 99.8 | 0.799 (0.602–0.996)f | |
| 51 | 6 | 6 | 5 | 2 | 99.7 | 0.832 (0.715–0.949)f | |
| 52 | 45 | 43 | 40 | 8 | 98.7 | 0.902 (0.868–0.936)f | |
| 56 | 7 | 7 | 7 | 0 | 100 | 1.000 (1.000–1.000)e | |
| 58 | 26 | 27 | 25 | 3 | 99.5 | 0.941 (0.907–0.975)f | |
| 59 | 1 | 1 | 1 | 0 | 100 | 1.000 (1.000–1.000)e | |
| 68 | 8 | 9 | 8 | 1 | 99.8 | 0.940 (0.881–0.999)f | |
| Low | 6 | 11 | 8 | 8 | 3 | 99.5 | 0.840 (0.749–0.931)f |
| 11 | 14 | 11 | 11 | 3 | 99.5 | 0.878 (0.808–0.948)f | |
| 42 | 0 | 0 | 0 | 0 | 100 | ||
| 43 | 0 | 0 | 0 | 0 | 100 | ||
| 44 | 1 | 2 | 1 | 1 | 99.8 | 0.666 (0.351–0.981)g | |
| 53 | 17 | 18 | 16 | 3 | 99.5 | 0.912 (0.861–0.963)f | |
| 66 | 8 | 8 | 7 | 2 | 99.7 | 0.873 (0.784–0.962)f | |
| 81 | 11 | 11 | 9 | 4 | 99.4 | 0.815 (0.724–0.906)f | |
The results for 168 concordant and 42 discordant samples after initial analysis are shown.
rtPCR, real-time PCR assay.
GA, GenoArray assay.
CI, confidence interval.
Strength of agreement considered perfect.
Strength of agreement considered very good.
Strength of agreement considered good.
By referring to the histopathological results, we observed that the real-time PCR assay had a higher rate of positive detection of high-risk HPV than the GenoArray method (74.8% versus 69.9%) in samples with cervical intraepithelial neoplasia (CIN) lesions. With CIN grade II (CIN II) or higher (CIN II+) in the histopathologic results as a clinical reference, the sensitivity for the real-time PCR method was 93.8% (95% confidence interval [CI], 89.1% to 98.5%], the specificity was 44.6% (95% CI, 34.9% to 54.3%), and the positive predictive value was 30.9%, whereas the sensitivity of the comparison method was 90.6% (95% CI, 84.9% to 96.3%), the specificity was 47.9% (95% CI, 38.1% to 57.7%), and the positive predictive value was 31.5%. When the number was summarized for each genotype, HPV-16 and HPV-52 were found to be the two most common high-risk genotypes.
Among all HPV types, only the HPV-16 load was reportedly relevant to the CIN status of cervical cancer (8, 9, 28). Among the 647 samples, 30 HPV-16-positive samples had CIN lesion information, and their viral loads were thus detected. The results showed that the viral loads in samples with CIN I lesions ranged from 2.73 to 5.41 log copies/μl (median, 4.6 log copies/μl). The viral loads were more concentrated at higher levels in samples with CIN II lesions (range, 3.68 to 5.32 log copies/μl; median, 4.7 log copies/μl). The highest viral loads were detected in samples with CIN III lesions, and a more condensed distribution of high viral loads was observed (range, 5.26 to 5.84 log copies/μl; median, 5.6 log copies/μl). To examine the difference of HPV-16 loads among the three CIN statuses, an independent-samples t test was performed. The results demonstrated that HPV-16 loads had no significant difference between CIN I and CIN II states (P = 0.236), while HPV-16 loads in the CIN III state were significantly higher than those in both the CIN I and CIN II states (P < 0.001).
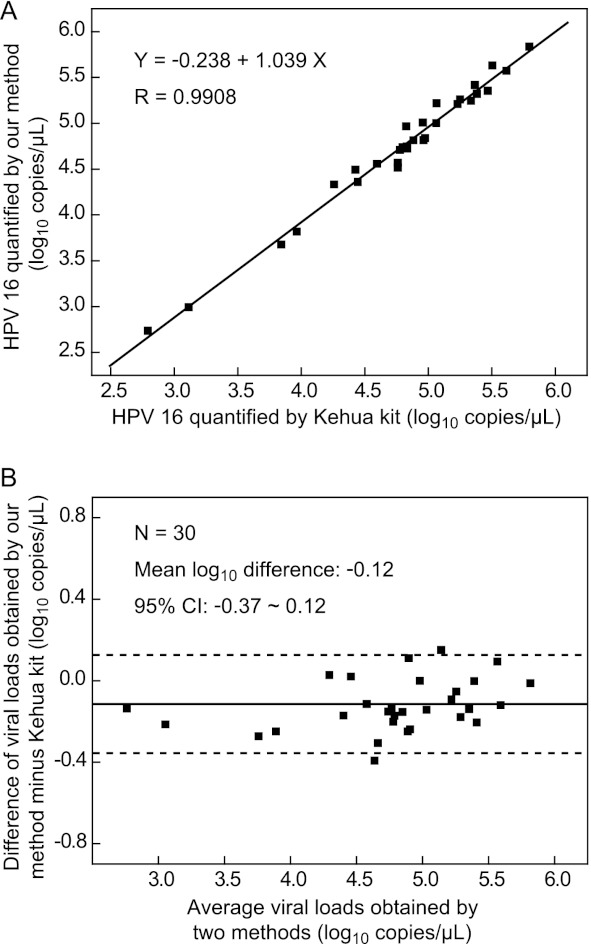
To validate the quantification results presented above, the same samples were analyzed using a commercial real-time PCR-based assay. Comparison between the results of the two methods was carried out by both regression analysis (29) and Bland-Altman analysis (30). Regression analysis showed that the slope approached 1 (1.039; 95% confidence interval, 0.987 to 1.091) and the y intercept approached 0 (−0.238; 95% confidence interval, −0.493 to 0.017), and the correlation coefficient (R) between the two methods was 0.9908 (P < 0.001) (Fig. 2A). Bland-Altman analysis gave a mean difference of −0.12 log10 unit (95% confidence interval, −0.37 log10 unit to 0.12 log10 unit) of the viral loads between the two methods, while the loads for all 30 samples differed by less than 0.5 log10 unit (Fig. 2B). The results presented above together demonstrated that our method and the comparison method had no significant difference in the quantification of HPV-16 loads.
Fig 2.
Comparison of the HPV-16 quantification results obtained by the new method and the commercial Kehua kit. (A) Regression analysis of the two methods; (B) Bland-Altman analysis for the two methods. Solid lines, mean log10 unit difference; dotted lines, borders of the 95% confidence interval.
DISCUSSION
We developed a real-time PCR-based assay that could simultaneously detect, differentiate, and quantify 30 HPV types in three reactions, enabling multilayer information to be retrieved from a sample with no extra manipulations. Such an advantage is attributed to a mode of detection of one channel and multiple genotypes realized by the combined use of multicolor real-time PCR and melting curve analysis. By comparison, existing real-time PCR assays use a mode of detection of either one channel and one genotype or one channel and mixed genotypes. The former requires extra reactions for additional HPV types, while the latter, though it can detect more HPV genotypes in one channel, is unable to differentiate them.
The key feature of the described assay is the use of multiple type-specific probes with predefined Tm values in one reaction to identify the respective HPV genotypes. This was achieved by the use of the dually labeled, self-quenched probes (24), which allowed us to design probes with finely adjusted and well-separated Tms in one channel. The Tm values obtained through melting curve analysis proved to be highly reproducible under constant reaction conditions (31). Our results from 168 positive samples showed that all the Tm values obtained were concordant with their predefined values regardless of the HPV concentrations. This concordance in Tm value was also seen when we tested the same samples on other real-time PCR machines like the Stratagene 3005p (La Jolla, CA) and Bio-Rad CFX-96 (Hercules, CA) machines. A systematic Tm shift of 0.5 to 1.5°C was observed for certain HPV types due to the varied melting programs, as previously reported (32). By simply adjusting the predefined Tm values in these machines, genotyping could be performed as reproducibly as it could with the ABI 7500 machine.
There is a potential problem, in that an HPV type may be falsely identified if cross-reactivity between probes occurs or if intratype variants in HPV exist (18). To avoid any possible cross-reactivity, all probes were designed to have at least 4 to 5 bases different from each other so that no cross-hybridization could be measured at temperatures above 40°C. To overcome the destabilization effect caused by intratype variants, the probe binding region was chosen to make the variant sites close to the terminus. Otherwise, inosine was used to base pair with the variant site. The experiment with mimic targets demonstrated that no crossover with those intratype variant-containing targets took place, in spite of somewhat of a reduction in Tm. The clinical validation study showed that 3 samples had intratype variants, and none of them was misidentified.
Although the value of viral load is still controversial, inclusion of its measurement could improve the effectiveness of HPV-based screening and triage strategies (7). Moreover, a comprehensive method for quantification of common pathogenic HPV types is helpful to clarify the type-specific association between viral load and CIN lesions (8, 9, 28). As a real-time PCR assay, our method enabled quantification of HPV regardless of whether the sample was infected with single or multiple types. At the current stage, the quantification procedure for multiple infections seemed complex, but it could be simplified or even automated through a computer program with preinstalled calibration curves.
The clinical validation study was based on a large clinical sample consecutively enrolled from a population-based screening program. The results confirmed that the new method was comparable to the GenoArray method in HPV genotyping, even though our method had a higher positive detection rate. One reason for the higher positive rate of our method should be that our assay tested for more HPV types than the GenoArray method (30 versus 21 types). For example, among the 54 discrepant samples, 12 samples were found to be infected with HPV types not included in the GenoArray technique. Another reason might be that our assay has a higher analytical sensitivity than GenoArray. In the 48 discrepant typing results, 41 results agreed with those of our method. By looking back at their quantification data, we noticed that 23 of 41 samples with discrepant results had copy numbers between 50 and 3,000 copies per reaction; nevertheless, all of the samples were detected as negative by the GenoArray method. The limit of detection of our method was 5 to 50 copies per reaction for all the 30 HPV types, while that of the GenoArray was reported to be 10 to 50 copies per reaction for HPV-16 and HPV-18 only (15). In fact, GenoArray is based on the color intensity judged by the naked eyes, while our method is through fluorescence detection by real-time PCR. The subjective judgment of the GenoArray result might vary with the observer, and therefore, the exact limit of detection can differ from the reported one. Indeed, the user of GenoArray was often confused by the weak dot blots found in some samples. Nevertheless, there were 7 discrepant samples whose results turned out to be inconsistent with those of our method but agreed with those of sequencing. Among them, 4 of the samples had at least 1 concordant HPV type and therefore would not have been misclassified as HPV negative in a screening situation. The other 3 samples were all detected as positive by both GenoArray and sequencing but negative by our method. These false-negative results reported by our method could be due to DNA degradation during long-term storage (3 to 4 months) or some potential interference.
Compared with reported PCR-based methods, our method had the combined advantages of the real-time PCR and solid-phase hybridization-based methods, while it eliminated their respective shortcomings. As a real-time PCR-based method, it has advantages such as ease of use, high throughput, and the ability of quantitative detection. On the other hand, the melting curve analysis step offers full genotyping that so far has been achieved only by solid-phase hybridization. Currently, all real-time PCR-based commercial systems detect HPV-16/18 and non-16/18 types as well as an internal control in a single reaction. Thus, all those non-16/18 types are classified as one type, and therefore, the genotype information is lost (20–22). While type-specific real-time PCR can independently target one or a few types in one reaction, it will need multiple reactions when detecting dozens of HPV types. Consequently, when testing a large number of samples, the throughput would be substantially decreased, while the labor and the overall cost would be increased (23).
One important advantage of the new assay is the adjustability of its throughput in different clinical settings. For example, 30 samples could be tested per run in a standard 96-well real-time thermocycler, and 120 samples could be tested per run if the system is changed to a 384-well model. As most of the time is spent in the amplification stage, the combined use of extra standard PCR machines can further increase the throughput if quantification is omitted. In our experience, as many as 360 samples could be analyzed within 8 h by the combined use of 4 standard PCR machines and 1 real-time PCR machine. Such flexibility in throughput could satisfy clinical requirements of different levels. The throughput could be further increased if we used four instead of three detection channels. In this assay, our aim was to develop an assay that can be used on mainstream real-time PCR machines, and we therefore chose the three most commonly used channels, i.e., FAM, ROX, and Cy5. It is easy to add one channel to increase the number of HPV types that can be detected in one reaction. For example, if only 14 high-risk HPV types are detected, a single reaction can be designed to cover all 14 HPV types by using the principle of our current method on a 4-color real-time PCR thermocycler. The only constraint of this assay is that the Tm gap between two neighboring types should be large enough to avoid the influence of temperature fluctuation caused by the machine and the reaction system. Such a gap restricts the overall number of HPV types that may be differentiated within one channel. Lastly, the consumable cost of this assay is approximately $3 per sample, which is much cheaper than most existing commercial assays.
In summary, the described assay enabled simultaneous detection, typing, and quantification of 30 HPV types in 3 reactions. The probe-based detection nature ensured the reliability of this method, while the closed-tube operation mode significantly reduced post-PCR manipulations, time requirements, and carryover contamination. Furthermore, the high throughput and low cost would also facilitate its acceptance in clinical settings.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Key Project of Science and Technology Program of Fujian no. 2009D024 (to J.L. and Q.L).
We thank Qiuying Huang for manuscript revision. We also thank Lingyuan Kong and Liangkai Zheng for sample collection.
Footnotes
Published ahead of print 21 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02115-12.
REFERENCES
- 1. Walboomers JMM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijder PJF, Peto J, Meijer CJLM, Munoz N. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12–19 [DOI] [PubMed] [Google Scholar]
- 2. Woodman CBJ, Collins SI, Young LS. 2007. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer 7:11–22 [DOI] [PubMed] [Google Scholar]
- 3. De Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27 [DOI] [PubMed] [Google Scholar]
- 4. Muñoz N, Bosch FX, De Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJF, Meijer CJLM. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527 [DOI] [PubMed] [Google Scholar]
- 5. Roden R, Wu TC. 2006. How will HPV vaccines affect cervical cancer? Nat. Rev. Cancer 6:753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Josefsson AM, Magnusson PKE, Ylitalo N, Sørensen P, Qwarforth-Tubbin P, Andersen PK, Melbye M, Adami H, Gyllensten UB. 2000. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 355:2189–2193 [DOI] [PubMed] [Google Scholar]
- 7. Lörincz AT, Castle PE, Sherman ME, Scott DR, Glass AG, Wacholder S, Rush BB, Gravitt PE, Schussler JE, Schiffman M. 2002. Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet 360:228–229 [DOI] [PubMed] [Google Scholar]
- 8. Schlecht NF, Trevisan A, Duarte-Franco E, Rohan TE, Ferenczy A, Villa LL, Franco EL. 2003. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int. J. Cancer 103:519–524 [DOI] [PubMed] [Google Scholar]
- 9. Van Duin M, Snijders PJF, Schrijnemakers HFJ, Voorhorst FJ, Rozendaal L, Nobbenhuis MAE, van den Brule AJC, Verheijen RHM, Merhorst TJ, Meuer CJLM. 2002. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int. J. Cancer 98:590–595 [DOI] [PubMed] [Google Scholar]
- 10. Molijn A, Kleter B, Quint W, Doorn LJ. 2005. Molecular diagnosis of human papillomavirus (HPV) infections. J. Clin. Virol. 32:43–51 [DOI] [PubMed] [Google Scholar]
- 11. Trofatter KF. 1997. Diagnosis of human papillomavirus genital tract infection. Am. J. Med. 102:21–27 [DOI] [PubMed] [Google Scholar]
- 12. Lörincz AT. 1996. Hybrid Capture™ method for detection of human papillomavirus DNA in clinical specimens: a tool for clinical management of equivocal Pap smears and for population screening. J. Obstet. Gynaecol. Res. 22:629–636 [DOI] [PubMed] [Google Scholar]
- 13. Gravitt P, Peyton C, Apple R, Wheeler C. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kleter B, Van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, Ter Schegget J, Lindeman J, Harmsel BT, Burger M, Quint W. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu SS, Leung RCY, Chan KKL, Cheung ANY, Ngan HYS. 2010. Evaluation of a newly developed GenoArray human papillomavirus (HPV) genotyping assay and comparison with the Roche Linear Array HPV genotyping assay. J. Clin. Microbiol. 48:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong SP, Shin SK, Lee EH, Kim EO, Ji SI, Chung HJ, Park SN, Yoo W, Folk WR, Kim S. 2008. High-resolution human papillomavirus genotyping by MALDI-TOF mass spectrometry. Nat. Protoc. 3:1476–1484 [DOI] [PubMed] [Google Scholar]
- 17. Söderlund-Strand A, Dillner J, Carlson J. 2008. High-throughput genotyping of oncogenic human papilloma viruses with MALDI-TOF mass spectrometry. Clin. Chem. 54:86–92 [DOI] [PubMed] [Google Scholar]
- 18. Barzon L, Militello V, Lavezzo E, Franchin E, Peta E, Squarzon L, Trevisan M, Pagni S, Bello FD, Toppo S, Palù G. 2011. Human papillomavirus genotyping by 454 next generation sequencing technology. J. Clin. Virol. 52:93–97 [DOI] [PubMed] [Google Scholar]
- 19. Gharizadeh B, Kalantari M, Garcia CA, Johansson B, Nyrén P. 2001. Typing of human papillomavirus by pyrosequencing. Lab. Invest. 81:673–679 [DOI] [PubMed] [Google Scholar]
- 20. Heideman D, Hesselink A, Berkhof J, van Kemenade F, Melchers W, Daalmeijer NF, Verkuijten M, Meijer CJLM, Snijders PJF. 2011. Clinical validation of the Cobas 4800 HPV test for cervical screening purposes. J. Clin. Microbiol. 49:3983–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cuzick J, Ambroisine L, Cadman L, Austin J, Ho L, Terry G, Liddle S, Dina R, McCarthy J, Buckley H, Bergeron C, Soutter WP, Lyons D, Szarewski A. 2010. Performance of the Abbott real time high-risk HPV test in women with abnormal cervical cytology smears. J. Med. Virol. 82:1186–1191 [DOI] [PubMed] [Google Scholar]
- 22. Hwang Y, Lee M. 2012. Comparison of the AdvanSure human papillomavirus screening real-time PCR, the Abbott real time high risk human papillomavirus test, and the Hybrid Capture human papillomavirus DNA test for the detection of human papillomavirus. Ann. Lab. Med. 32:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Micalessi IM, Boulet GA, Bogers JJ, Benoy IH, Depuydt CE. 2012. High-throughput detection, genotyping and quantification of the human papillomavirus using real-time PCR. Clin. Chem. Lab. Med. 50:655–661 [DOI] [PubMed] [Google Scholar]
- 24. Huang Q, Liu Z, Liao Y, Chen X, Zhang Y, Li Q. 2011. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS One 6:e19206 doi:10.1371/journal.pone.0019206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Roda Husman AM, Walboomers JMM, van den Brule AJC, Meijer CJLM, Snijders PJF. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057–1062 [DOI] [PubMed] [Google Scholar]
- 26. Zhou L, Palais RA, Smith GD, Anderson D, Rowe LR, Wittwer CT. 2010. Enrichment and detection of rare alleles by means of snapback primers and rapid-cycle PCR. Clin. Chem. 56:814–822 [DOI] [PubMed] [Google Scholar]
- 27. Van den Brule AJ, Meijer CJ, Bakels V, Kenemans P, Walboomers J. 1990. Rapid detection of human papillomavirus in cervical scrapes by combined general primer-mediated and type-specific polymerase chain reaction. J. Clin. Microbiol. 28:2739–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swan DC, Tucker RA, Tortolero-Luna G, Mitchell MF, Wideroff L, Unger ER, Nisenbaum RA, Reeves WC, Icenogle JP. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J. Clin. Microbiol. 37:1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Draper NR, Smith H, Pownell E. 1966. Applied regression analysis, vol 3 John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 30. Bland MJ, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327:307–310 [PubMed] [Google Scholar]
- 31. Lyon E, Millson A, Phan T, Wittwer CT. 1998. Detection and identification of base alterations within the region of factor V Leiden by fluorescent melting curves. Mol. Diagn. 3:203–210 [DOI] [PubMed] [Google Scholar]
- 32. Lay MJ, Wittwer CT. 1997. Real-time fluorescence genotyping of factor V Leiden during rapid-cycle PCR. Clin. Chem. 43:2262–2267 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.