Abstract
We evaluated three different PCR-based capsular gene typing methods applied to 312 human and bovine Streptococcus agalactiae (group B Streptococcus [GBS]) isolates and compared the results to serotyping results obtained by latex agglutination. Among 281 human isolates 27% could not be typed by latex agglutination. All 312 isolates except 5 could be typed by the three PCR methods combined. Two of these methods were multiplex assays. Among the isolates that were typeable by both latex agglutination and capsular gene typing, 94% showed agreement between the two methods. However, each of the PCR methods showed limitations. One of the methods did not include all 10 recognized serotypes, one misidentified eight isolates of serotypes Ib and IV as serotype Ia, and one did not distinguish between serotypes VII and IX. For five isolates that showed aberrant patterns in the capsular gene typing, long-range PCR targeting the cps operon disclosed large insertions or deletions affecting the cps gene cluster. A sensitive flow cytometric assay based on serotype-specific antibodies applied to 76 selected isolates that were nontypeable by latex agglutination revealed that approximately one-half of these did express capsular polysaccharide. A procedure for convenient and reliable capsular gene typing to be included in epidemiological and surveillance studies of S. agalactiae is proposed.
INTRODUCTION
Streptococcus agalactiae (group B Streptococcus [GBS]) emerged as an important human pathogen in the 1960s and has become the leading cause of neonatal invasive infections (1–3). More recently, S. agalactiae has also been recognized as a common cause of infections in immunocompromised patients and in the elderly (4). In addition, S. agalactiae is a notable cause of contagious mastitis in cattle (5). Some evolutionary lineages of the species are exclusively adapted to either humans or cattle and have spread globally, whereas others are genetically more diverse and may colonize both hosts (6). In addition, S. agalactiae may be isolated from other species including other mammals, amphibians, and fish. Subpopulations of S. agalactiae originating from these animal species may occasionally cause infections in humans (7, 8).
The polysaccharide capsule is a major virulence factor in invasive disease caused by S. agalactiae, enabling the bacterium to evade host innate defense mechanisms (9). The most commonly used method for serotyping of S. agalactiae is latex agglutination based on polyclonal antibodies specific for the 10 recognized capsular polysaccharides, i.e., serotypes Ia, Ib, and II to IX. Serological methods have limitations, as they may fail to type an isolate due to lack of or low expression of capsular polysaccharide under the experimental conditions. Furthermore, these methods are highly dependent on the quality of the antibodies used and on the experience of the laboratory (10). Therefore, capsular gene typing is an attractive alternative. Recently, several methods for typing of S. agalactiae based on PCR targeting genes in the cps operon were published. In this study, we compared three different PCR-based capsular gene typing methods (11–14) with serotyping by latex agglutination. In addition, the expression of capsular polysaccharides in strains that were nontypeable by latex agglutination was evaluated by a sensitive flow cytometric analysis (15) using monoclonal or polyclonal antibodies specific for the five most common serotypes.
MATERIALS AND METHODS
S. agalactiae isolates.
A total of 312 S. agalactiae isolates were included in the study. Among these, 281 were vaginorectal isolates obtained from pregnant women (n = 251) and clinical isolates from neonatal infections (n = 30) in Belgium, Bulgaria, the Czech Republic, Denmark, Germany, Great Britain, Italy, and Spain. These isolates were collected during the DEVANI project (Design of a Vaccine against Neonatal Infections) supported by the European Commission Seventh Framework. Among these DEVANI isolates, 77 were received from other participating laboratories as being problematic in serotyping by latex agglutination. In addition to these human isolates, 31 bovine S. agalactiae strains, nontypeable by serological means, were included from our previous global study (6). These strains were isolated in Australia (n = 5), Europe (n = 15), and the United States (n = 11).
All included strains were confirmed as S. agalactiae by a positive Remel Streptex Latex Group B test (Thermo Fisher Scientific), a positive PCR using primers dltS-F and dltS-R as described previously (14), and a positive CAMP reaction. Isolates were cultured on blood agar plates at 37°C in air plus 5% CO2 and in Todd-Hewitt broth (Oxoid).
Serotyping by latex agglutination.
The S. agalactiae strains were cultured on blood agar plates. A heavy suspension of the test organism harvested from the blood agar plate was prepared in 250 μl phosphate-buffered saline (PBS), pH 7.4. For each of the 10 serotypes, a 20-μl aliquot of the bacterial suspension was applied to a Wellcogen disposable reaction card and mixed with 1 μl of latex suspension (reagents Ia, Ib, and II to IX; Strep-B-Latex kit; Statens Serum Institut, Copenhagen, Denmark) as recently described (10). The reaction card was rotated slowly and observed for agglutination, and a positive reaction was scored when clear-cut agglutination appeared within 30 s. Notably, false-positive reactions may occur if the reaction time exceeds 30 s. Serotyping by latex agglutination and the capsular gene typing described below were performed in the same laboratory at Aarhus University.
Antibodies.
Mouse monoclonal antibodies (MAbs) were generated by Areta International (Varese, Italy) using standard protocols. Hybridoma clones were screened by enzyme-linked immunosorbent assay (ELISA). Positive clones were then tested for binding to the surface of S. agalactiae by flow cytometry, and the MAbs were purified by protein G affinity chromatography.
A polyclonal antiserum specific for capsular polysaccharide type II was produced by immunizing CD1 mice with the purified tetanus toxoid-conjugated serotype II polysaccharide. Animal treatments were performed in compliance with the Italian laws and approved by the institutional review board (Animal Ethical Committee) of Novartis Vaccines and Diagnostics, Siena, Italy.
Serotyping by flow cytometry.
Fluorescence-activated cell analysis was performed as described elsewhere (15) by using MAbs specific for S. agalactiae capsular polysaccharides Ia, Ib, III, and V and a polyclonal serum specific for the polysaccharide type II (see above). Briefly, mid-exponential-phase S. agalactiae cells were fixed in 0.08% (wt/vol) paraformaldehyde and incubated for 1 h at 37°C. Fixed bacteria were then washed once with PBS, resuspended in newborn calf serum (Sigma), and incubated for 20 min at 25°C. The cells were then incubated for 1 h at 4°C in the presence of MAbs in preimmune or immune sera, diluted 1:200 in dilution buffer (PBS, 20% newborn calf serum, 0.1% bovine serum albumin [BSA]). Cells were washed in PBS–0.1% BSA and incubated for a further 1 h at 4°C with a 1:100 dilution of R-phycoerythrin-conjugated F(ab)2 goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.). After washing, cells were resuspended in PBS and analyzed with a fluorescence-activated cell sorting (FACS) CANTO II apparatus (Becton, Dickinson, Franklin Lakes, NJ) using FlowJo Software (Tree Star, Ashland, OR).
Capsular gene typing.
Genomic DNA was prepared using the Nucleo-Spin Tissue kit (Macherey-Nagel). Approximately 1 ng of DNA was used in the PCRs with primers as described elsewhere (11–14). For the one-set multiplex PCR described by Imperi et al. (11), we used the 5 Prime HotMaster Mix kit (5 Prime); for the two-set multiplex PCR described by Poyart et al. (14), we used AmpliTaq Gold with GeneAmp (Roche); and for the serotype-specific PCRs described by Kong et al. (12, 13), we used PuReTaq Ready-To-Go PCR beads (GE Healthcare). The amplicons were analyzed by agarose gel electrophoresis, and the presence of DNA fragments of the same size as in one of the strains with known serotype was used to establish the capsular gene type. For the multiplex PCR described by Imperi et al. (11), we used 1.5% and for the other methods 1% agarose gels (SeaKem ME agarose; Lonza).
Long-range PCR.
The primers GBS-cpsA-F, 5′-TGGCTCTATATCAACAGTATCAAGATCTGA-3′, and GBS-neuA-R, 5′-CTCAATATCTGCATTATTGACAGTTTCTCT-3′, targeting the cpsA gene (the first gene in the cps operon) and the neuA gene (the last gene in the cps operon), respectively, were used in long-range PCR with the Advantage Genomic LA (Clonetech). The samples were amplified by a denaturation step for 2 min at 94°C, followed by 30 cycles of 94°C for 10 s, 55°C for 30 s, and 68°C for 20 min and a final elongation of 68°C for 10 min.
RESULTS
Serotyping by latex agglutination.
The study included 281 human and 31 bovine isolates of S. agalactiae. Among the human S. agalactiae strains, latex agglutination assigned 205 to one of the 10 known serotypes, 74 did not react with any of the reagents and were scored as nontypeable, and 2 showed autoagglutination. Being selected as previously nonserotypeable, none of the bovine isolates included were typeable by latex agglutination.
Capsular gene typing.
The 312 strains were all examined by the three capsular gene typing methods as described in Materials and Methods. Each of the three methods showed limitations. The two-set multiplex PCR developed by Poyart et al. (14) was published before the discovery of serotype IX (16), and serotype IX strains were found to yield a PCR product of the same size as those of serotype VII strains in this test (Fig. 1). In the one-set multiplex PCR developed by Imperi et al. (11), the middle band in six and two strains of serotypes Ib and IV, respectively, was very weak or not detectable, and therefore, strains of these serotypes may be incorrectly typed as serotype Ia, which is characterized by the lack of the middle band (Fig. 2). The serotype-specific PCRs described by Kong et al. (12, 13), which we used to confirm the results of the two multiplex PCRs described above, do not include a PCR specific for serotypes II, VII, or VIII.
Fig 1.
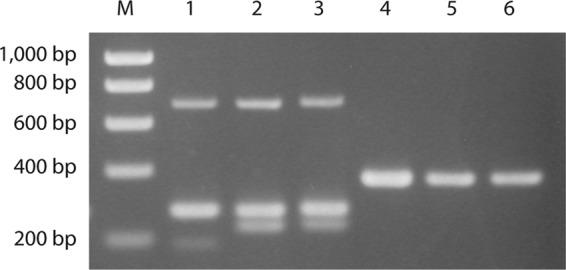
Serotype IX strains are identified as serotype VII in the two-set multiplex PCR (14). Lanes 1 to 3, amplicons from the multiplex PCR performed as described by Imperi et al. (11); lanes 4 to 6, amplicons from the multiplex PCR 2 performed as described by Poyart et al. (14). M, molecular weight marker; lanes 1 and 4, strain CZ-PW-093 (serotype VII); lanes 2 and 5, strain CZ-PW-032 (serotype IX); lanes 3 and 6, DE-PW-092 (serotype IX).
Fig 2.
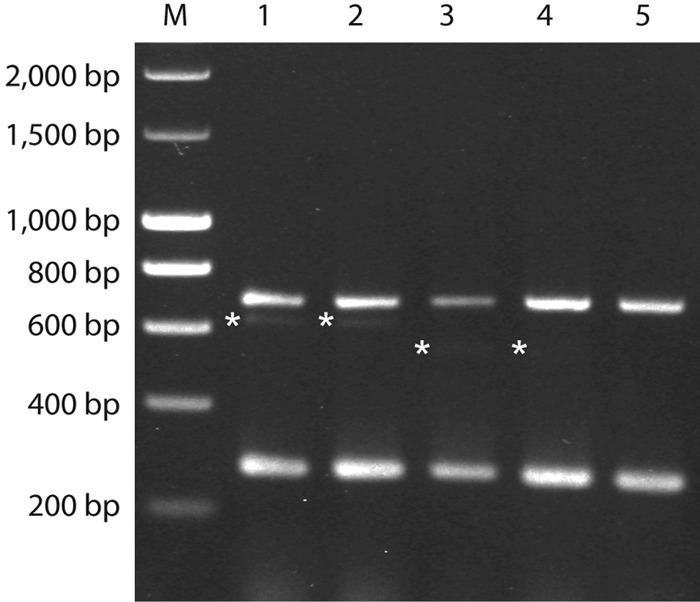
Examples of weak bands in the multiplex PCR performed as described by Imperi et al. (11). M, molecular weight marker; lane 1, DE-PW-096 (serotype Ib); lane 2, DE-PW-203 (serotype Ib); lane 3, DE-PW-196 (serotype IV); lane 4, ES-PW-167 (serotype IV); lane 5, DE-PW-091 (serotype Ia). The weak bands are marked by asterisks.
By combining all results, 307 strains were unambiguously assigned to a capsular gene type, showing full agreement between the three PCR methods used except for the limitations pointed out above. Five strains only, four of human and one of bovine origin, showed an aberrant amplicon pattern or lack of amplicons in at least one of the PCRs and could not be unambiguously typed. The sizes of amplicons obtained in the long-range PCR targeting the cps operon indicated that insertions or deletions (indels) had affected the cps gene cluster in these strains (Fig. 3). Among these five strains, four were nontypeable by latex agglutination, whereas one was positive for serotype IX (Table 1), which most likely is a mistake because of the problems with the serotype IX reagent in the Strep-B-Latex kit (see below).
Fig 3.
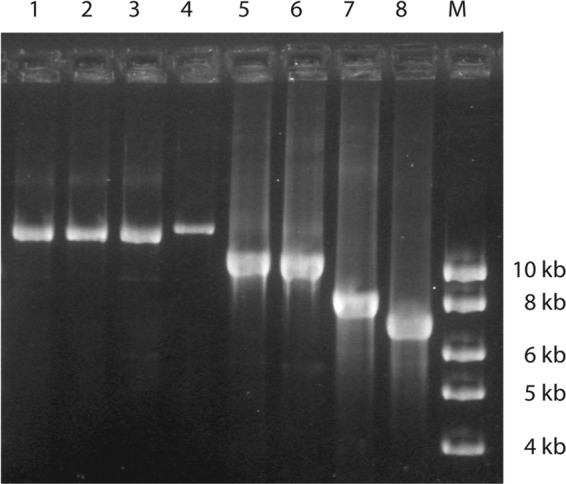
Long-range PCR with primers cpsA-F and neuA-R flanking the cps locus on the five strains that were nontypeable by the molecular capsular gene typing method. For comparison, three strains that could be typed by both serology and PCR are included. Lane 1, CZ-PW-123 (serotype Ia); lane 2, DE-PW-121 (serotype III); lane 3, DE-PW-119 (serotype Ib); lane 4, GB-PW-013 (nontypeable); lane 5, USS215 (nontypeable, bovine strain); lane 6, IT-PW-0085 (nontypeable); lane 7, IMMI266 (nontypeable); lane 8, GB-PW-075 (nontypeable); M, molecular weight marker.
Table 1.
Typing of 281 human GBS isolates by latex agglutination and a combination of PCR methods
| Serotype assigned by latex agglutination | No. of isolates assigned by molecular capsular gene typing to serotype: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VII | VIII | IX | NTa | |
| Ia | 32 | |||||||||
| Ib | 15 | |||||||||
| II | 33 | |||||||||
| III | 64 | |||||||||
| IV | 9 | |||||||||
| V | 2 | 2 | 30 | |||||||
| VII | 1 | |||||||||
| VIII | 1 | |||||||||
| IX | 3 | 1 | 2 | 1 | 1 | 7 | 1 | |||
| NT | 22 | 3 | 13 | 18 | 4 | 10 | 1 | 3 | ||
| Autoaggregation | 2 | |||||||||
NT, nontypeable.
Of the 204 human strains that were typeable by both latex agglutination and capsular gene typing, 191 strains (94%) showed agreement between the two methods (Table 1). The 13 strains that showed disagreement were assigned to serotypes V, VIII, or IX by latex agglutination and to Ia, Ib, II, III, IV, or V by molecular capsular gene typing (Table 1). Among the 31 bovine strains that were nontypeable by latex agglutination, 4, 3, 16, 2, 4, and 1 strains were assigned to serotypes Ia, Ib, II, III, IV, and V, respectively, by the capsular gene typing method. One bovine strain was nontypeable by all methods. No serotype VIII strain was detected by any of the PCR methods.
Detection of capsule expression by flow cytometry.
As a negative result with the latex reagents may be due to lack of gene expression, a sensitive flow cytometric assay was used to test for lack of versus low expression of capsular polysaccharide in selected strains that were typeable by capsular gene typing only and not by latex agglutination. Monoclonal antibodies against serotype Ia, Ib, III, and V polysaccharides and a polyclonal antibody against serotype II were available for the analysis, and strains exhibiting other capsular gene types were excluded. Among 50 human and 26 bovine strains tested, 28 and 11, respectively, could be typed by flow cytometry, indicating that these strains expressed amounts of capsular polysaccharide undetectable by the latex agglutination test, as confirmed by the complete agreement of the reactions with the molecular typing results. The 22 human and 15 bovine strains negative in the flow cytometric assay presumably do not express capsular polysaccharides. This explanation is supported by the finding that the five strains that were nontypeable by gene typing due to disruptive deletions or insertions in the cps locus were all nontypeable by flow cytometry. Besides, the flow cytometric analyses confirmed the capsular gene typing for three strains that showed discrepant results in the latex agglutination assay.
Capsule gene locus.
Long-range PCR on the cps operon was performed on a selection of 41 strains that could be typed by the molecular methods and were negative in latex agglutination. Among these, 33 strains had capsular gene types that were covered by the antibodies used in the flow cytometric assay, and they were selected because they were negative in this assay, whereas the remaining 8 strains had capsular gene types not covered by the flow cytometric assay. One human strain showed no amplicon in this PCR analysis, presumably due to strain variation in the primer-binding sites. Among the remaining 23 human and 17 bovine S. agalactiae strains tested, the amplified fragment was of a size compatible with an intact cps operon, indicating that the presence of larger indels in the cps operon was not the molecular mechanism causing lack of capsule expression.
DISCUSSION
Like current vaccines against Streptococcus pneumoniae and Haemophilus influenzae serotype b, future vaccines against S. agalactiae infections are presumed to include capsular polysaccharides from the most common types associated with disease. The capsule is the major known protective antigen, and capsular conjugate vaccines are in clinical trials (17). Therefore, correct serotyping of clinical isolates is essential to predict vaccine coverage. Available methods are based either on agglutination with serotype-specific reagents or on detection of serotype-specific sequences in the capsular polysaccharide biosynthesis gene loci. For strains that were typeable by both latex agglutination and capsular gene typing, our study showed a high degree of agreement (94%) between results obtained by the two methods. However, the results identify a problem with the serotype V and IX latex reagents (Statens Serum Institute) used for the agglutination tests (Table 1). Moreover, the accuracy of the results is highly dependent on experience (10), and the latex agglutination assay is less sensitive than the flow cytometric method as demonstrated by our observation that approximately one-half of the strains scored as nontypeable in this assay were found to express type-specific polysaccharides when examined by the flow cytometric method. The exact proportion of unselected isolates that would present this problem in clinical microbiology is not clear, as the strain collection examined in this study was enriched with strains that were difficult to type in other laboratories. The clinical significance, if any, of the apparently limited capsule expression under laboratory conditions is also unknown.
With few exceptions, the PCR-based capsular gene typing methods accurately identified the strains except for serotypes not covered by two of the assays. However, in our hands the multiplex PCR described by Imperi et al. (11) misidentified a few serotype Ib and IV strains as serotype Ia, and the assay described by Poyart et al. (14) does not distinguish between serotypes VII and IX. The latter serotype was identified after publication of the Poyart method. Thus, the multiplex PCR by Poyart et al. (14) has to be combined with another PCR in order to distinguish between serotypes VII and IX in strains assigned to serotype VII in the assay. We suggest to reexamine strains assigned to capsular gene type VII in this PCR by using primers cpsI-Ia-6-7-F and cpsI-7-R, which amplify a 179-bp fragment of cpsI in serotype VII strains, together with primers cpsI-7-9-F and cpsI-9-R, which amplify a 229-bp fragment of the gene in serotype IX strains as described by Imperi et al. (11). This combination of two PCR methods provides a convenient and reliable typing system (Fig. 4).
Fig 4.
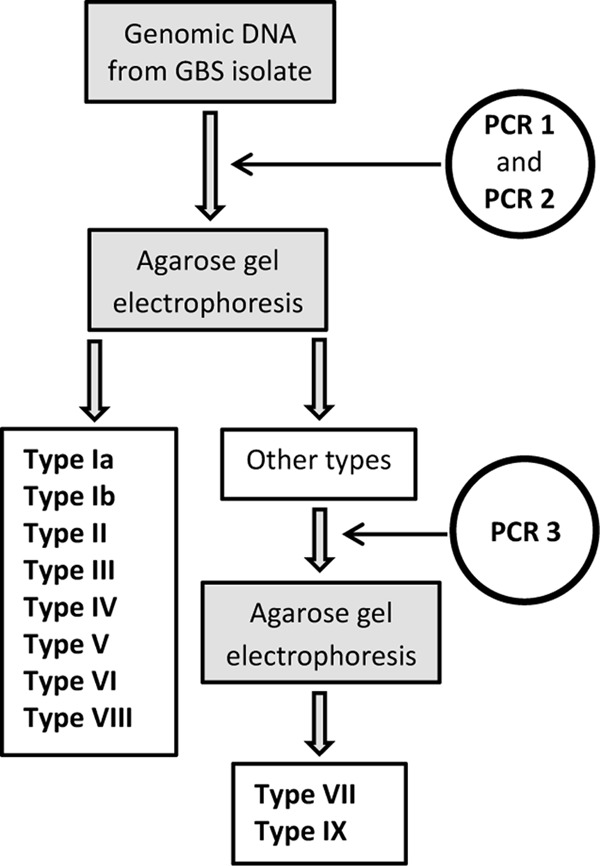
Diagram illustrating the proposed procedure for capsular gene typing of S. agalactiae. PCR 1 and PCR 2 are the two multiplex PCRs 1 and 2 described by Poyart et al. (14) and analyzed by 1% agarose gel electrophoresis. PCR 3 is a multiplex PCR using primers cpsI-Ia-6-7-F, cpsI-7-R, cpsI-7-9-F, and cpsI-9-R and analyzed by 1.5% agarose gel electrophoresis as described by Imperi et al. (11).
An inherent disadvantage of capsular gene typing is that the PCR-based methods do not reveal if the detected cps locus is expressed as a polysaccharide capsule. Different genetic mechanisms of capsule loss have been described for S. agalactiae, including insertion or deletion of DNA fragments in the cps region (13, 18, 19) and variation in the number of nucleotides in homopolymeric tracts in cps genes (13, 19, 20). The latter is presumably caused by slipped-strand mispairing events resulting in frameshift mutations, which enable phase variation in capsule expression (21). Among the 312 strains included in our study, 5 showed evidence of indels in the cps locus. However, the vast majority of the strains recorded as noncapsulated by both latex agglutination and flow cytometry did not show larger indels in the cps region as detected by a long-range PCR covering the region. Smaller indels of up to a few hundred nucleotides as described by Kong et al. (13) would not be revealed by this method due to limitations in resolution of large DNA fragments by agarose gel electrophoresis.
In conclusion, available serological and gene-based methods for capsular typing of S. agalactiae all have shortcomings that may result in lack of or erroneous results. Apart from misidentification of certain serotypes, the latex agglutination method depends on experience and is less sensitive than the flow cytometry method. We suggest that capsular gene typing using the described combination of existing methods should be included in epidemiological and surveillance studies of S. agalactiae in order to correctly identify the full spectrum of serotypes or to confirm serotyping results obtained by latex agglutination.
ACKNOWLEDGMENTS
This study was supported by the European Commission Seventh Framework (grant agreement number 200481).
We thank Tove Findahl, Karin Gregersen, and Lise Hald Schultz for their technical help.
The contributing members of the DEVANI (Design of a Vaccine Against Neonatal Infections) Study Group were P. Melin (Belgium), A. Decheva and B. Petrunov (Bulgaria), P. Kriz (Czech Republic), R. Berner, A. Büchele, M. Hufnagel, and M. Kunze (Germany), R. Creti, I. Margarit, and G. Orefici (Italy), J. R. Granger and M. De La Rosa Fraile (Spain), and B. Afshar and A. Efstratiou (United Kingdom).
J.L.T., D.M., and C.D.R. are employees of Novartis Vaccines and Diagnostics s.r.l., and J.L.T. holds shares in the company.
Footnotes
Published ahead of print 28 November 2012
REFERENCES
- 1. Dermer P, Lee C, Eggert J, Few B. 2004. A history of neonatal group B streptococcus with its related morbidity and mortality rates in the United States. J. Pediatr. Nurs. 19:357–363 [DOI] [PubMed] [Google Scholar]
- 2. Rodriguez-Granger J, Alvargonzalez JC, Berardi A, Berner R, Kunze M, Hufnagel M, Melin P, Decheva A, Orefici G, Poyart C, Telford J, Efstratiou A, Kilian M, Krizova P, Baldassarri L, Spellerberg B, Puertas A, Rosa-Fraile M. 2012. Prevention of group B streptococcal neonatal disease revisited. The DEVANI European project. Eur. J. Clin. Microbiol. Infect. Dis. 31:2097–2104 [DOI] [PubMed] [Google Scholar]
- 3. Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sendi P, Johansson L, Nørrby-Teglund A. 2008. Invasive group B streptococcal disease in non-pregnant adults. A review with emphasis on skin and soft-tissue infections. Infection 2:100–111 [DOI] [PubMed] [Google Scholar]
- 5. Barkema HW, Green MJ, Bradley AJ, Zadoks RN. 2009. The role of contagious disease in udder health. J. Dairy Sci. 92:4717–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sørensen UBS, Poulsen K, Ghezzo C, Margarit I, Kilian M. 2010. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1:e00178-10 doi:10.1128/mBio.00178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elliott JA, Facklam RR, Richter CB. 1990. Whole-cell protein patterns of nonhemolytic group B, type Ib, streptococci isolated from humans, cattle, frogs, and fish. J. Clin. Microbiol. 28:628–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans JJ, Bohnsack JF, Klesius PH, Whiting AA, Garcia JC, Shoemaker CA, Takahashi S. 2008. Phylogenetic relationships among Streptococcus agalactiae isolated from pischine, dolphin, bovine and human sources: a dolphin and pischine lineage associated with a fish epidemic in Kuwait is also associated with human neonatal infections in Japan. J. Med. Microbiol. 57:1369–1376 [DOI] [PubMed] [Google Scholar]
- 9. Rubens CE, Wessels MR, Heggen LM, Kasper DL. 1987. Transposon mutagenesis of type III group B Streptococcus: correlation of capsule expression with virulence. Proc. Natl. Acad. Sci. U. S. A. 84:7208–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Afshar B, Broughton K, Creti R, Decheva A, Hufnagel M, Kriz P, Lambertsen L, Lovgren M, Melin P, Orefici G, Poyart C, Radtke A, Rodriguez-Granger J, Sørensen UB, Telford J, Valinsky L, Zachariadou L, Members of the DEVANI Study Group, Efstratiou A. 2011. International external quality assurance for laboratory identification and typing of Streptococcus agalactiae (Group B streptococci). J. Clin. Microbiol. 49:1475–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R. 2010. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 80:212–214 [DOI] [PubMed] [Google Scholar]
- 12. Kong F, Gowan S, Martin D, James G, Gilbert GL. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kong F, Lambertsen LM, Slotved Ko H-CD, Wang H, Gilbert GL. 2008. Use of phenotypic and molecular serotype identification methods to characterize previously nonserotypeable group B streptococci. J. Clin. Microbiol. 46:2745–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poyart C, Tazi A, Réglier-Poupet H, Billoët A, Tavares N, Raymond J, Trieu-Cuot P. 2007. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J. Clin. Microbiol. 45:1985–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, Tettelin H, Brettoni C, Iacobini ET, Rosini R, D'Agostino N, Miorin L, Buccato S, Matiani M, Galli G, Nogarotto R, Nardi Dei V, Vegni F, Fraser C, Mancuso G, Teti G, Madoff LC, Paoletti LC, Rappuoli R, Kasper DL, Telford JL, Grandi G. 2005. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309:148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. 2007. A proposed new Streptococcus agalactiae serotype, serotype IX. J. Clin. Microbiol. 45:2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heath PT. 2011. An update on vaccination against group B streptococcus. Expert Rev. Vaccines 10:685–694 [DOI] [PubMed] [Google Scholar]
- 18. Creti R, Imperi M, Pataracchia M, Alfarone G, Recchia S, Baldassarri L. 2012. Identification and molecular characterization of a S. agalactiae strain lacking the capsular locus. Eur. J. Clin. Microbiol. Infect. Dis. 31:233–235 [DOI] [PubMed] [Google Scholar]
- 19. Ramaswamy SV, Ferrieri P, Madoff LC, Flores AE, Kumar N, Tettelin H, Paoletti LC. 2006. Identification of novel cps locus polymorphisms in nontypeable group B Streptococcus. J. Med. Microbiol. 55:775–783 [DOI] [PubMed] [Google Scholar]
- 20. Janulczyk R, Masignani V, Maione D, Tettelin H, Grandi G, Telford J. 2010. Simple sequence repeats and genome plasticity in Streptococcus agalactiae. J. Bacteriol. 192:3990–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sellin M, Håkansson S, Norgren M. 1995. Phase-shift of polysaccharide capsule expression in group B streptococci, type III. Microb. Pathog. 18:401–415 [DOI] [PubMed] [Google Scholar]


