Abstract
Enterovirus 68 was detected in 10 respiratory specimens from pediatric patients hospitalized for acute wheezing or bronchitis during 2009 in the northeast of France. Viral loads ranged from 2 × 105 to 7.2 × 107 copies/ml. Alignment of 5′ nontranslated regions and phylogenetic analysis of partial VP1 gene sequences show that these viruses clustered and belonged to clade C.
TEXT
Human enterovirus 68 (HEV-D68) was isolated from respiratory specimens for the first time in 1962 in the United States in children with pneumonia and bronchiolitis (1). Unlike other enteroviruses, HEV-D68 is acid labile and biologically more similar to human rhinoviruses in being mainly associated with respiratory diseases; however, until recently, reports of respiratory infections due to HEV-D68 were rare (2, 3). Over the past 3 years, outbreaks in Japan, the Philippines, and the Netherlands as well as epidemic clusters in the United Kingdom have implicated HEV-D68 as an emerging respiratory pathogen (2, 4–9). The clinical presentation of HEV-D68 infections during these outbreaks ranged from mild illness to rare severe pneumonia cases requiring admission in intensive care units (2, 4–9). Among these epidemic strains, novel genetic variants were described (2, 4–8). The present classical enterovirus molecular assays used in routine virological diagnosis can fail to detect HEV-D68 strains in respiratory samples, leading to an underestimation of the prevalence and the role of HEV-D68 infections in pediatric acute airway diseases (10). We report here the phylogenetic characterization of HEV-D68 strains detected in a cohort of pediatric patients hospitalized for acute airway diseases.
The study.
From September 2009 to June 2010, 651 respiratory specimens (nasopharyngeal aspirate [NPA]) were obtained from 651 consecutive pediatric patients (median age [months], 13 [range, 0 to 176]; sex ratio [male/female], 1.8) admitted to the pediatric department of the University Hospital Centre of Reims (Champagne Ardenne, France) for a diagnosis of acute airway disease. The hospital's ethics committee (institutional review board of the Reims University Hospital Center) approved the present study.
Total nucleic acid extraction was performed using the NucliSens EasyMAG instrument (bioMérieux, Lyon, France), according to the manufacturer's instructions. A quantitative real-time reverse transcription (RT)-PCR targeting the 5′ nontranslated region (5′NTR) of the Enterovirus genus (EV; including enteroviruses HEV A to D and rhinoviruses HRV A to C) was carried out to perform a pan-enterovirus and rhinovirus detection and to quantify the viral load. Briefly, 2 μl total nucleic acid extract was amplified in a 10-μl RT-PCR mixture containing 5 μl 2× reaction mix, 2 U SuperScript III RT/Platinum Taq mix (Invitrogen, Life Technologies, Saint-Aubin, France), and 0.4 μM each primer and probe. The primer sequences used were 5′-AGCCTGCGTGGCKGCC-3′ and 5′-GAAACACGGACACCCAAAG-3′, and the probe was 5′-6-carboxyfluorescein (FAM)-CTCCGGCCCCTGAATGYGGCTAA-3′ (10). The cycling conditions included initial incubations at 55°C for 30 min and 94°C for 2 min, followed by 40 cycles of 94°C for 15 s and 63°C for 1 min. Full-length CVB3-Nancy RNA transcripts containing the target sequences were synthesized and used as a positive control for copy number calculation. Sensitivity of the PCR assay was estimated to 100 copies per PCR well reaction by 10-fold serial limit dilutions of these RNA transcripts (11).
To discriminate HEV from HRV strains, we performed a specific HEV real-time RT-PCR using the primers NC1M-F and E2-R and the probe OL27LC targeting in the 5′NTR (12).
HEV-positive specimens were subjected to genetic typing in a part of the translated genomic region: VP1 amplification of HEV-D68 was performed using the primers VP1-2547-F and VP1-2772-R (5). The primers DK001-164-F and DK004-559-R (13) targeting the 5′NTR region were used to identify clades and variants of HEV-D68 (14).
EV strains were detected by a quantitative real-time RT-PCR in 257 (39.5%) (median age [months], 11 [range, 0 to 176]; sex ratio [male/female], 1.9) NPA specimens taken from 651 hospitalized children with acute airway diseases. EV viral loads in NPA specimens ranged from 2.1 × 104 to 6.5 × 1010 copies/ml, with a median value of 6.3 × 107 copies/ml of NPA specimen. Among the 257 EV-positive strains, the HEV-specific RT-PCR differentiated 16 (6.3%) HEV from 241 (93.6%) HRV strains.
HEV-infected cases were older (median age [years], 3 versus 0.9; P = 0.003) and were more frequently associated with a respiratory distress (11 HEV versus 62 HRV; P = 0.01) and a need for oxygen therapy at the time of admission (8 HEV versus 45 HRV; P = 0.01) than cases infected by HRV strains (not shown). Moreover, respiratory viral load levels appeared to be lower in HEV-positive patients than those infected by HRV (median values, 7.4 × 106 versus 4.9 × 107 copies/ml; P = 0.005) (data not shown). Taken together, our findings suggested that respiratory HEV strains might be more pathogenic than HRV strains and could induce more severe respiratory symptoms in hospitalized acute airway illness cases.
Among the 16 HEV strains, 10 (62.5%) were identified as the HEV-D68 genotype by alignment and phylogenetic analysis on part of the 5′NTR, VP1 (Fig. 1). The HEV-D68-positive samples accounted for 1.5% (10/651) of all respiratory samples tested and for 4% (10/257) of all EV-positive respiratory samples detected over 10 months. The positive HEV-D68 specimens were collected only from September to November 2009 (Table 1). HEV-D68 viral loads in NPA specimens ranged from 2 × 105 to 7.2 × 107 copies/ml (median, 1.2 × 107 copies/ml). The HEV-D68-positive patients were 6 months to 10 years old (median age [years], 3.8), with a sex ratio (male/female) of 1.5. The majority of these patients (8/10) were suffering from underlying pathologies. Six of these 10 HEV-D68-infected patients required oxygen supplementation. HEV-D68 infection was only associated with acute wheezing and febrile bronchitis among the 651 hospitalized cases (Table 1). None of the 10 HEV-D68-positive patients required an intensive care admission, and their clinical outcomes were good at 12 months after the time of hospital discharge.
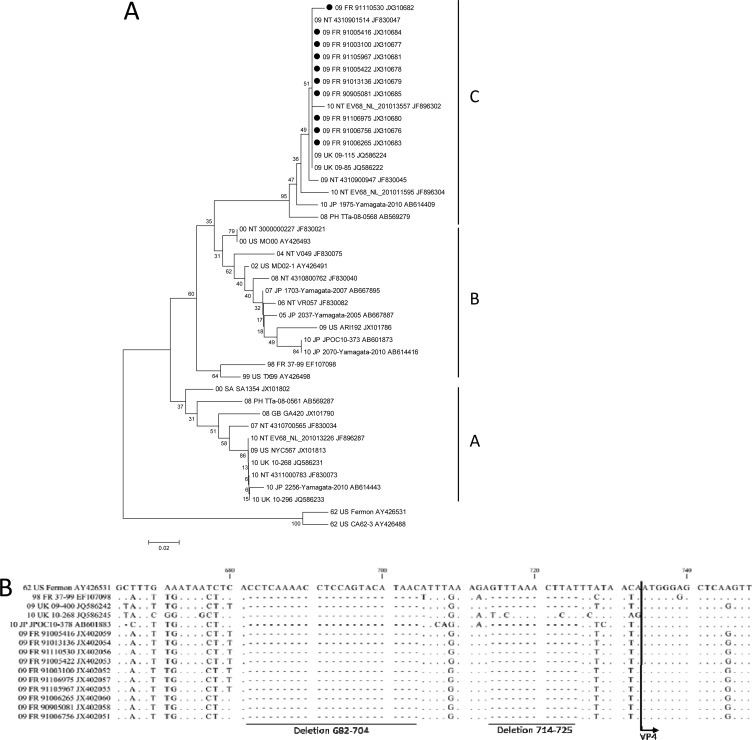
Fig 1.
Molecular characterization of HEV-D68 clinical isolates based on phylogenetic analyses of nucleotide sequences of the partial VP1 genomic region (A) and alignment of nucleotide sequence 5′ nontranslated regions (5′NTR) (B). Sequences were aligned using Clustal W version 1.81. (www.clustal.org/). Genetic distances between sequences were calculated using the Kimura 2-parameter method. Trees were constructed using the neighbor-joining method as implemented in MEGA 5 software (www.megasoftware.net). Bootstrap values from 1,000 replicates are shown at the nodes. Scale bar indicates number of nucleotide substitutions per site. HEV-D68 Fermon is used as the reference strain (GenBank accession no. AY426531). Black circles indicate HEV-D68 strains analyzed in this study (GenBank accession no. for VP1, JX310677 to JX310685; for 5′NTR, JX310686 to JX310695).
Table 1.
Demographic, clinical, and virological characteristics of 10 children with acute respiratory diseases caused by HEV-D68, northeastern France, September 2009 to June 2010c
| Patient no. | Age/sex | Mo, yr sampled | Viral load copies/ml specimens (NPA) | Clinical diagnosisa | % SaO2b | O2 supplement (length) | Length of hospitalization (no. of days) | Bacterial superinfection |
|---|---|---|---|---|---|---|---|---|
| 90905081 | 10 yr 0 mo 23 days/male | September 2009 | 7.20 × 107 | Bronchitis | 92 | Life | 2 | – |
| 91006265 | 4 yr 5 mo 17 days/male | October 2009 | 3.84 × 106 | Acute asthma | 90 | 12 h | 2 | – |
| 91005416 | 2 yr 3 mo 29 days/male | October 2009 | 5.40 × 107 | Acute asthma | NA | 24 h | 5 | – |
| 91006756 | 6 yr 3 mo 29 days/male | October 2009 | 5.05 × 107 | Asthma | 97 | – | 2 | – |
| 91003100 | 5 mo 28 days/female | October 2009 | 3.93 × 107 | Bronchiolitis | NA | – | 5 | – |
| 91005422 | 6 mo 8 days/female | October 2009 | 2.01 × 105 | Bronchiolitis | NA | – | 1 | – |
| 91013136 | 3 yr 0 mo 17 days/female | October 2009 | 1.42 × 107 | Febrile bronchitis | NA | – | 1 | Streptococcus pneumoniae |
| 91106975 | 9 mo 17 days/male | November 2009 | 9.20 × 106 | Bronchiolitis | 95 | – | 4 | – |
| 91105967 | 4 yr 7 mo 21 days/male | November 2009 | 6.10 × 106 | Acute asthma | 94 | 48 h | 5 | – |
| 91110530 | 6 yr 11 mo 5 days/female | November 2009 | 8.60 × 106 | Acute asthma | 93 | 48 h | 3 | Streptococcus pneumoniae |
Clinical diagnosis was established at the time of hospital discharge.
SaO2 (%) measured at time of hospital admission.
NA, none available; –, none.
Recent phylogenetic analyses of all known HEV-D68 strains revealed that over the past decades, these viruses have emerged and they are now divided into three major evolutionary clades. These clades are distinguishable by specific signatures of nucleotide substitutions and deletions relative to the Fermon strain (15). Phylogenetic analysis of our HEV-D68 clinical isolates was based on the nucleotide sequence partial VP1 genomic region and alignment of the nucleotide sequence 5′NTR. The phylogenetic analyses of our nucleotide sequences evidenced that they grouped into the same genetic cluster (Fig. 1). Our French 2009 VP1 sequences were close to those obtained from the United Kingdom 2009 strains and from some Japanese 2010 strains that belonged to clade C (6, 7, 14) (Fig. 1). Our phylogenetic findings indicated that some Japanese and European strains belonging to the same genetic clade might have the same ancestor strain and that these viral strains could be responsible for mild respiratory illnesses in pediatric patients (15). Moreover, we observed variations of the 5′NTR spacer region between the end of the internal ribosome entry site (IRES) and the beginning of the polyprotein open reading frame (ORF) (Fig. 1). The NTR between 164 to 623 nucleotides is highly conserved and has more than 95% nucleotide similarities across HEV-D68 strains (15). Our French strains showed the same NTR pattern that was shared by all the known clade C sequences recently described (15). Within 624 and 730 nucleotide positions, we distinguished 2 deletions consisting of 23 and 12 nucleotides at positions 682 to 704 and 714 to 725, respectively (Fig. 1). This second deletion, firstly reported by Kaida et al. (6), might have a significant effect on the initiation of viral translation. Although it was established that variations within the IRES could have major effects on virulence, little is known about this spacer region and its role in HEV-D68 fitness. These deletions may affect the virulence of the virus by enhancing translational efficiency and may be correlated with the recent increase in HEV-D68 cases worldwide (15). Therefore, the HEV-D68 clade C strains could be characterized by an enhanced viral fitness resulting in high respiratory viral loads (Table 1) that could allow a rapid epidemiological spread in some pediatric populations.
In conclusion, we provide evidence that in France during the fall of 2009, HEV-D68 strains were responsible for a low proportion of pediatric cases hospitalized for acute airway diseases including acute wheezing and bronchitis. Our findings provide further insights into the potential specific lower respiratory tract tropism of HEV-D68 belonging to the clade C and increase our awareness of the clinical role of these strains.
ACKNOWLEDGMENTS
This work was supported by a clinical research grant from the Reims University Medical Centre (EA4684-CardioVir). Fanny Renois is supported by an official grant from the French Army Department (Bourse DGA [Délégation Générale de l'Armement], Ministère de la Défense; topic: microbiology, infectious diseases).
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Schieble JH, Fox VL, Lennette EH. 1967. A probable new human picornavirus associated with respiratory diseases. Am. J. Epidemiol. 85:297–310 [DOI] [PubMed] [Google Scholar]
- 2. Ikeda T, Mizuta K, Abiko C, Aoki Y, Itagaki T, Katsushima F, Katsushima Y, Matsuzaki Y, Fuji N, Imamura T, Oshitani H, Noda M, Kimura H, Ahiko T. 2012. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol. Immunol. 56:139–143 [DOI] [PubMed] [Google Scholar]
- 3. Oberste MS, Maher K, Schnurr D, Flemister MR, Lovchik JC, Peters H, Sessions W, Kirk C, Chatterjee N, Fuller S, Hanauer JM, Pallansch MA. 2004. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J. Gen. Virol. 85:2577–2584 [DOI] [PubMed] [Google Scholar]
- 4. Imamura T, Fuji N, Suzuki A, Tamaki R, Saito M, Aniceto R, Galang H, Sombrero L, Lupisan S, Oshitani H. 2011. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg. Infect. Dis. 17:1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacobson LM, Redd JT, Schneider E, Lu X, Chern Oberste S-WWMS, Erdman DD, Fischer GE, Armstrong GL, Kodani M, Montoya J, Magri JM, Cheek JE. 2012. Outbreak of lower respiratory tract illness associated with human enterovirus 68 among American Indian children. Ped. Infect. Dis. J. 31:309–312 [DOI] [PubMed] [Google Scholar]
- 6. Kaida A, Kubo H, Sekiguchi J, Kohdera U, Togawa M, Shiomi M, Nishigaki T, Iritani N. 2011. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg. Infect. Dis. 17:1494–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meijer A, van der Sanden S, Snijders BEP, Jaramillo-Gutierrez G, Bont L, van der Ent CK, Overduin P, Jenny SL, Jusic E, van der Avoort HGAM, Smith GJD, Donker GA, Koopmans MPG. 2012. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology 423:49–57 [DOI] [PubMed] [Google Scholar]
- 8. Rahamat-Langendoen J, Riezebos-Brilman A, Borger R, van der Heide R, Brandenburg A, Schölvinck E, Niesters HGM. 2011. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J. Clin. Virol. 52:103–106 [DOI] [PubMed] [Google Scholar]
- 9. Xiang Z, Gonzalez R, Wang Z, Ren L, Xiao Y, Li J, Li Y, Vernet G, Paranhos-Baccalà G, Jin Q, Wang J. 2012. Coxsackievirus A21, enterovirus 68, and acute respiratory tract infection, China. Emerg. Infect. Dis. 18:821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tapparel C, Cordey S, Van Belle S, Turin L, Lee Regamey W-MN, Meylan P, Muhlemann K, Gobbini F, Kaiser L. 2009. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J. Clin. Microbiol. 47:1742–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lévêque N, Renois F, Talmud D, Nguyen Y, Lesaffre F, Boulagnon C, Bruneval P, Fornes P, Andréoletti L. 2012. Quantitative genomic and antigenomic enterovirus RNA detection in explanted heart tissue samples from patients with end-stage idiopathic dilated cardiomyopathy. J. Clin. Microbiol. 50:3378–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petitjean J, Vabret A, Dina J, Gouarin S, Freymuth F. 2006. Development and evaluation of a real-time RT-PCR assay on the LightCycler for the rapid detection of enterovirus in cerebrospinal fluid specimens. J. Clin. Virol. 35:278–284 [DOI] [PubMed] [Google Scholar]
- 13. Kiang D, Kalra I, Yagi S, Louie JK, Boushey H, Boothby J, Schnurr DP. 2008. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J. Clin. Microbiol. 46:3736–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lauinger IL, Bible JM, Halligan EP, Aarons EJ, MacMahon E, Tong CYW. 2012. Lineages, sub-lineages and variants of enterovirus 68 in recent outbreaks. PLoS One 7:e36005 doi:10.1371/journal.pone.0036005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tokarz R, Firth C, Madhi SA, Howie SRC, Wu WY, Sall AA, Haq S, Briese T, Lipkin WI. 2012. Worldwide emergence of multiple clades of enterovirus 68. J. Gen. Virol. 93:1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]