Abstract
Strain comparison is important to population genetics and to evaluate relapses in patients with Mycobacterium avium complex (MAC) lung disease, but the “gold standard” of pulsed-field gel electrophoresis (PFGE) is time-consuming and complex. We used variable-number tandem repeats (VNTR) for fingerprinting of respiratory isolates of M. intracellulare from patients with underlying bronchiectasis, to establish a nonsequence-based database for population analysis. Different genotypes identified by PFGE underwent species identification using a 16S rRNA gene multiplex PCR. Genotypes of M. intracellulare were confirmed by internal transcribed spacer 1 (ITS1) sequencing and characterized using seven VNTR primers. The pattern of VNTR amplicon sizes and repeat number defined each specific VNTR type. Forty-two VNTR types were identified among 84 genotypes. PFGE revealed most isolates with the same VNTR type to be clonal or exhibit similar grouping of bands. Repetitive sequence-based PCR (rep-PCR) showed minimal pattern diversity between VNTR types compared to PFGE. Fingerprinting of relapse isolates from 31 treated patients using VNTR combined with 16S multiplex PCR unambiguously and reliably distinguished different genotypes from the same patient, with results comparable to those of PFGE. VNTR for strain comparison is easier and faster than PFGE, is as accurate as PFGE, and does not require sequencing. Starting with a collection of 167 M. intracellulare isolates, VNTR distinguished M. intracellulare into 42 clonal groups. Comparison of isolates from different geographic areas, habitats, and clinical settings is now possible.
INTRODUCTION
The ability to compare different strains of microorganisms, including nontuberculous mycobacteria (NTM) of the same species, is essential for understanding sources of disease, clinical outbreaks, disease relapses, and population dynamics. This need is especially important for environmental species where exposure to individual strains may occur frequently.
Efforts to compare isolates of Mycobacterium intracellulare have been difficult, as the species has no known unique, multicopy insertional elements, and until recently the genome had not been sequenced (1–3). Strain comparison of this species has been dependent upon pulsed-field gel electrophoresis (PFGE) (4, 5), which is expensive, technically difficult, and time-consuming. Other methods such as repetitive sequence-based PCR (rep-PCR) (6, 7) and gene sequencing have also been employed.
In 2008, sequences of the type strain of M. intracellulare ATCC 13950T became publicly available as 353 contiguous consensus sequences (http://www.ncbi.nlm.nih.gov/nuccore/NZ_ABIN00000000.1), but the sequence was only published as a complete sequence in 2012 (1). Variable-number tandem (i.e., consecutive) repeats (VNTR) of the minisatellite class (10 to 100 bp) were identified. VNTR typing has been used for strain comparison and population studies of other species of mycobacteria, including Mycobacterium ulcerans, Mycobacterium avium, and Mycobacterium tuberculosis (8–15). Subsequently, studies of the use of VNTR for M. intracellulare isolates were reported in 2009 and 2010 from Japan (16) and (Bordeaux) France (17), and the candidate markers were described as mycobacterial interspersed repetitive unit–variable-number tandem repeats (MIRU-VNTR) (17). Those studies looked at approximate VNTR sizes based on sequencing of M. intracellulare populations from the two countries. They did not utilize estimated amplicon sizes as an additional strategy, did not look at isolates of the same VNTR type for strain relatedness using other typing techniques, and did not use VNTR typing for strain comparison except for a small number of patients.
To expand upon their work, we utilized M. intracellulare MIRU-VNTR for strain comparison in isolates from the same patients, and we utilized the composite data to create a database of local isolates VNTR type, amplicon size, and alleles generated by using all of the above data.
MATERIALS AND METHODS
Patients.
The University of Texas Health Science Center at Tyler (UTHSCT) patients were being monitored in the bronchiectasis and NTM clinic. Patient isolates were candidates for study if the patient had nodular bronchiectasis that met current American Thoracic Society/Infectious Diseases Society of America (ATS/IDSA) 2007 guidelines for clinical disease (18). Some patients and their initial isolates had been included in previous clinical studies from this institution (4, 5). Clinical information was extracted from medical records. This study was approved by the Institutional Review Board of the UTHSCT.
Isolates.
Patients with nodular bronchiectasis whose isolates were submitted for strain comparison because of microbiologic relapse during or after drug treatment of their infection were chosen for the fingerprint portion of this study. Current isolates were compared to previous isolates stored at −70°C or in their original broth culture vial, which had been stored at room temperature following primary isolation.
Sputum cultures from patients had been submitted and processed by the clinical mycobacteriology lab at the UTHSCT, as previously described (4, 5).
A recent study compared patient and household water isolates of M. avium complex (MAC) isolates (6) using rep-PCR. Selected patient isolates from that study were also chosen for the population portion of this study. The type strains of M. intracellulare, ATCC 13950T, and M. chimaera, DSM 44623T, were included as controls.
Species identification.
Isolates for study were identified as MAC using a commercial hybridization assay (AccuProbe) (Hologic Gen Probe, Inc., San Diego, CA). They were then identified as M. avium, M. intracellulare, or MAC “X” using the multiplex 16S rRNA gene PCR technique described by Wilton and Cousins (18a). This method does not separate M. intracellulare from the closely related M. chimaera (19). To separate these two species, sequencing of the internal transcribed spacer (ITS) region was performed (19, 20).
PFGE.
PFGE was performed as previously described using restriction enzymes XbaI and AseI (4, 5, 21). Isolates from the same patient were compared and categorized as “indistinguishable,” “probably related,” or “not related” using differences in band number and position by methods originally described by Tenover et al. for bacterial outbreaks (22) and then modified for M. intracellulare (4). Strains in the first two categories were considered clonal. Different (not related) genotypes from the same patient were given different letter designations starting with A, then B, and then C, etc. Numbers were added to letter designations (e.g., A2) if fewer than seven band differences with each restriction enzyme were observed.
VNTR.
All different PFGE genotypes (“not related” patterns) from individual patients (range, 2 to 5) and patients with a single isolate or genotype underwent VNTR analysis.
Mycobacterial DNA was obtained with PrepMan Ultra (Applied Biosystems, Carlsbad, CA) according to the manufacturer's protocol. The DNA from the supernatant was directly used as a template for identification of M. intracellulare isolates using multiplex PCR with designed primers (18a).
The size of PCR products was estimated by electrophoresis in 2% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer using a 100-bp ladder (Life Technologies, formerly Invitrogen, Carlsbad, CA).
rep-PCR.
Primers previously described by Versalovic et al. (23) were used for rep-PCR (9).
Visual strain comparison of rep-PCR band patterns utilized definitions previously published for the use of random primers with isolates of Mycobacterium abscessus (21). Isolates were considered “indistinguishable” if all major bands were the same, with four or more major bands run on the same gel. They were called “probably related” if they differed by one major band and “not related” if they differed by two or more major bands. As with random primers (21), isolates in the “indistinguishable” and “probably related” categories with rep-PCR could be found to be unrelated by PFGE, as some isolates yielded a low number of rep-PCR bands. (This definition was based on the use of a minimum of three random primers. The validity of its use with the single rep-PCR has not been studied without the use of molecular software.)
DNA fingerprinting of isolates with the same VNTR type using PFGE and rep-PCR.
M. intracellulare isolates from different patients subsequently shown to belong to the same VNTR type were compared to each other using PFGE, rep-PCR, and ITS sequencing. The same three PFGE categories of relatedness were used as with intrapatient isolates, with a fourth category of “similar grouping” describing isolates from different patients whose grouping of DNA fragments on the sizing gel looked similar, but the number of individual band differences exceeded seven.
DNA fingerprinting of relapse isolates of M. intracellulare from the same patient.
Patients who underwent relapse after or during drug treatment and who had at least one isolate of M. intracellulare had the initial and relapse isolates compared by multiplex 16S rRNA gene PCR, PFGE, and VNTR fingerprinting.
Isolates were considered different if they belonged to different species or met the PFGE criteria for “not related” (4, 5). Isolates were considered the same if they belonged to the same species and were “indistinguishable” or “probably related” by PFGE. Results were then compared to the VNTR fingerprinting results for isolates of M. intracellulare.
DNA sequencing.
Sequencing of the ITS region (263 bp) of each M. intracellulare isolate was performed as previously described (19, 20, 24). The sequences were compared to GenBank sequences, including the Min-A sequence for M. intracellulare ATCC 13950T (AB026691) and the MAC-A sequence for the type strain of M. chimaera DSM 44623T (EF521902) (19, 20, 24). Examples of each of the different-size alleles for each of the seven primers were also sequenced to determine the exact tandem-repeat size.
Comparisons with pulmonary isolates from patients from the Bordeaux region of France.
The Texas isolates were numbered by rounding each tandem-repeat size up to the next whole number. This was the technique for strain comparison used for the 52 isolates from France reported by Dauchy et al. (17) and the same seven VNTRs. ATCC 13950T was included as a control, as it was sized in both studies.
Nucleotide sequence accession numbers.
The ITS1 rRNA gene sequences for the newly encountered Min-E and Min-F found in M. intracellulare have been deposited in GenBank with accession numbers KC018476 and KC018477, respectively.
RESULTS
Patients.
A total of 56 patients (including the 2 patients who yielded the ATCC isolates) contributed isolates to the study. The 48 patients from the UTHSCT were typical of older patients with nodular bronchiectasis and comparable to previous descriptions (25). All met the American Thoracic Society/Infectious Diseases Society of America guidelines for NTM lung disease (18).
Species identification.
A total of 167 isolates were identified as M. intracellulare by different means and were utilized in the study. The human type strain of M. intracellulare ATCC 13950T was included as a control.
PFGE.
Isolates of M. intracellulare from the same patient were compared using PFGE to determine the number of unrelated genotypes. PFGE revealed 84 genotypes among the 167 isolates of M. intracellulare. Individual patients had up to four different genotypes. The average time for completion of PFGE from receipt of the initial and relapse cultures was approximately 3 weeks (S. McNulty, unpublished observations).
VNTR.
A range of between 1 and 7 isolates (mean, 1.9 isolates) shared the same PFGE genotype from the same patient and underwent VNTR typing (167 isolates). All 84 PFGE genotypes of M. intracellulare produced VNTR amplicons satisfactorily for gel sizing and DNA sequencing. Examples of nine different-size amplicons with different copy number tandem repeats for MIN22 is shown in Fig. 1.
Fig 1.
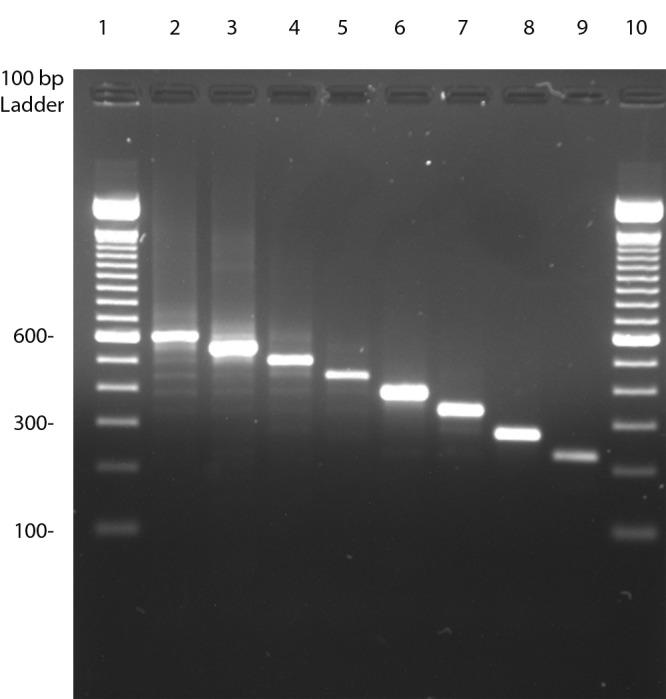
Examples of eight different amplicon sizes associated with eight different tandem-repeat copy numbers for MIN22. The amplicons from lanes 2 to 8 are from isolates of M. intracellulare, while that in lane 9 is from another MAC species (MAC “X”). The gel was performed with 2% agarose in 1× TAE buffer. Lane 1, 100-bp ladder; lane 2, strain 3068 (6.93 copies) (600 bp); lane 3, strain 1760 (5.9 copies) (550 bp); lane 4, strain 07-3369 (4.94 copies) (500 bp); lane 5, ATCC 13950T (3.93 copies) (450 bp); lane 6, strain 2788 (3.04 copies) (400 bp); lane 7, strain 0416 (2.3 copies) (340 bp); lane 8, strain 289-0316 (0.93 copies) (275 bp); lane 9, water isolate LH-Sw-11-2 (0.16 copy) (220 bp); lane 10, 100-bp ladder.
The different copy numbers and amplicon sizes observed for each tandem repeat were given allele numbers according to the size that reflected their complete copy number (partial copies were not included) (17). Amplicons from each primer set were grouped together as a single allele if their base pair size difference did not allow separation on the sizing gel (generally <20 bp) (Table 1). This resulted in 42 VNTR types (Table 2). Most types (28/42 [67%]) consisted of a single genotype from one patient. Only 14 VNTR types included multiple PFGE genotypes, and only 5 VNTR types (types 5, 11, 24, 37, and 40) (11.9%) included four or more isolates from different patients. The average time for completion of VNTR typing from receipt of initial and relapse isolates was 6 to 8 h (E. Iakhiaeva, unpublished observations).
Table 1.
VNTR copy number, amplicon sizes, and allele designation seen in isolates of M. intracellulare with seven M. intracellulare tandem repeats from Dauchy et al. (17)a
| (Allele) copy no./amplicon size (bp) for each tandem repeat | ||||||
|---|---|---|---|---|---|---|
| MIN22 (57 bp) | MIN20 (55 bp) | MIN18 (56 bp) | MIN19 (53 (bp) | MIN33 (54 bp) | MIN31 (57 bp) | MIRU3 (53 bp) |
| (7) 7.9/635 | ||||||
| (6) 6.93/600 | ||||||
| (5) 5.9/550 | (5) 5.13/620 | |||||
| (4) 4.94/500 | (4) 4.24/560 | (4) 4.13/550 | (4) 4.7/450 | (4) 4.13/595 | ||
| (3a) 3.93/450 | ||||||
| (3) 3.04/400 | (3) 3.24/500 | (3) 3.13/500 | (3) 3.74/390 | (3) 3.13/530 | (3) 3.43/400 | |
| (2) 2.2/445 | ||||||
| (2) 2.61/490 | (2) 2.11/445 | |||||
| (2) 2.3/340 | (2) 2.24/475 | (2) 2.7/330 | (2) 2.17/480 | (2) 2.05/440 | (2) 2.43/350 | |
| (1a) 1.96/425 | (1) 1.11/450 | |||||
| (1) 1.25/400 | (1) 1.13/380 | (1) 1.74/220 | (1) 1.13/420 | (1) 1.05/375 | (1) 1.43/290 | |
| (0a) 0.93/275 | (0a) 0.24/350 | (0a) 0.74/220 | (0a) 0.42/335 | (0a) 0.69/250 | ||
| (0) 0/200 | (0) 0/320 | (0) 0/200 | (0) 0/200 | (0) 0/350 | (0) 0.280 | (0) 0/200 |
The allele number in parentheses represents the tandem-repeat complete copy number, with partial values not included. Consecutive alleles generally differ by one complete copy number (e.g., for MIN22, copy numbers are 7.9, 6.9, 5.9, 4.9, etc.).An “a” indicates a second amplicon size that contains the same number of complete copies. The last row contains copy numbers/amplicon sizes that contained no tandem-repeat sequences in group 0 that were reported by Dauchy et al. (17) but were not seen in the current study.
Table 2.
VNTR types among M. intracellulare isolates from patients with nodular bronchiectasis using the seven tandem repeats described by Dauchy et al. (17)a
| VNTR type | Complete tandem repeat no. |
ITS1 (100% identity) | No. of isolates | No. of patientsb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIN22 | MIN20 | MIN18 | MIN19 | MIN33 | MIN31 | MIRU3 | ||||
| 1 | 7 | 2 | 3 | 2 | 2 | 2 | 1 | Min-A | 1 | 1 |
| 2 | 6 | 2 | 3 | 0 | 5 | 2 | 4 | Min-A | 2 | 2 |
| 3 | 5 | 2 | 1 | 1 | 1 | 2 | 2 | Min-A | 7 | 1 |
| 4 | 4 | 4 | 3 | 4 | 3 | 2 | 4 | Min-A | 1 | 1 |
| 5 | 5 | 4 | 3 | 3 | 3 | 2 | 3 | Min-A | 10 | 5 |
| 6 | 4 | 4 | 3 | 3 | 3 | 2 | 2 | Min-A | 4 | 1 |
| 7 | 4 | 4 | 3 | 2 | 3 | 2 | 3 | Min-A | 2 | 2 |
| 8 | 4 | 4 | 1 | 3 | 4 | 1 | 3 | Min-A | 1 | 1 |
| 9 | 4 | 2 | 3 | 3 | 3 | 2 | 3 | Min-A | 4 | 1 |
| 10 | 3a | 4 | 3 | 3 | 4 | 2 | 1 | Min-A | 4 | 3 |
| 11c | 3a | 4 | 3 | 3 | 3 | 2 | 3 | Min-A | 15 | 7 |
| 11a | 3a | 4 | 3 | 2 | 5 | 2 | 1 | Min-A | 1 | 1 |
| 12 | 3a | 4 | 1 | 3 | 3 | 2 | 3 | Min-A | 1 | 1 |
| 13 | 3a | 3 | 3 | 3 | 5 | 2 | 2 | Min-A | 1 | 1 |
| 14 | 3a | 3 | 3 | 3 | 4 | 2 | 3 | Min-A | 5 | 2 |
| 15 | 3a | 3 | 3 | 2 | 3 | 2 | 3 | Min-A | 4 | 1 |
| 16 | 3a | 2 | 3 | 2 | 4 | 2 | 1 | Min-A | 9 | 3 |
| 17 | 3a | 2 | 3 | 2 | 4 | 1 | 3 | Min-A | 1 | 1 |
| 18 | 3a | 2 | 3 | 2 | 3 | 1 | 0 | Min-A | 1 | 1 |
| 19 | 3a | 2 | 1 | 3 + 2 | 3 + 4 | 2 | 2 | Min-A | 3 | 1 |
| 20 | 3a | 2 | 1 | 3 | 3 | 1 | 1 | Min-E (new) (all 3 patients) | 5 | 3 |
| 21 | 3a | 2 | 1 | 1 | 4 | 2 | 0 | Min-A | 4 | 1 |
| 22 | 3a | 1 | 4 | 2 | 3 | 1 | 1 | Min-A | 3 | 1 |
| 23 | 3a | 1 | 3 | 3 | 4 | 2 | 0 | Min-A | 1 | 1 |
| 24 | 3a | 1 | 1 | 3 | 4 | 1 | 2 | Min-A | 9 | 5 |
| 25 | 3a | 12 | 1 | 3 | 4 | 0 | 2 | Min-A | 2 | 1 |
| 26 | 3a | 1 | 1 | 2 | 3 | 1 | 2 | Min-A | 9 | 3 |
| 27 | 3a | 1 | 1 | 1 | 3 | 1 | 2 | Min-A | 1 | 1 |
| 27a | 3a | 0a | 3 | 3 | 4 | 2 | 2 | Min-A | 1 | 1 |
| 28 | 3 | 2 | 1a | 0 | 3 | 2 | 0 | Min-B | 1 | 1 |
| 29 | 2 | 1 | 1 | 3 | 4 | 1 | 2 | Min-A | 1 | 1 |
| 30 | 2 | 1 | 1 | 2 | 4 | 2 | 0 | Min-F (new) | 1 | 1 |
| 31 | 0a | 4 | 3 | 2 | 3 | 2 | 2 | Min-A | 1 | 1 |
| 32 | 0a | 3 | 3 | 3 | 3 | 1 | 0 | Min-A | 1 | 1 |
| 33 | 0a | 3 | 1a | 2 | 4 | 2 | 0 | Min-A | 1 | 1 |
| 34 | 0a | 2 | 3 | 1 | 3 | 2 | 0 | Min-A | 2 | 1 |
| 35 | 0a | 2 | 3 | 3 | 3 | 1 | 0 | Min-A | 2 | 1 |
| 36 | 0a | 2 | 1a | 2 | 2 | 2 | 0 | Min-A | 10 | 3 |
| 37 | 0a | 2 | 1a | 0 | 2 | 2 | 0 | Min-A | 13 | 7 |
| 38 | 0a | 2 | 1 | 1 | 4 | 2 | 0 | Min-A | 13 | 3 |
| 39 | 0a | 0a | 1a | 1 | 1 | 0 | 1 | Min-A | 1 | 1 |
| 40 | 0a | 0a | 1a | 0 | 1 | 0 | 1 | Min-A | 15 | 6 |
| Totals | 176 | 82 | ||||||||
Some patients had more than one isolate tested. All VNTR types were validated as M. intracellulare by sequencing of the ITS1 region.
Some patients are included in more than one VNTR type.
The VNTR type and ITS1 sequence match M. intracellulare ATCC 13950T.
rep-PCR.
rep-PCR was very reproducible, with both transparent and opaque colony variants from a single genotype done on three separate occasions with different growth times and DNA preparation showing indistinguishable patterns (data not shown).
Because little comparison between the results of rep-PCR and other typing techniques has been published, we performed rep-PCR on nine different VNTR types and compared the results to those obtained by PFGE. There were five major bands with the M. intracellulare isolates, and all bands were present with most VNTR types. Figure 2A shows the striking similarity of isolates of M. intracellulare (lanes 2 to 9 and 12) despite the fact that each pattern came from a different VNTR type. Lane 3 (type 7), lane 4 (type 11), lane 5 (type 16), lane 7 (type 38), lane 8 (type 36), and lane 9 (type 37) have “indistinguishable” patterns based on the five major bands. Single major band differences are seen in lane 2 (type 5) and lane 12 (type 34), with two major band differences in lane 6 (type 24). The two isolates with six major band differences from all other isolates (lanes 10 and 11) both belonged to VNTR type 41. The differences between different VNTR types of this same isolate collection were much greater using PFGE (Fig. 2B).
Fig 2.
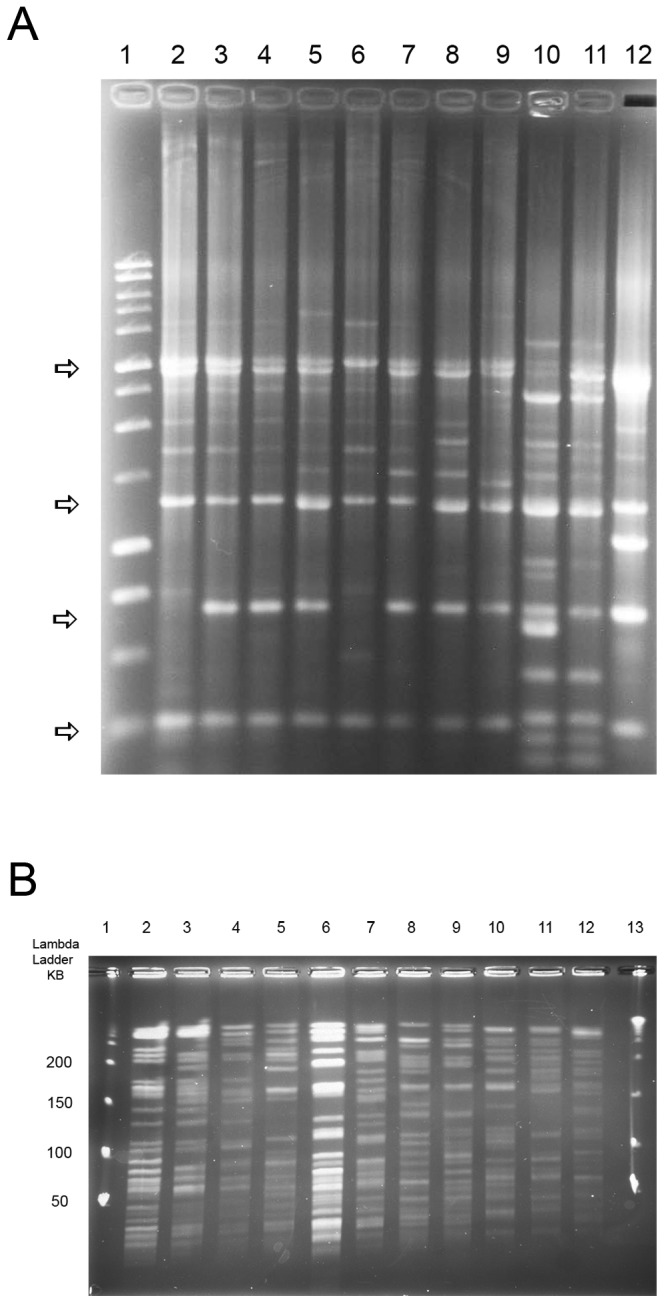
(A) rep-PCR of representative isolates from 9 different VNTR types of M. intracellulare. Note the overall similarity of the four major bands of M. intracellulare (marked by the arrows) despite isolates from multiple different VNTR types. Lane 1, 1-kb ladder; lane 2, type 5; lane 3, type 7; lane 4, type 11; lane 5, type 16; lane 6, type 24; lane 7, type 38; lane 8, type 36; lane 9, type 37; lanes 10 and 11, M. chimaera controls; lane 12, type 34. Lanes 2 to 9 and lane 12 are M. intracellulare. (B) PFGE using XbaI of representative isolates from the same 9 VNTR types of M. intracellulare and the two M. chimaera controls studied by rep-PCR for panel A. Note the overall difference of isolates from different VNTR types which were not as apparent with rep-PCR (A). Lanes 1 and 13, λ ladder; lane 2, type 5; lane 3, type 7; lane 4, type 11; lane 5, type 16; lane 6, type 24; lane 7, type 38; lane 8, type 36; lane 9, type 37; lanes 10 and 11, M. chimaera; lane 12, type 34. Lanes 2 to 9 and lane 12 are M. intracellulare.
DNA fingerprinting of relapse isolates of MAC from the same patient using multiplex 16S rRNA gene PCR, PFGE, and VNTR.
A total of 31 patients who relapsed with positive cultures one or more times after sputum conversion to negative cultures (4) and who had one or more isolates of M. intracellulare among both the initial and relapse isolates were studied. For ease of comparison, the first M. intracellulare genotype was considered the initial isolate and designated PFGE pattern A (see Table S1 in the supplemental material). Among the 31 isolates of M. intracellulare in both the initial and relapse categories, there were 11 episodes in which the isolates met the definition of indistinguishable (“relapse”) and 19 episodes in which the isolates met the PFGE definition of “unrelated” (“new infection”), and two patients with both unrelated and relapse genotypes. There was one patient for whom the results of PFGE and VNTR typing were discrepant, with the same VNTR type but an unrelated PFGE pattern. The PFGE and VNTR typing were done by different individuals who were unaware of the other results until the fingerprinting was complete. (The results are shown in Table S1 in the supplemental material.)
DNA fingerprinting of isolates with the same VNTR type.
PFGE and ITS1 sequencing was performed on the 14 VNTR types with two or more isolates from different patients (range, 2 to 10). Almost all isolates within a VNTR type were clonal by PFGE (Fig. 3; see also Table S2 in the supplemental material).
Fig 3.
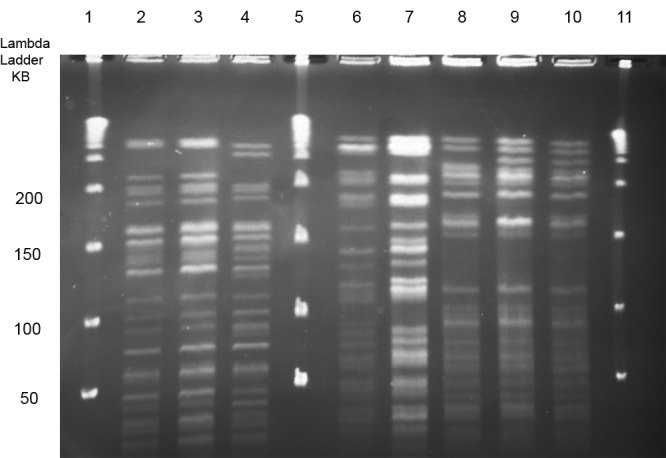
PFGE using Xba of three M. intracellulare VNTR types (types 10, 14, and 16) with isolates from different patients. Note the identity or close similarity (clonality) of the patterns within the three types but the unrelated band patterns between the different types. Lanes 1, 5, and 11 contain DNA ladder; lanes 2, 3, and 4, VNTR type 10; lanes 6 and 7; VNTR type 14; and lanes 8, 9, and 10, VNTR 16.
Sequencing.
All 84 genotypes of M. intracellulare that fell into the 42 VNTR types underwent ITS1 region sequencing. The two control strains of M. chimaera (including the type strain) matched the MAC-A ITS1 sequence (20) of the M. chimaera type strain, DSM 44623T (GenBank sequence no. LO7847). Of the remaining 82 PFGE genotypes of M. intracellulare, 77 (93.9%) had 100% sequence identity to Min-A (i.e., ATCC 13950T; GenBank sequence no. L07859), one genotype in VNTR 28 (1.2%) matched Min-B (GenBank sequence no. Z46423), three patient isolates (3.1%) in VNTR 20 belonged to a new sequence designated Min-E (GenBank sequence no. KC018476), and a single patient isolate in VNTR 30 (1.2%) belonged to a second new sequence designated Min-F (GenBank sequence no. KC018477). No isolates with Min-C or Min-D were identified.
Comparison of VNTR results of Texas isolates with those of isolates from France.
VNTR results of the Texas isolates described herein were also compared with those of a group of isolates from France (17) (Table 3). The type strain ATCC 13950T was used as a control and gave the same tandem-repeat copy number for all seven alleles in both studies. Overall, the same tandem-repeat sizes were found in isolates from both collections, except that isolates with no partial or complete tandem repeats (the amplicon consisted only of the flanking regions) were more common among the French strains (54/357 alleles [15.1%]) than among the Texas isolates (0/357 alleles [<0.3%]). A number of sequenced tandem repeats gave base pair sizes that differed from a whole copy number by only a few bases (Table 1), making an error in rounding up or rounding down a concern. This was especially true because only one control was used. The use of multiple controls of isolates whose sequence length varied from a whole number by only 1 or 2 bp would have provided strong confirmation that measured differences in each rounded-up copy number were real.
Table 3.
Number of isolates with each VNTR allele based on rounding off each patient copy number to the next whole number and then comparing the current isolates with previously reported French isolates (17)a
| Rounded-up VNTR copy no. | VNTR size (rounded-off next whole no.) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIN22 |
MIN20 |
MIN18 |
MIN19 |
MIN33 |
MIN31 |
MIRU3 |
||||||||
| Fr | Tx | Fr | Tx | Fr | Tx | Fr | Tx | Fr | Tx | Fr | Tx | Fr | Tx | |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 9 | 0 | 0 | 0 | 1 |
| 5 | 0 | 11 | 4b | 23b | 0 | 2 | 0 | 1 | 6 | 20 | 0 | 0 | 0 | 0 |
| 4 | 13b | 39b | 19 | 7 | 15b | 38b | 8b | 37b | 12b | 33b | 0 | 0 | 0 | 3 |
| 3 | 10 | 2 | 7 | 32 | 7 | 0 | 21 | 20 | 5 | 12 | 29b | 58b | 13b | 36b |
| 2 | 24 | 0 | 2 | 13 | 19 | 41 | 0 | 7 | 6 | 7 | 15 | 17 | 17 | 11 |
| 1 | 2 | 25 | 19 | 6 | 1 | 0 | 1 | 16 | 0 | 0 | 3 | 5 | 13 | 31 |
| 0 | 3 | 0 | 1 | 0 | 10 | 0 | 22 | 0 | 4 | 0 | 5 | 0 | 9 | 0 |
| Total | 52 | 81 | 52 | 81 | 52 | 81 | 52 | 81 | 52 | 81 | 52 | 81 | 52 | 81 |
These values may not be predictive of the number or nature of the VNTR types from the two locations, as the French isolates were not characterized as to type. Fr, French isolates from the Bordeaux region (17). Tx, U.S. isolates from Northeast Texas region (current study).
Copy number of ATCC 13950T determined in both studies.
DISCUSSION
VNTR typing has major advantages over PFGE. Typing can be done in a single day using only PCR and a sizing agarose gel. With PFGE, the average time to complete a gel in our laboratory is 3 weeks, and one or more PFGE instruments (about $20,000 each) are required. PFGE requires much more operator experience to produce reproducible results and interpretable gels.
We studied respiratory tract isolates from patients with nodular bronchiectasis and MAC lung disease who are being monitored by one of us (R.J.W.). Most isolates were recovered in the UTHSCT mycobacterial laboratory, and most patients lived within a 200-mi radius of Northeast Texas. We identified 42 MIRU-VNTR types based on the combination of the seven loci. Fourteen MIRU-VNTR types (33%) contained isolates from multiple patients (range, 2 to 10), while 28 types (67%) were unique to only a single patient.
All but five isolates of M. intracellulare (171/176 [97.2%]) contained the proven or presumptive Min-A ITS1 sequence of the M. intracellulare type strain, ATCC 13950T. (Of multiple isolates from the same patient of the same VNTR type, only one was sequenced and the remainder were considered presumptive.) Similar ITS1 sequence findings in the same patient population were reported by Stout et al. (14). Of the five isolates of M. intracellulare with different ITS1 sequences, one belonged to the previously described Min-B (GenBank accession number 246423), and four belonged to two new ITS1 sequences designated Min-E and Min-F.
In 2010, Dauchy et al. published an MIRU-VNTR study of 61 M. intracellulare isolates collected from 51 French patients between 2001 and 2008, as well as the M. intracellulare type strain, ATCC 13950T (17). Six isolates were from Brittany, while the remainder were recovered from patients at Bordeaux University Hospital (France). Multiple isolates over time were collected from eight patients. The majority of isolates (51/61 isolates [84%]) were from the respiratory tract. Fifty-one of the Bordeaux isolates and the ATCC type strain met the ATS criteria for NTM clinical disease (18). The second method used for strain comparison (PCR-restriction fragment length polymorphism [PCR-RFLP]), described by Picardeau and Vincent (26), was not generally successful with M. intracellulare isolates (17). The designation of VNTR types was not done; rather, the authors reported the number of isolates for each VNTR primer combination.
The current data are similar to those obtained in the study by Dauchy et al. (17) that identified 44 VNTR using the same primers, but they differ in that the French study found a much higher frequency of types having a single isolate (40/44 [91%], compared to 28/43 [65.9%] in the current study) (17). Further, the current study had no VNTR alleles without any partial tandem-repeat sequence (0/644 [<0. 2% of all alleles among 92 genotypes]), in contrast to the French study, in which all primer combinations had some alleles or isolates with no tandem-repeat sequence (54/364 [14% of all alleles among 52 separate isolates]).
In 2010, a second study of the use of MIRU-VNTR for M. intracellulare from Nagoya University Hospital (Japan) was published by Ichikawa et al. (16). These authors utilized 16 different VNTR loci to separate 74 clinical isolates into 50 MIRU-VNTR types (16). These results are not comparable to those obtained in the current study or the study from France (17), as different VNTRs were examined. These VNTRs were not chosen for the current study because the individual repeats were less discriminatory than those characterized by Dauchy et al. (17), and the number of primers (16 versus 7) would have made this study more complex.
There are two motivations for performing comparisons of M. intracellulare isolates. The first is to determine the basis for a drug relapse in a single patient. Second, comparison of environmental and clinical isolates is essential to understanding disease sources with an eye toward preventing or limiting exposure for high risk patients. Comprehensive, large-scale studies have not yet been possible because strain comparisons by PFGE are too expensive or unproven (rep-PCR). Understanding the basis for relapse is critical to patient care. Treatment of a true relapse (same genotype) usually requires addition of new drugs (especially an injectable such as streptomycin) and longer therapy. Treatment of a new infection requires the same standard therapy and treatment duration.
Such strain comparison is relatively easy using two PCR techniques. The first is the 16S rRNA gene multiplex PCR using the primers of Wilton and Cousins (18a) to determine the grouping of the initial and relapse isolates. If different MAC species, subspecies, or types are identified, no additional testing is needed. If both initial and relapse isolates (or, preferred, multiple isolates from both periods given the frequent polymicrobial nature of these infections) (5) are M. intracellulare, then VNTR fingerprinting using the current primers with the original and relapse isolates will provide an answer. In most cases, sequencing is not needed, as the amplicon sizes of the original and relapse isolates can be run side by side on a gel. It will be evident whether the isolates are the same or different. Species identification is important, as some of these primers will not amplify other species or subspecies in the M. avium complex.
This study demonstrates that M. intracellulare strain comparison by rep-PCR lacked a high level of discrimination, as isolates of M. intracellulare all had similar major band patterns despite belonging to different VNTR types and having unrelated PFGE patterns (Fig. 2). Similar problems were noted with random amplified polymorphic DNA primed-PCR (RAPD-PCR) using random primers with Mycobacterium abscessus (21), and RAPD-PCR became useful only if isolates were clearly “different” based on major band patterns or if related; this was seen with at least three different random primers (21). Different mycobacterial rep-PCR primers are available commercially as the Diversilab mycobacterial typing kit (Bacterial Barcodes, Inc., Houston, TX, now subsidiary of bioMérieux, Inc., Durham, NC). This kit utilizes a software package to determine strain pattern relatedness (7). Whether this system would provide more discriminating results was not evaluated.
The creation of a database on a national or international scale is within reach for the first time for M. intracellulare with the availability of MIRU-VNTR typing. The current technique does not require sequencing, only PCR. The seven VNTR loci described by Dauchy et al. in 2010 were used in the current study and produced separation of isolates into multiple groups comparable to PFGE while reducing time to results (17). Multiple isolates of M. intracellulare from different patients shared the same VNTR type both by amplicon size and by complete VNTR sequence, including the flanking regions (data not shown). The study by Ichikawa et al., from Nagoya University Hospital (Japan), used 16 different VNTR loci, but again, multiple isolates from different patients shared the same VNTR type (16). The finding that different patient isolates with the same VNTR type are mostly clonal by PFGE demonstrates that clonal groups do exist within M. intracellulare and that additional tandem repeats for strain separation may not be helpful. A major objective of developing an international system will be agreement regarding which MIRU-VNTR loci should be tested and what strains are to be used as references. (We did not determine if the 16 primers characterized by Ichikawa et al. produced groupings [type] similar to those produced by the primers utilized in the current study.) The presence of a typing system that identifies specific clones of M. intracellulare should allow for comparison with isolates from other M. intracellulare-associated diseases (e.g., disseminated disease and cavitary lung disease), from isolates from other geographic areas (e.g., North and South America), and from different environmental compartments (e.g., water and soil).
In summary, separation of M. intracellulare into distinct groups can now be done worldwide as has been done for multiple other genera and species, including M. tuberculosis (8–15). This characterization should be done to allow us to begin to understand the significance and virulence of different groups and species in the large family currently known as M. avium complex. It will also allow for determination of recurrence of M. intracellulare disease. The clinical/therapeutic approach to new infections is often less aggressive than for relapse of the original genotype.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by institutional funds, MAC patient donors, and the Amon G. Carter Foundation.
We acknowledge the NTM-IR private foundation whose grant to Marcel Behr allowed for the genomic sequencing of the type strain of M. intracellulare (ATCC 13950T) that made this study possible.
Footnotes
Published ahead of print 21 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02443-12.
REFERENCES
- 1. Kim B-J, Choi B-S, Lim J-S, Choi I-Y, Lee J-H, Chun J, Kook Y-H, Kim B-J. 2012. Complete genome sequence of Mycobacterium intracellulare strain 13950T. J. Bacteriol. 194:2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim B-J, Choi B-S, Lim J-S, Choi I-Y, Lee J-H, Chun J, Kook Y-H, Kim B-J. 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-02. J. Bacteriol. 194:2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim B-J, Choi B-S, Lim J-S, Choi I-Y, Kook Y-H, Kim B-J. 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-64, belonging to the INT1 genotype. J. Bacteriol. 194:3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wallace RJ, Jr, Zhang Y, Brown-Elliott BA, Yakrus MA, Wilson RW, Mann L, Couch L, Girard WM, Griffith DE. 2002. Repeat positive cultures in Mycobacterium intracellulare lung disease after macrolide therapy represent new infections in patients with nodular bronchiectasis. J. Infect. Dis. 186:266–273 [DOI] [PubMed] [Google Scholar]
- 5. Wallace RJ, Jr, Zhang Y, Brown BA, Dawson D, Murphy DT, Wilson R, Griffith DE. 1998. Polyclonal Mycobacterium avium complex infections in patients with nodular bronchiectasis. Am. J. Respir. Crit. Care Med. 158:1235–1244 [DOI] [PubMed] [Google Scholar]
- 6. Falkinham JO. 2011. Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg. Infect. Dis. http://dx.doi.org/10.3201/eid1703.1015 [DOI] [PMC free article] [PubMed]
- 7. Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, Goldberg SV. 2004. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacteria avium complex strains. J. Clin. Microbiol. 42:2685–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bull TJ, Sidi-Boumedine K, McMinn EJ, Stevenson K, Pickup R, Hermon-Taylor J. 2003. Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 17:157–164 [DOI] [PubMed] [Google Scholar]
- 9. Ablordey A, Swings J, Hubans C, Chemlal K, Locht C, Portaels F, Supply P. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inagaki T, Nishimori K, Yagi T, Ichikawa K, Moriyama M, Nakagawa T, Shibayama T, Uchiya K-I, Nikai T, Ogawa K. 2009. Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J. Clin. Microbiol. 47:2156–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martín A, Herranz M, Serrano MJ, Bouza E, Garcia de Viedma D. 2007. Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol. 7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Radomski N, Thibault VC, Karoui C, de Cruz K, Cochard T, Gutiérrez C, Supply P, Biet F, Boschiroli ML. 2010. Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J. Clin. Microbiol. 48:1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romano MI, Amadio A, Bigi F, Klepp L, Etchechoury I, Llana MN, Morsella C, Paolicchi F, Pavlik I, Bartos M, Leão SC, Cataldi A. 2005. Further analysis of VNTR and MIRU in the genome of Mycobacterium avium complex, and application to molecular epidemiology of isolates from South America. Vet. Microbiol. 110:221–237 [DOI] [PubMed] [Google Scholar]
- 14. Stout JE, Hopkins GW, McDonald JR, Quinn A, Hamilton CD, Reller LB, Frothingham R. 2008. Association between 16S-23S internal transcribed spacer sequence groups of Mycobacterium avium complex and pulmonary disease. J. Clin. Microbiol. 46:2790–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thibault VC, Grayon M, Boschiroli ML, Hibbans C, Overduin P, Stevenson K, Gutierrez MC, Supply P, Biet F. 2007. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS12456 restriction fragment length polymorphism typing. J. Clin. Microbiol. 45:2404–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ichikawa K, Yagi T, Inagaki T, Moriyama M, Nakagawa T, Uchiya KI, Nikal T, Ogawa K. 2010. Molecular type of Mycobacterium intracellulare using multilocus variable-number of tandem-repeat analysis: identification of loci and analysis of clinical isolates. Microbiology 156(Part 2):496–504 [DOI] [PubMed] [Google Scholar]
- 17. Dauchy F-A, Dégrange S, Charron A, Dupon M, Xin Y, Bébéar C, Maugein J. 2010. Variable-number tandem-repeat markers for typing Mycobacterium intracellulare strains isolates in humans. BMC Microbiol. 10:93 doi:10.1186/1471-2180-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment and prevention of nontuberculous mycobacterial diseases. American Thoracic Society Statement. Am. J. Respir. Crit. Care Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 18a. Wilton S, Cousins D. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. Genome Res. 1:269–273 [DOI] [PubMed] [Google Scholar]
- 19. Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int. J. Syst. Evol. Microbiol. 54:1277–1285 [DOI] [PubMed] [Google Scholar]
- 20. Frothingham R, Wilson KH. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J. Infect. Dis. 169:305–312 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y, Rajagopalan M, Brown BA, Wallace RJ., Jr 1997. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J. Clin. Microbiol. 35:3132–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turenne CY, Semret M, Cousins DV, Collins DM, Behr MA. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44:433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim RD, Greenberg DE, Ehrmantraut ME, Guide SV, Ding L, Shea Y, Brown MR, Chernick M, Steagall WK, Glasgow CG, Lin JP, Jolley C, Sorbara L, Raffeld M, Hill S, Avila N, Sachdev V, Barnhart LA, Anderson VL, Claypool R, Hilligoss DM, Garofalo M, Fitzgerald A, Anaya-O'Brien S, Darnell D, DeCastro R, Menning HM, Ricklefs SM, Porcella SF, Olivier KN, Moss J, Holland SM. 2008. Pulmonary nontuberculous mycobacterial disease. Prospective study of a distinct preexisting syndrome. Am. J. Respir. Crit. Care Med. 178:1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Picardeau M, Vincent V. 1996. Typing of Mycobacterium avium isolates by PCR. J. Clin. Microbiol. 34:389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


