Abstract
Campylobacter antigen detection by enzyme immunoassay (EIA) provides rapid results compared to traditional culture. However, concern exists regarding specificity. Verification studies of an EIA compared to culture revealed a positive predictive value (PPV) of 91%, whereas PPV fell to 42% during routine diagnostic testing. We suggest all positive EIA results be confirmed via culture.
TEXT
Campylobacter enteritis is a food- and waterborne zoonotic illness and one of the most common causes of infectious diarrhea in the United States (1, 2). While identification of the etiological agent does not typically affect treatment outcomes, as the majority of these infections are self-limited, laboratory diagnosis is essential for epidemiological studies and outbreak tracking through strain identification and typing. Conventional laboratory diagnosis of campylobacteriosis is based on the recovery of the organism from stool specimens by microaerophilic culture. Current recommendations for the recovery of Campylobacter stipulate that cultures be held for a minimum of 72 h prior to signing out a negative result (3); however, a recent laboratory surveillance by the Centers for Disease Control and Prevention found that, of laboratories surveyed, 66% reported negative results at 48 h, while only 33% reported negative results after 72 h (4). In contrast to traditional culture methods, more rapid methods for the detection of Campylobacter antigens in stool, including enzyme immunoassay (EIA) and lateral flow systems, require only 1 to 2 h until results (5). Herein, we describe the verification and subsequent implementation of a Campylobacter EIA at a large university clinical laboratory that serves both acute and tertiary care hospitals and a network of outpatient clinics.
Campylobacter cultures were performed on stool specimens transported in C&S medium (Medical Chemical Corporation, Torrance, CA), streaked for isolation on Campy CVA agar plates (BBL, Sparks, MD), and incubated for 48 h in a microaerophilic environment at 42°C. All morphotypes growing on Campy CVA agar plates were Gram stained for microscopic morphology and tested for oxidase reactivity to evaluate for the presence or absence of Campylobacter spp. The rate of recovery of Campylobacter by culture is tracked through laboratory electronic records. Campylobacter EIA was performed using Meridian Biosciences Premier CAMPY (Meridian Biosciences, Cincinnati, OH), per the manufacturer's instructions. Results of EIA testing were read both visually and spectrophotometrically using a dual wavelength of 450/630 nm (ELX800; Biotek Instruments Inc., Winooski, VT). All protocols for this study were reviewed and approved by the UCLA institutional review board.
Prior to implementation of the Premier CAMPY, laboratory verification of the assay's performance characteristics included testing 60 remnant, deidentified stool specimens submitted to the clinical laboratory for Campylobacter culture between August and November 2011. Campylobacter culture results were recorded prior to freezing (10 positive for Campylobacter, 50 negative for Campylobacter) (Table 1), and the specimens were deidentified and stored in C&S at −20°C for further testing by EIA. The EIA demonstrated 98% concordance, 98% specificity, and 98% sensitivity compared to the culture results, similar to what was previously reported (5). One discordant sample was found to be positive by EIA but negative by culture. Chart review revealed gastroenteritis-like symptoms, including diarrhea with abdominal pain, fever, and nausea, which is consistent with Campylobacter infection. Furthermore, culture results for both Salmonella and Shigella were negative, suggesting the causative agent may have been Campylobacter. Based on these results, the laboratory replaced traditional culture for Campylobacter with the EIA for routine testing.
Table 1.
Accuracy of study results for Premier CAMPY EIA compared to those for the reference method
| Study | EIA result | No. of samples |
% (range) |
||||
|---|---|---|---|---|---|---|---|
| Culture positive | Culture negative | Sensitivity | Specificity | PPV | NPV | ||
| Initial verification study | Positive | 10 | 1 | 100 (66–100) | 98 (88–100) | 91 (57–100) | 100 (91–100) |
| Negative | 0 | 49 | |||||
| Study following implementationa | Positive | 3 | 4 | 75 (22–98) | 96.5 (92–99) | 42.9 (12–80) | 99 (94–100) |
| Negative | 1 | 111 | |||||
Discrepant results resolved by PCR analysis.
In the first week of clinical testing with the EIA, a positivity rate of 11.3% (n = 9/75 specimens; 6/9 were outpatients) was observed, in stark contrast to the 1.7% positivity observed by culture in the previous 4 weeks. Although an increased number of positive stools were expected (due to the increased sensitivity of the EIA compared to that of culture) (5), this increase was higher than what others have reported when converting to the EIA (5, 6). Further, three clinicians caring for different patients voiced concern of false-positive Campylobacter EIA results to the laboratory director. All three patients were on the pediatric transplant service and had no risk factors for Campylobacter exposure; two of these patients were on total parenteral nutrition. At this point, the laboratory returned to Campylobacter cultures for clinical testing in order to further investigate the specificity of the EIA.
Stool samples submitted for Campylobacter culture (n = 119) in the subsequent 2 weeks were stored at −20°C and batch tested by the EIA. To ensure that the EIA performed consistently between users, batch testing was divided equally among the five clinical laboratory scientists who typically perform Campylobacter testing. In order to resolve discrepant results, 16S rRNA gene sequence analysis (7) was performed on isolates recovered from specimens that were EIA negative, and culture-negative, EIA-positive samples and a random sample of culture/EIA concordant samples (n = 12) were sent to the Wadsworth Center (New York State Department of Health) for PCR analysis.
Over the 2 weeks, 5.98% of the EIA results were positive (n = 7), whereas 2.6% (n = 3) of culture results were positive. Four samples negative by culture but positive by EIA tested negative by PCR, indicating 3.4% false-positive (FP) results by the EIA. Patients with FP results ranged in age from 55 to 89 years, and 75% were from an outpatient setting (3 of 4). No enteric pathogens were identified via aerobic bacterial stool culture for any of these samples; however, other uncommon Campylobacter species causing gastroenteritis cannot be ruled out. The single EIA-negative, culture-positive isolate was determined to be a Campylobacter jejuni/Campylobacter coli strain with 98.6% identity by MicroSeq sequencing of the 16S rRNA subunit, suggesting that this was a false negative (FN) by EIA; however, since only 500 bp are sequenced, the possibility of the isolate being Campylobacter lari could not be ruled out (8). Importantly, the positive predictive value (PPV) of the EIA went from 91% (95% confidence interval [CI], 57 to 100%) during initial evaluation to 43% (95% CI, 42 to 80%) in the subsequent studies (Table 1). Negative predictive values (NPV) remained high, at 99 to 100% (Table 1).
Analysis of the positivity rates pre- and post-EIA implementation show Campylobacter prevalence to be 1.74% in the 4 weeks preceding EIA testing and 1.71% in the 4 weeks following EIA testing, suggesting that the increased positivity rate was not due to an increased number of positives but rather an increase in FP reads by the EIA. Likewise, in our small sample set of 117 samples where culture results were reported and EIA was batch tested, 3 positives were reported via culture and 5 positives were reported via EIA, for a 2.6 and 4.3% positivity rate, respectively. While these numbers are small, the in vivo testing of the Premier EIA in our laboratory suggests that EIA should not be used to replace traditional culture methods but may function as a screening tool in high-volume laboratories.
The discrepant results of the EIA between the initial verification and the subsequent clinical testing are disconcerting. The original small sampling size (n = 60) of the verification assay may have contributed to the variance between the testing. Determining the number of specimens to include in a verification study is a challenge faced by all clinical laboratories, and this report highlights the necessity for a thorough verification, which includes review of all discrepant results and careful evaluation of patient demographics included in the assay's clinical trials for FDA approval. For the EIA, FDA performance claims were based entirely on outpatient specimens, whereas specimens received in our laboratory encompass both outpatients and many inpatients with complicated medical histories. As such, a more thorough verification of the assay's performance for patients not included in the FDA trials, such as inpatients on TPN, could have been performed. Many laboratories perform verifications based on convenience sampling rather than on an informed cross-section of patients, and results such as observed herein are the risk of such limited verification studies. In addition, inappropriate inpatient testing for gastroenteritis seems to have a role in the false positivity rate, as three of our initial cases that gave a positive EIA result were from patients hospitalized for greater than 72 h. Reeducation of hospital house staff on the importance of pretest predictive value was performed following completion of this study.
Ultimately, our initial verification results were similar to those previously published on the Campylobacter EIAs and lateral flow systems (5). Our results show that while the EIA performed well in our initial verification study, in vivo clinical testing demonstrated a high number of FP (Fig. 1 and 2). The Campylobacter EIAs provide a rapid alternative to laboratory culture for identification of C. jejuni and C. coli, however, laboratories are urged to verify positive results with culture.
Fig 1.
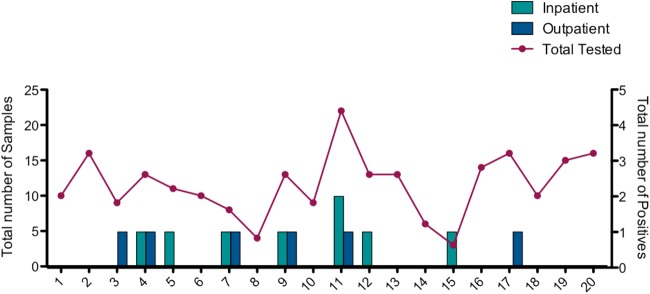
Campylobacter-positive samples during Campylobacter EIA testing. The total number of specimens tested is compared to the total number of positive Campylobacter EIA specimens. Positive specimens were plotted by patient location; light blue bars represent number of positive Campylobacter samples from inpatients, while dark blue bars represent positive samples from outpatients.
Fig 2.
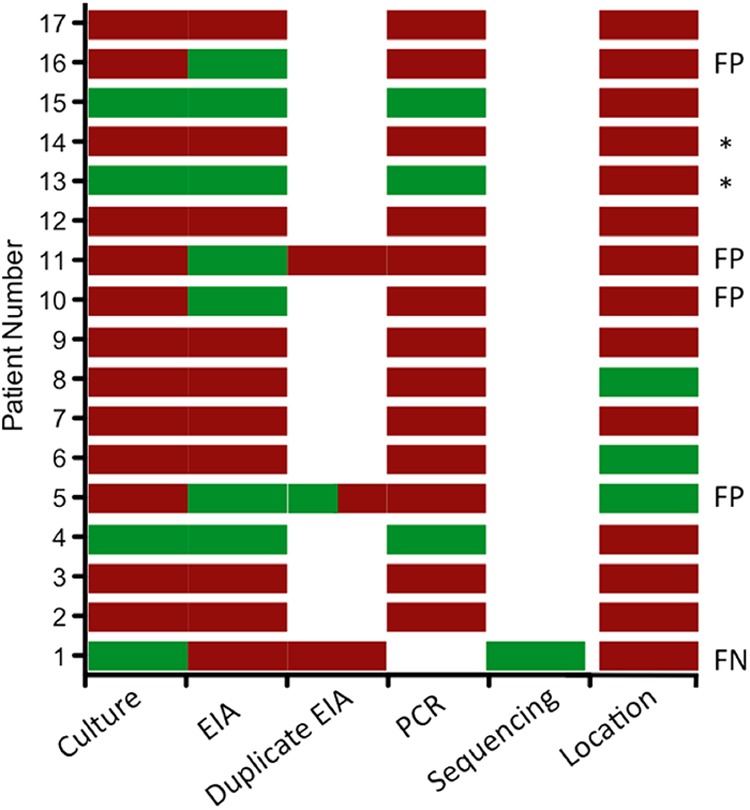
Comparison of culture and EIA results with PCR as an arbitrator test. On 20 April 2012, EIA testing was replaced by culture to report patient results. All stool samples received between 16 and 27 April were collected and batch tested via EIA. All positive culture or EIA specimens and a random sampling of samples negative for both culture and EIA were sent to the Wadsworth Center for verification via PCR. Four FP and 1 FN were observed. Colored bars represent testing performed: red bars represent a negative test result (or outpatient for location), green represents a positive result (or inpatient for location), while a white box represents test not performed. *, pediatric patients.
ACKNOWLEDGMENTS
We thank Gary Rowley and Marissa Carvalho for their help compiling the raw data. Thanks to Kimberlee A. Musser and the Wadsworth Center for providing the PCR arbitrator test. We also thank Meridian Biosciences for their help with training and for providing the kits used for the assay verification.
Footnotes
Published ahead of print 21 November 2012
REFERENCES
- 1. Adedayo O, Kirkpatrick BD. 2008. Campylobacter jejuni infections: update on presentation, diagnosis, and management. Hosp. Phys. 44:9–15 [Google Scholar]
- 2. Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201–1206 [DOI] [PubMed] [Google Scholar]
- 3. Garcia LS. (ed). 2007. Fecal culture for Campylobacter and related species, p 3.8.2.5–3.8.2.14 In Clinical microbiology procedures handbook, 3rd ed LSG & Associates, Santa Monica, CA [Google Scholar]
- 4. M'ikanatha NM, Dettinger LA, Perry A, Rogers P, Reynolds SM, Nachamkin I. 2012. Culturing stool specimens for Campylobacter spp., Pennsylvania, U. S. A. Emerg. Infect. Dis. doi:10.3201/eid1803.111266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Granato PA, Chen L, Holiday I, Rawling RA, Novak-Weekley SM, Quinlan T, Musser KA. 2010. Comparison of Premier CAMPY enzyme immunoassay (EIA), ProSpecT Campylobacter EIA, and ImmunoCard STAT! CAMPY tests with culture for laboratory diagnosis of Campylobacter enteric infections. J. Clin. Microbiol. 48:4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hindiyeh M, Jense S, Hohmann S, Benett H, Edwards C, Aldeen W, Croft A, Daly J, Mottice S, Carroll KC. 2000. Rapid detection of Campylobacter jejuni in stool specimens by an enzyme immunoassay and surveillance for Campylobacter upsaliensis in the Greater Salt Lake City area. J. Clin. Microbiol. 38:3076–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelesidis T, Dien Bard J, Humphries RM, Ward K, Lewinski MA, Uslan DZ. 2010. First report of treatment of Anaerobiospirillum succiniciproducens bloodstream infection with levofloxacin. J. Clin. Microbiol. 48:1970–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorkiewicz G, Feierl G, Schober C, Dieber Köfer J, Zechner R, Zechner EL. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]


