Abstract
A method for rapid species identification of ticks may help clinicians predict the disease outcomes of patients with tick bites and may inform the decision as to whether to administer postexposure prophylactic antibiotic treatment. We aimed to establish a matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) spectrum database based on the analysis of the legs of six tick vectors: Amblyomma variegatum, Rhipicephalus sanguineus, Hyalomma marginatum rufipes, Ixodes ricinus, Dermacentor marginatus, and Dermacentor reticulatus. A blind test was performed on a trial set of ticks to identify specimens of each species. Subsequently, we used MALDI-TOF MS to identify ticks obtained from the wild or removed from patients. The latter tick samples were also identified by 12S ribosomal DNA (rDNA) sequencing and were tested for bacterial infections. Ticks obtained from the wild or removed from patients (R. sanguineus, I. ricinus, and D. marginatus) were accurately identified using MALDI-TOF MS, with the exception of those ticks for which no spectra were available in the database. Furthermore, one damaged specimen was correctly identified as I. ricinus, a vector of Lyme disease, using MALDI-TOF MS only. Six of the 14 ticks removed from patients were found to be infected by pathogens that included Rickettsia, Anaplasma, and Borrelia spp. MALDI-TOF MS appears to be an effective tool for the rapid identification of tick vectors that requires no previous expertise in tick identification. The benefits for clinicians include the more targeted surveillance of patients for symptoms of potentially transmitted diseases and the ability to make more informed decisions as to whether to administer postexposure prophylactic treatment.
INTRODUCTION
Ticks are obligate hematophagous parasites of the order Acari. These arthropods can feed on every known class of vertebrate and can bite people (1). Ticks are currently the second leading vector of human infectious diseases and can carry bacterial (1), viral (1a), and protozoan pathogens (2). However, only in 1982, with the identification of Borrelia burgdorferi as the etiological agent of Lyme disease, was the major effect of ticks on public health recognized, leading to an increased awareness of tick-borne diseases (3). Since then, more than 15 tick-borne rickettsioses have emerged throughout the world (4).
The removal of a tick from the human body is a common situation, and patients may visit a physician with an attached or removed tick. Certain tick species are well-known vectors of human diseases, such that identifying the species, which will alert the physician to the diseases that may have been transmitted, is clinically helpful if such information is obtained quickly (1). Indeed, recent studies confirm that the use of doxycycline prophylaxis following an Ixodes tick bite is useful for the prevention of Lyme disease (4). Similar postexposure regimens could also prevent tick-borne relapsing fever in areas of endemicity (5). However, for prophylactic treatment to be effective, it must be delivered shortly after potentially infectious ticks are removed from patients (6).
Ticks species can be morphologically identified using taxonomic keys for endemic species in several geographic regions (1). However, morphological identification can be difficult because it requires some entomological expertise, and it is difficult to identify a specimen that is damaged or at an immature stage of its life cycle (1). Molecular methods, such as the sequencing of the mitochondrial 12S (7), 18S (8), and 16S ribosomal DNAs (rDNAs) (7), mitochondrial cytochrome oxidase subunit 1 (COX1), and nuclear internal transcribed spacer 2 (ITS2), have been developed to identify arthropods, including ticks (9). However, there is currently no PCR assay that can distinguish tick species, and ideal PCR primer pairs that can amplify the relevant gene fragments are not available.
In addition to the technical and logistical drawbacks of PCR assays, this approach is further limited by the availability of gene sequences in GenBank. Protein profiling by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is now increasingly common for the routine identification of microorganisms in clinical microbiology (10). This revolutionary, reliable, and cost-effective technique is simpler and faster than conventional phenotypic and molecular methods for the identification of human pathogens (10).
The MALDI-TOF MS approach was first applied to arthropods for the differentiation of Drosophila species (11). It was found that protein extracts obtained from whole specimens generated reproducible spectra (11). Species-specific protein profiles have also been be used to differentiate three species of aphids (insects that feed on plants) (12). In 2011, a blind test in which 111 wild specimens were compared to the database profiles of Culicoides species showed that MALDI-TOF MS can differentiate species of Culicoides biting midges collected in the field (13). More recently, a MALDI-TOF MS study of seven ticks reported that whole ticks or body parts, excluding the legs, generate spectra that are sufficient for species identification (14).
The objective of the present study was to investigate the use of MALDI-TOF MS for the rapid differentiation of tick species using only their legs. Our goals were to establish a reference database, to evaluate the MALDI-TOF MS-based identification system in a blind test, and to evaluate this new identification tool using ticks removed from patients.
MATERIALS AND METHODS
Arthropods.
To establish a reference database, we used laboratory-reared hard ticks, fleas, lice, triatomines, mosquitoes, and bedbugs (Table 1). All specimens were fresh and nonengorged and maintained at room temperature for less than 1 week postmortem. The tick specimens included Amblyomma variegatum, Hyalomma marginatum rufipes, Rhipicephalus sanguineus, Ixodes ricinus, Dermacentor marginatus, and Dermacentor reticulatus. For the blind test, other specimens of the same species and specimens of other arthropod families were used as controls. Starved ticks collected in the field or removed from animals were characterized by MALDI-TOF MS (Table 1). A total of 14 ticks removed from 14 patients in France from June to August 2012 were morphologically characterized using standard taxonomic keys as D. marginatus, R. sanguineus, I. ricinus, Rhipicephalus sp., and Ixodes sp. (Table 2). One extensively damaged hard tick removed from a patient was also tested. After the morphological identification procedure, four legs were removed for MALDI-TOF MS (see below) and sequencing assays. DNA was extracted from tick body halves, and a 360-bp fragment of the mitochondrial 12S rDNA sequence was amplified by PCR and sequenced (14a). The sequences were analyzed using ChromasPro, version 1.34 (Technelysium Pty, Ltd., Tewantin, Queensland, Australia), and were compared with sequences from GenBank. The unused tick body parts were stored at −80°C for subsequent analyses.
Table 1.
Arthropods used to establish the reference database of MALDI-TOF spectra and arthropods used in the blind test
| Arthropod for database | Geographical origin | Sourcea | No. of specimens used to create the database |
No. of specimens used for the blind test procedure |
||
|---|---|---|---|---|---|---|
| Total | Sex and/or stageb | Total (score range) | Sex and/or stageb | |||
| Ticks | ||||||
| Amblyomma variegatum | Senegal | LC | 18 | 6 M, 5 F, 7 N | 2 (2.248–2.357) | 1 M, 1F |
| Rhipicephalus sanguineus | France | LC | 10 | 5 M, 5 F | 2 (1.995- 2.135) | 1 M, 1 F |
| France | Vegetation | 3 (1.715–1.745) | M | |||
| France | Vegetation | 2 (1.701–1.884) | F | |||
| Dermacentor marginatus | France | LC | 10 | 5 M, 5 F | 2 (2.232–2.519) | 1 M, 1 F |
| Dermacentor reticulatus | Croatia | LC | 10 | 5 M, 6 F | 3 (2.119- 2.281) | 1 M, 2 F |
| Hyalomma marginatum rufipes | Senegal | LC | 12 | 6 M, 6 F | 2 (1.992–2.352) | 1 M, 1 F |
| Ixodes ricinus | France | LC | 7 | N | 2 (1.706–1.831) | 1 M, 1 F |
| France | Vegetation | 7 (1.304–1.878) | 3 M, 2 F, 2 N | |||
| Rhipicephalus sulcatus | Senegal | Animal | 2 (1.009–1.166) | 1 M, 1 F | ||
| Haemaphysalis concinna | France | Vegetation | 2 (0.838–1.062) | 1 M, 1 F | ||
| Other arthropods | ||||||
| Ctenocephalides felis | England | LC | 17 | 11 F, 6 M | 2 (2.103–2.123) | 1 M, 1 F |
| Pediculus humanus corporis | USA | LC | 12 | 7 M, 6 F | 2 (1.788–1.797) | 1 M, 1 F |
| Triatoma infestans | Bolivia | LC | 12 | 6 M, 6 F | 2 (1.733–1.779) | 1 M, 1 F |
| Cimex lectularius | France | LC | 12 | 6 M, 6 F | 2 (2.145–2.429) | 1 M, 1 F |
| Culex pipiens | France | LC | 7 | F | 2 (1.655–1.749) | F |
| Apis mellifera | France | Field | 2 (0.732–0.778) | 1 M, 1 F | ||
| Pyrrhocoris apterus | France | Field | 2 (0.759–0.998) | 1 M, 1 F | ||
| Blaptica dubia | India | Field | 2 (0.707–1.066) | 1 M, 1 F | ||
| Tenibrio molitor | France | Field | 2 (0.755–0.958) | 1 M, 1 F | ||
LC, laboratory colonies.
M, male; F, female; N, nymph stage.
Table 2.
Identification of ticks removed from patients using morphological, MALDI-TOF MS, and molecular methods and the results of the detection of Rickettsia, Borrelia, Anaplasma, and Bartonella DNA
| Morphological identification | MALDI-TOF MS identification (score) | Molecular identification (% similarity with the indicated GenBank sequence) of:a |
|
|---|---|---|---|
| Ticke | Bacteria | ||
| D. marginatus | D. marginatus (1.83) | D. marginatus (99.3% AM410570.1) | |
| D. marginatus | D. marginatus (1.798) | D. marginatus (99.4% AM410570.1) | |
| D. marginatus | D. marginatus (1.799) | D. marginatus (99.4% AM410570.1) | R. slovacad, R. raoultiid |
| R. sanguineus | R. sanguineus (1.957) | R. sanguineus (99.1% JX206975.1) | R. massiliaed |
| I. ricinus | I. ricinus (1,737) | I. ricinus (99.5% JN248424.1) | |
| Damaged | I. ricinus (1,376) | Not obtainedc | |
| I. ricinus | I. ricinus (1.321) | I. ricinus (99.3% JN248424.1) | |
| Rhipicephalus sp. | Not identifiedb | R. bursa (100% AM410572.1) | |
| Ixodes sp. | I. ricinus (1.308) | I. ricinus (99.2% JM248424.1) | B. garinii (99.7% JN828689.1) |
| Rhipicephalus sp. | R. sanguineus (2.117) | R. sanguineus (99.2% AY559843.1) | R. conoriid |
| I. ricinus | I. ricinus (1.728) | I. ricinus (99.7% JN248424.1) | A. phagocytophilumd |
| I. ricinus | I. ricinus (1.823) | I. ricinus (99.2% JN248424.1) | |
| I. ricinus | I. ricinus (1.641) | I. ricinus (99.06% AY945481.1) | Incompletely described bacterium (97.6% AY776167.1), B. miyamotoi (100% FJ874925.1) |
| I. ricinus | I. ricinus (1.668) | I. ricinus (100% JN248424.1) | |
| D. marginatus | D. marginatus (1.83) | D. marginatus (99.3% AM410570.1) | |
| D. marginatus | D. marginatus (1.798) | D. marginatus (99.4% AM410570.1) | R. slovacad, R. raoultiid |
| D. marginatus | D. marginatus (1.799) | D. marginatus (99.4% AM410570.1) | R. massiliaed |
DNA extracted from laboratory colonies of uninfected ticks was used as a negative control. DNA extracted from Rickettsia montanensis, Bartonella elizabethae, Borrelia crocidurae, and Anaplasma phagocytophilum was used as a positive control.
No R. bursa spectrum was available in our MALDI-TOF MS database.
After two attempts.
qPCR.
By 12S RNA sequencing.
MALDI-TOF procedure. (i) Preparation of samples.
Each specimen was placed in a 1.5-ml microcentrifuge tube and immobilized or anesthetized at −20°C for 30 min (11). Subsequently, the specimens were rinsed once with distilled water. After the specimens were dried with paper, four legs were removed with scalpels. One of two different homogenization solutions was used depending on the specimen's size (11). The tick legs were manually homogenized in 60 μl of 70% formic acid and 60 μl of 50% acetonitrile in 1.5-ml microcentrifuge tubes using pellet pestles (Fischer Scientific). The legs of mosquitoes, fleas, and lice were also homogenized in 20 μl of formic acid and 20 μl of acetonitrile. Triatomine legs were homogenized in 100 μl of 70% formic acid and 100 μl of 50% acetonitrile. All homogenates were centrifuged at 10,000 rpm for 20 s, and 1 μl of each supernatant was spotted onto a steel target plate (Bruker Daltonics) in quadruplicate (13). One microliter of a CHCA matrix suspension composed of saturated α-cyano-4-hydroxycinnamic acid (Sigma), 50% acetonitrile, 10% trifluoroacetic acid, and high-performance liquid chromatography (HPLC)-grade water was directly spotted onto each sample on the target plate to allow cocrystallization (15). The target plate was dried for several minutes at room temperature before insertion into the MALDI-TOF MS instrument.
(ii) MALDI-TOF MS parameters.
Protein mass profiles were acquired using a Microflex MALDI-TOF mass spectrometer (Bruker Daltonics) with Flex Control software (Bruker Daltonics). We performed measurements in the linear positive-ion mode (15) between 2 and 20 kDa, and each spectrum corresponded to ions obtained from 240 laser shots performed in six regions of the same spot. The obtained spectra were processed using Flex Analysis, version 3.3, and MALDI Biotyper, version 3.0, software.
(iii) Spectral analysis and reference database creation.
To study the reproducibility of the species spectra, the average spectral profiles obtained from the four spots for each specimen of the same species were analyzed and compared using the ClinProTools, version 2.2, program (Bruker Daltonics). Using MALDI Biotyper, version 3.0, a database containing the studied arthropod groups was assembled from at least five specimens of each sex for all of the studied arthropod species, excluding mosquitoes, for which the majority of tested specimens were females. The reproducible spectra were used in the database.
(iv) Study validation using a blind test and identification of ticks from the field and patients.
An intercomparison of the reference spectra of each species was performed to interrogate the database (14, 16). The “start identification” function in the MALDI Biotyper allows the identification of each specimen to be tested, and the results were presented as log score values. A blind test was then performed with specimens from our laboratory colonies for which there were corresponding reference spectra in our database. For each species, two new specimens were used. We also tested two specimens of arthropods not included in our database, including bees, firebugs, roaches, and beetles. Identification scores were obtained for each spectrum of the tested sample. All ticks collected in the field and removed from patients were tested against the database. When each tick was tested, particular attention was paid to determine if the spectral profile matched profiles for the reference samples of the same sex as the specimen. The Mantel Haenszel test (Epi Info, version 7, program) was used to evaluate the quality of the sex identification.
(v) Cluster analysis.
We performed hierarchical clustering of the mass spectra of all tested species that were present in or absent from the database using the mean spectrum projection (MSP) dendrogram function of MALDI Biotyper, version 3.0. The objective was to determine whether the ticks could be clustered using this approach.
Molecular detection of bacteria in ticks removed from patients.
All DNA samples of ticks removed from patients were screened for Rickettsia spp. by quantitative real-time PCR (qPCR) analysis of a fragment of the gltA gene (17). To screen for Bartonella spp., an internally transcribed spacer was targeted, whereas a fragment of the 16S rRNA gene was targeted for Borrelia spp. (18). Rickettsia massiliae-specific qPCR and Rickettsia conorii-specific qPCR were performed on positive Rhipicephalus sanguineus DNA samples (19). Rickettsia slovaca- and Rickettsia raoultii-specific qPCRs were performed on positive Dermacentor DNA samples (17). Borrelia-positive samples were confirmed by Borrelia-specific qPCR amplification of the internal transcribed spacer (ITS) (19), and identification was performed by sequencing a 750-bp gene fragment of the flaB gene (20).
RESULTS
MALDI-TOF MS spectrum analysis and database assembly.
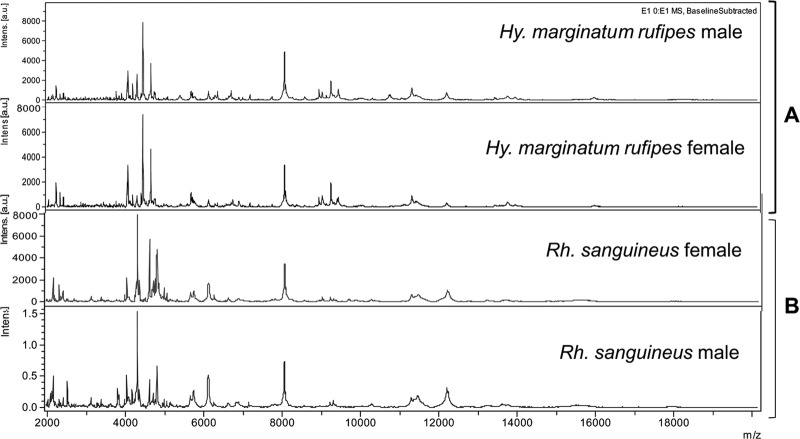
A total of 120 arthropod specimens, including 67 ticks representing six species, were subjected to MALDI-TOF MS analysis. The protein spectral profiles of all tested arthropod species were very similar between specimens of the same species within the mass range of 2 to 20 kDa, and the signal intensities were very strong. The alignment of the spectra of several specimens of the same species showed that the major identified protein peaks were present in each specimen of the same species (Fig. 1). The analysis in ClinProTools of the tick protein profiles yielded spectra comprising between 60 and 136 peaks in the mass range of 2 to 20 kDa, with an average of 106 peaks per spectrum. The spectra corresponding to the legs of other arthropods (Culex pipiens, Triatoma infestans, Ctenocephalides felis, Pediculus humanus, and Cimex lectularius) comprised 47 to 64, 94 to 120, 75 to 93, 46 to 76, and 87 to 132 peaks, respectively, in the mass range of 2 to 20 kDa.
Fig 1.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectral profiles for the legs of different tick species. (A) Hyalomma (Hy) marginatum rufipes male and female. (B) Rhipicephalus (Rh) sanguineus male and female in the range of 2 to 20 kDa. Intens, signal intensity; au, arbitrary units.
The spectra for each species of ticks were shown to be reproducible (Fig. 2) for all tested groups after spectral analysis and alignment. This reproducibility was observed for both the male and female specimens. Subsequently, a database was created and loaded with all of the spectra for these species.
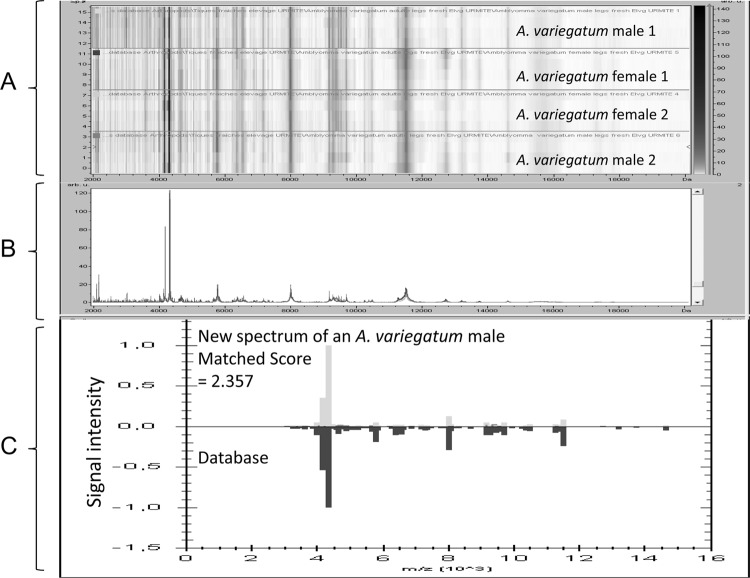
Fig 2.
Comparison of the A. variegatum spectral profiles to assess reproducibility and to identify an A. variegatum specimen. (A) Alignment of the spectra of several specimens of A. variegatum (gel view) showing that the major identified protein peaks were present in each specimen of this species. Arb u, arbitrary units. (B) Average peak list for the A. variegatum spectral profiles. (C) Results obtained for the blind testing of an A. variegatum specimen against the database using MALDI Biotyper, version 3.0, software (correct identification with a score of 2.357).
Study validation using a blind test.
Querying spectra in the database yielded satisfactory results, with scores between 2.224 and 2.764. The blind test of the reference database, in which two specimens of each of the six tick species maintained in the laboratory as well as the remaining arthropod groups were assayed, successfully identified all species groups. The majority (69%) of specimens were identified to the species level with scores of ≥2. All specimens tested had spectra that matched the reference spectra with scores between 1.749 and 2.519. The spectra of bees, firebugs, roaches, and beetles, for which references were not included in the database, scored lower, between 0.778 and 1.066. Regarding the sex of the specimen, it was not possible to definitively discriminate male and female ticks based on their MS spectra. However, the ticks' spectral profiles more frequently matched those of the correct sex than the incorrect sex, with 16/22 (72%) matches for males and 19/29 (65%) matches for females (P = 0.007).
MALDI-TOF MS using ticks from the field and patients.
The field-collected ticks for which the species were represented in the database (I. ricinus and R. sanguineus) were identified correctly by MALDI-TOF MS (scores of ≥1.8). R. sulcatus and Haemaphysalis concinna ticks from the field (absent from our MALDI-TOF MS database) did not match any known spectra. Eleven morphologically identified adult ticks (I. ricinus, R. sanguineus, and D. marginatus) removed from patients were correctly identified by MALDI-TOF MS (identification scores between 1.321 and 2.117). An Ixodes sp. nymph removed from a patient was identified as I. ricinus using MALDI-TOF MS. Additionally, the tick that was extensively damaged was identified as I. ricinus based on its four spectra, with a score of 1.376. However, one specimen (Rhipicephalus sp.) removed from a patient could not be identified by MALDI-TOF MS, as the spectrum did not match that of any specimens in the database.
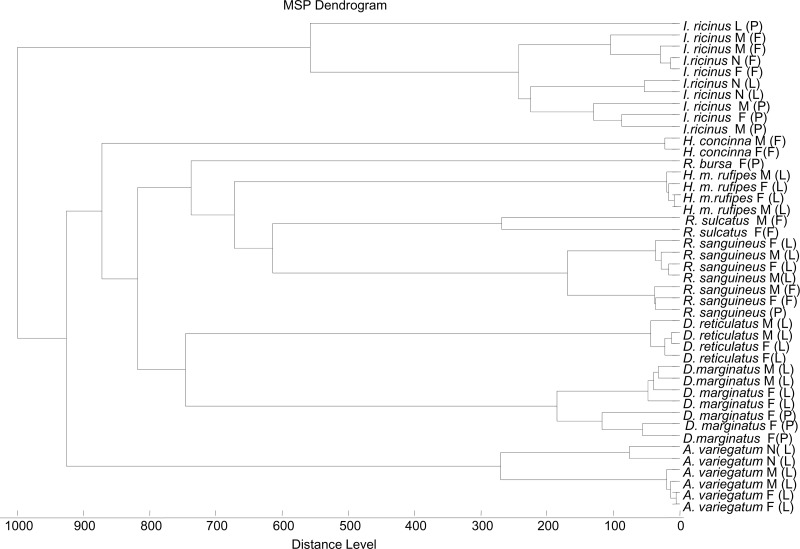
For the phylogenetic analysis, we performed hierarchical clustering of the mass spectra of different species of tested ticks that were present in or absent from our reference database. In the generated dendrogram, specimens of the same tick species, including R. sanguineus, I. ricinus, and D. marginatus from the laboratory, field, and patients, clustered together with the exception of one specimen (Fig. 3).
Fig 3.
Dendrogram obtained by cluster analysis of spectra obtained from laboratory-reared ticks, field-collected ticks, and ticks removed from patients. The ticks were analyzed using MALDI Biotyper software. M, male; F, female; N, nymph; L, ticks from laboratory colonies; F, ticks collected in the field; P, ticks collected from patients.
Molecular identification of ticks and the detection of bacteria.
12S gene sequencing was performed on 13 of 14 specimens removed from patients. The results corroborated those obtained by morphological analysis and MALDI-TOF MS for most samples (Table 1). R. sulcatus and H. concinna collected from the field were not identified by MALDI-TOF MS (no reference spectra in our database). After two attempts, we were unable to obtain a high-quality 12S RNA gene sequence for the damaged tick that was removed from a patient. This specimen was identified as I. ricinus by MALDI-TOF MS. Finally, the tick that was morphologically identified as Rhipicephalus sp. and was not identified by MALDI-TOF MS was identified by sequencing as Rhipicephalus bursa (100% agreement with AM410572 in GenBank), for which no spectrum was available in the MALDI-TOF database. Six of the 14 ticks removed from patients were found to be infected by bacteria, including Rickettsia massiliae, R. conorii, R. slovaca, R. raoultii, Borrelia garinii, Borrelia miyamotoi, and Anaplasma phagocytophilum (Table 1). DNA sequences closely related to those of an incompletely described bacterium (21) were also detected in one specimen of I. ricinus.
DISCUSSION
Although other factors should be considered, the identification of ticks that have bitten or been removed from patients is the first step in assessing the risk of infection (1). In this study, all ticks removed from patients were potential vectors of pathogens that depend on the involved tick species for propagation. In Europe, D. marginatus is one of the primary vectors of R. slovaca and R. raoultii, two spotted fever group rickettsiae responsible for tick-borne lymphadenopathy (TIBOLA), also called Dermacentor-borne necrosis erythema and lymphadenopathy (DEBONEL) or SENLAT, which is defined as scalp eschar and neck lymphadenopathy after a tick bite (21a). I. ricinus is a major vector of several bacterial agents, including the causative agent of Lyme borreliosis, Borrelia garinii (3), B. miyamotoi, which causes relapsing fever (21b), A. phagocytophilum, the causative agent of human granulocytic anaplasmosis (22), and Rickettsia helvetica, an emerging pathogen (4).
Rhipicephalus sanguineus, the brown dog tick, is a primary vector of Rickettsia conorii, the causative agent of life-threatening Mediterranean spotted fever (23), Rickettsia rickettsii is the vector of Rocky Mountain spotted fever in southern regions of the United States (23a), and R. massiliae is an emerging pathogen (23b). Rhipicephalus bursa is a known vector of several cattle parasites as well as a putative vector of Crimean-Congo hemorrhagic fever in certain regions of the world, such as Turkey (24). The potential for these ticks to transmit disease agents was illustrated in this study by the detection of several pathogens in the ticks that had bitten patients. This finding highlights the clinical need for the species identification of ticks.
Our results suggest that the MALDI-TOF MS spectra of protein extracts from tick legs are a suitable tool for identifying ticks. The results obtained using this method corroborated those from morphological and molecular identification methods.
Overall, 63% of the laboratory tick specimens were identified by MALDI-TOF MS with scores of >2, which are considered to be reliable scores for the identification of bacterial species (15).
Ticks collected in the field or removed from patients were reliably identified with lower score values, but each specimen's spectrum matched a reference spectrum. Only one tick was not identified by MALDI-TOF MS, R. bursa (identification confirmed by 12S sequencing). This tick was not identified because the corresponding spectrum was not present in our database. The species not represented in our database, which were tested as controls (fleas, mosquitoes, bugs, bees, and beetles), had low scores, less than 1.1, and none of their spectra matched the reference spectra in the database.
The establishment of a cutoff score for accurate identification would be ideal. When ticks removed from patients were tested, most (76%) were identified with scores of >1.7, and all were identified with scores of >1.3. However, it is not known if a score of 1.3 can be used as a definitive cutoff in other regions. The ticks used to construct the database are representative of the tick species in France and correspond to the species that have been removed from French patients, including returned travelers, and sent to our reference centers in the past 15 years (P. Parola, unpublished observations). In addition, the vectors included in this study are not closely related, and it must be confirmed that this MALDI-TOF MS method can differentiate closely related species, e.g., Ixodes species. However, when D. marginatus specimens removed from patients were tested against the database, which contained reference spectra for this species and for the closely related species D. reticulatus, the identification of the specimens was unequivocal.
Using the MSP dendrogram function of MALDI Biotyper, version 3.0, all but one tick (R. bursa) clustered according to their genera, species, and strain. A similar discrepancy was observed in a preliminary study using the entire body of the tick (14). Therefore, although MALDI-TOF MS has opened new doors for the phylogenetic study of arthropods and bacteria, additional data are still needed, and detailed studies should be conducted.
Contrary to a recent report (14), we found that the use of tick legs appears to be an effective means to identify tick vectors if a reference database is available. The remainder of the body can be reserved for other purposes, such as the detection of pathogens, as performed in this study. However, various storage conditions (10, 25) and chemical extraction methods (26) appear to influence the results of the MALDI-OF MS analysis of arthropod specimens. To expand our database, we will test a large number of tick species. Some species are available in collections. However, it seems that long-term storage in 70% ethanol reduces the reproducibility of the MALDI-TOF MS spectra (13, 25). It will also be useful to evaluate the use of MALDI-TOF MS for the identification of frozen specimens.
In conclusion, we showed in this study that MALDI-TOF MS is an efficient approach for the rapid identification of tick vectors. This method was used for the first time to identify ticks removed from patients. The results were obtained rapidly relative to the time required for molecular methods, and the completion of this assay does not require any specific entomological expertise. One damaged tick removed from a patient was successfully identified using MALDI-TOF MS as I. ricinus, a vector of Lyme disease. The knowledge of the tick species can be used to inform the clinician's decision as to whether to prescribe prophylactic doxycycline treatment. At the very least, the identity of the tick species can tell physicians which specific clinical signs they should look for in their patients. The rapid identification of ticks, and most likely of other arthropod vectors, is now possible in any laboratory with a MALDI-TOF MS system. In our unit, results are now available for clinicians in less than 1 h, with no requirement for entomological expertise. Our database contains reference spectra for ticks removed from humans in our area. This database can be shared and used directly by any clinical microbiology laboratory equipped with a MALDI Biotyper system. Because MALDI-TOF studies with bacteria and yeast have demonstrated significant variation in the protein profile based on geographical region (27), it will be interesting to test ticks of the same species from a variety of geographical regions to determine if this technique can be used as a regional or global tool. We will continue to add new reference spectra to our database to test the ability of this MS method to discriminate closely related species and to define a definitive cutoff score for species identification. Finally, it will be informative to determine whether MALDI-TOF MS can be used to identify not only tick vectors but also the microorganisms with which the ticks are infected.
ACKNOWLEDGMENTS
We thank Arnaud Canet for providing ticks collected in the field, Nicolas Armstrong for technical assistance, and Guillaume Lacour for providing mosquito specimens.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Parola P, Raoult D. 2001. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin. Infect. Dis. 32:897–928 [DOI] [PubMed] [Google Scholar]
- 1a. Hubalek Z, Rudolf I. 2012. Tick-borne viruses in Europe. Parasitol. Res. 111:9–36 [DOI] [PubMed] [Google Scholar]
- 2. Gray J, Zintl A, Hildebrandt A, Hunfeld KP, Weiss L. 2010. Zoonotic babesiosis: overview of the disease and novel aspects of pathogen identity. Ticks Tick-Borne Dis. 1:3–10 [DOI] [PubMed] [Google Scholar]
- 3. Stanek G, Wormser GP, Gray J, Strle F. 2012. Lyme borreliosis. Lancet 379:461–473 [DOI] [PubMed] [Google Scholar]
- 4. Parola P, Paddock CD, Raoult D. 2005. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin. Microbiol. Rev. 18:719–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balicer RD, Mimouni D, Bar-Zeev Y, Levine H, Davidovitch N, Ankol OH, Zarka SS. 2010. Post exposure prophylaxis of tick-borne relapsing fever. Eur. J. Clin. Microbiol. Infect. Dis. 29:253–258 [DOI] [PubMed] [Google Scholar]
- 6. Piesman J, Hojgaard A. 2012. Protective value of prophylactic antibiotic treatment of tick bite for Lyme disease prevention: an animal model. Ticks Tick-Borne Dis. 3:193–196 [DOI] [PubMed] [Google Scholar]
- 7. Norris DE, Klompen JS, Keirans JE, Black WC. 1996. Population genetics of Ixodes scapularis (Acari: Ixodidae) based on mitochondrial 16S and 12S genes. J. Med. Entomol. 33:78–89 [DOI] [PubMed] [Google Scholar]
- 8. Mangold AJ, Bargues MD, Mas-Coma S. 1998. 18S rRNA gene sequences and phylogenetic relationships of European hard-tick species (Acari: Ixodidae). Parasitol. Res. 84:31–37 [DOI] [PubMed] [Google Scholar]
- 9. Song S, Shao R, Atwell R, Barker S, Vankan D. 2011. Phylogenetic and phylogeographic relationships in Ixodes holocyclus and Ixodes cornuatus (Acari: Ixodidae) inferred from COX1 and ITS2 sequences. Int. J. Parasitol. 41:871–880 [DOI] [PubMed] [Google Scholar]
- 10. Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754 [DOI] [PubMed] [Google Scholar]
- 11. Feltens R, Gorner R, Kalkhof S, Groger-Arndt H, von Bergen M. 2010. Discrimination of different species from the genus Drosophila by intact protein profiling using matrix-assisted laser desorption ionization mass spectrometry. BMC Evol. Biol. 10:95 doi:10.1186/1471-2148-10-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perera MR, Vanstone VA, Jones MG. 2005. A novel approach to identify plant parasitic nematodes using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 19:1454–1460 [DOI] [PubMed] [Google Scholar]
- 13. Kaufmann C, Schaffner F, Ziegler D, Pfluger V, Mathis A. 2012. Identification of field-caught Culicoides biting midges using matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasitology 139:248–258 [DOI] [PubMed] [Google Scholar]
- 14. Karger A, Kampen H, Bettin B, Dautel H, Ziller M, Hoffmann B, Suss J, Klaus C. 2012. Species determination and characterization of developmental stages of ticks by whole-animal matrix-assisted laser desorption/ionization mass spectrometry. Ticks Tick-Borne Dis. 3:78–89 [DOI] [PubMed] [Google Scholar]
- 14a. Beati L, Keirans JE. 2001. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 87:32–48 [DOI] [PubMed] [Google Scholar]
- 15. Fournier PE, Couderc C, Buffet S, Flaudrops C, Raoult D. 2009. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J. Med. Microbiol. 58:1154–1159 [DOI] [PubMed] [Google Scholar]
- 16. Sauer S, Freiwald A, Maier T, Kube M, Reinhardt R, Kostrzewa M, Geider K. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843 doi:10.1371/journal.pone.0002843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bechah Y, Socolovschi C, Raoult D. 2011. Identification of rickettsial infections by using cutaneous swab specimens and PCR. Emerg. Infect. Dis. 17:83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parola P, Diatta G, Socolovschi C, Mediannikov O, Tall A, Bassene H, Trape JF, Raoult D. 2011. Tick-borne relapsing fever borreliosis, rural Senegal. Emerg. Infect. Dis. 17:883–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Socolovschi C, Reynaud P, Kernif T, Raoult D, Parola P. 2012. Rickettsiae of spotted fever group, Borrelia valaisiana, and Coxiella burnetii in ticks on passerine birds and mammals from the Camargue in the south of France. Ticks Tick-Borne Dis. 3:355–360 [DOI] [PubMed] [Google Scholar]
- 20. Assous MV, Wilamowski A, Bercovier H, Marva E. 2006. Molecular characterization of tickborne relapsing fever Borrelia, Israel. Emerg. Infect. Dis. 12:1740–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skarphedinsson S, Jensen PM, Kristiansen K. 2005. Survey of tickborne infections in Denmark. Emerg. Infect. Dis. 11:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a. Parola P, Rovery C, Rolain JM, Brouqui P, Davoust B, Raoult D. 2009. Rickettsia slovaca and R. raoultii in tick-borne rickettsioses. Emerg. Infect. Dis. 15(7):1105–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b. Platonov AE, Karan LS, Kolyasnikova NM, Makhneva NA, Toporkova MG, Maleev VV, Fish D, Krause PJ. 2011. Humans infected with relapsing fever spirochete Borrelia miyamotoi, Russia. Emerg. Infect. Dis. 17:1816–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dumler JS. 2012. The biological basis of severe outcomes in Anaplasma phagocytophilum infection. FEMS Immunol. Med. Microbiol. 64:13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rovery C, Raoult D. 2008. Mediterranean spotted fever. Infect. Dis. Clin. North Am. 22:515–530 [DOI] [PubMed] [Google Scholar]
- 23a. Demma LJ, Traeger MS, Nicholson WL, Paddock CD, Blau DM, Eremeeva ME, Dasch GA, Levin ML, Singleton J, Jr, Zaki SR, Cheek JE, Swerdlow DL, McQuiston JH. 2005. Rocky Mountain spotted fever from an unexpected tick vector in Arizona. N. Engl. J. Med. 353:587–594 [DOI] [PubMed] [Google Scholar]
- 23b. Parola P, Socolovschi C, Jeanjean L, Bitam I, Fournier PE, Sotto A, Labauge P, Raoult D. 2008. Warmer weather linked to tick attack and emergence of severe rickettsioses. PLoS Negl. Trop. Dis. 2:e338 doi:10.1371/journal.pntd.0000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Estrada-Pena A, Bouattour A, Camicas A, Walker AR. 2004. Ticks of domestic animals in the Mediterranean region: a guide to identification of species. University of Zaragoza, Zaragoza, Spain [Google Scholar]
- 25. Kaufmann C, Ziegler D, Schaffner F, Carpenter S, Pfluger V, Mathis A. 2011. Evaluation of matrix-assisted laser desorption/ionization time of flight mass spectrometry for characterization of Culicoides nubeculosus biting midges. Med. Vet. Entomol. 25:32–38 [DOI] [PubMed] [Google Scholar]
- 26. Carbonnelle E, Mesquita C, Bille E, Day N, Dauphin B, Beretti JL, Ferroni A, Gutmann L, Nassif X. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 27. Cassagne C, Ranque S, Normand AC, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. 2011. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One 6(12):e28425 doi:10.1371/journal.pone.0028425 [DOI] [PMC free article] [PubMed] [Google Scholar]