Abstract
The TaqMan Array Card (TAC) system is a 384-well singleplex real-time PCR format that has been used to detect multiple infection targets. Here we developed an enteric TaqMan Array Card to detect 19 enteropathogens, including viruses (adenovirus, astrovirus, norovirus GII, rotavirus, and sapovirus), bacteria (Campylobacter jejuni/C. coli, Clostridium difficile, Salmonella, Vibrio cholerae, diarrheagenic Escherichia coli strains including enteroaggregative E. coli [EAEC], enterotoxigenic E. coli [ETEC], enteropathogenic E. coli [EPEC], and Shiga-toxigenic E. coli [STEC]), Shigella/enteroinvasive E. coli (EIEC), protozoa (Cryptosporidium, Giardia lamblia, and Entamoeba histolytica), and helminths (Ascaris lumbricoides and Trichuris trichiura), as well as two extrinsic controls to monitor extraction and amplification efficiency (the bacteriophage MS2 and phocine herpesvirus). Primers and probes were newly designed or adapted from published sources and spotted onto microfluidic cards. Fecal samples were spiked with extrinsic controls, and DNA and RNA were extracted using the QiaAmp Stool DNA minikit and the QuickGene RNA Tissue kit, respectively, and then mixed with Ag-Path-ID One Step real-time reverse transcription-PCR (RT-PCR) reagents and loaded into cards. PCR efficiencies were between 90% and 105%, with linearities of 0.988 to 1. The limit of detection of the assays in the TAC was within a 10-fold difference from the cognate assays performed on plates. Precision testing demonstrated a coefficient of variation of below 5% within a run and 14% between runs. Accuracy was evaluated for 109 selected clinical specimens and revealed an average sensitivity and specificity of 85% and 77%, respectively, compared with conventional methods (including microscopy, culture, and immunoassay) and 98% and 96%, respectively, compared with our laboratory-developed PCR-Luminex assays. This TAC allows fast, accurate, and quantitative detection of a broad spectrum of enteropathogens and is well suited for surveillance or clinical purposes.
INTRODUCTION
Diarrhea is the second cause of death worldwide in children under the age of five, with the majority of cases occurring in developing countries (1). Diarrheal diseases are also a public health problem in developed countries, particularly in the context of the food supply. Understanding the etiology of diarrhea is important to guide public health efforts and can be important for individual patient care. The conventional diagnosis for diarrheal diseases encompasses a variety of methods including bacterial culture, immunoassay for toxins and antigens of bacteria, viruses, and protozoa, and microscopy for parasites. PCR amplification has emerged as a useful tool to detect pathogen DNA or RNA. It can be performed in either singleplex or multiplex mode in combination with gel electrophoresis, probe hybridization, or real-time fluorescence for detection. Real-time PCR is found to be a sensitive and quantitative amplification/detection method, but its multiplexing capacity has been limited by the availability of fluorescent dyes and platforms. Recently, Life Technologies has started offering a TaqMan Array Card (TAC) platform enabling spatial multiplexing of up to 384 targets, which was originally intended for gene expression studies. The CDC pioneered the use of TAC for detection of 21 respiratory pathogens (2), while recently the United Kingdom's Defense Science and Technology Laboratory also demonstrated the feasibility of TAC for pathogen detection with an example of five biothreat agents (3). Here we report on a TaqMan Array card that we developed for simultaneous detection of 19 diarrhea-causing enteropathogens, including viruses (rotavirus, norovirus GII, adenovirus, astrovirus, and sapovirus), bacteria including diarrheagenic Escherichia coli (enterotoxigenic E. coli [ETEC], enteropathogenic E. coli [EPEC], enteroaggregative E. coli [EAEC], and Shiga-toxigenic E. coli [STEC]), Shigella/enteroinvasive E. coli (EIEC), Salmonella, Campylobacter jejuni/Campylobacter coli, Vibrio cholerae, and Clostridium difficile, and parasites (Cryptosporidium, Giardia lamblia, Entamoeba histolytica, Ascaris lumbricoides, and Trichuris trichiura).
MATERIALS AND METHODS
Specimens.
Reference strains purchased from the American Type Culture Collection (ATCC, Manassas, VA) or clinical isolates were used for analytical testing for Salmonella, Shigella, Campylobacter, Vibrio cholerae, EAEC, STEC, EPEC, ETEC, EIEC, C. difficile, Entamoeba histolytica, and Giardia lamblia. Cryptosporidium parvum oocysts were purchased from Waterborne Inc. (New Orleans, LA). For RNA targets including rotavirus, norovirus GII, astrovirus, and sapovirus, in vitro transcripts were generated as described earlier and used for analytical testing (4). For adenovirus, Ascaris lumbricoides, and Trichuris trichiura, amplicons covering the targeted regions were generated. One hundred nine stool samples were selected from studies in Haydom, Tanzania (2010 to 2011), and from the Mirpur region of Dhaka, Bangladesh (2008 to 2009), to validate the clinical performance of TAC with a goal of 15 samples positive by conventional methods for most pathogens. Tanzanian samples were tested through an ongoing 5-year study, Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED). Bangladeshi samples were selected from a birth cohort study (5). Stool samples were delivered to the laboratories within 3 h of collection while maintaining a cold chain, shipped to the University of Virginia on dry ice, and stored at −80°C prior to assays.
Conventional tests.
Clinical samples were tested by enzyme-linked immunosorbent assay (ELISA) for Cryptosporidium spp., Giardia lamblia, E. histolytica (all from TechLab, Blacksburg, VA), Campylobacter, rotavirus, adenovirus, and astrovirus (all from Oxoid, Hampshire, United Kingdom). Samples were cultured on MacConkey, xylose lysine deoxycholate (XLD), and thiosulfate-citrate-bile salts-sucrose (TCBS) agars. Salmonella, Shigella, and Vibrio cholerae were identified biochemically. Colonies were pooled from MacConkey agar and tested for diarrheagenic E. coli strains including EAEC (targeting aaiC and aatA), EIEC (targeting ipaH), EPEC (targeting eae and bfpA), ETEC (targeting the heat-stable and heat-labile enterotoxin [ST and LT] genes), and STEC (targeting stx1 and stx2) using a 9-plex assay described previously (6). Samples were examined for Ascaris and Trichuris by microscopy.
Nucleic acid extraction.
DNA was extracted from 200 mg of stool with a modified QiaAmp stool DNA extraction protocol (Qiagen, Valencia, CA). In brief, stool was first lysed with QiaAmp ASL buffer, beaten for 3 min with 212- to 300-μm glass beads (Sigma, St. Louis, MO), and boiled for 5 min, and we then proceeded according to the manufacturer's instructions. RNA was extracted from 50 mg of stool with the QuickGene RNA Tissue kit automated with QuickGene-810 (Fujifilm, Tokyo, Japan) (4). As extrinsic controls, 106 copies of phocine herpesvirus (PhHV; a gift from Martin Schutten, Erasmus MC, Department of Virology, Rotterdam, The Netherlands) and 107 MS2 bacteriophage per sample were spiked to the lysis buffers to monitor the efficiency/inhibition of extraction and amplification. The elution volume for DNA was 200 μl; for RNA, it was 50 μl, which was then supplemented with 50 μl of RNA storage solution (Life Technologies, Carlsbad, CA). A no-template extraction control was included every week to exclude lab contamination.
TAC design and procedure.
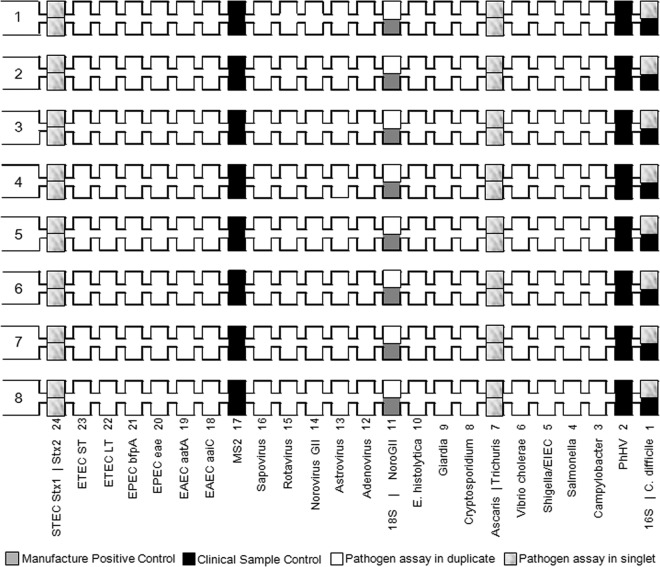
TAC assays were taken from our published assays and adapted from published sources when possible (Table 1). A single target was selected for 15 pathogens, while two targets were used for EPEC, STEC, ETEC, and EAEC. Assays were validated on plates based on the TaqMan array universal formula of a final primer concentration of 900 nM and a probe concentration of 250 nM. The Ag-Path-ID One-Step RT-PCR kit was used, and the cycling conditions were as follows: 45°C for 20 min and 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The assay mixtures were spotted onto the microfluidic card as laid out in Fig. 1. Twenty assay mixtures were spotted in duplicate and six in singlet. For each sample, equal volumes (20 μl each) of DNA and RNA extracts were combined and then mixed with 50 μl of Ag-Path-ID RT-PCR buffer, 4 μl of enzyme mix, and 6 μl of water to a 100-μl final volume. After thorough mixing, the reaction mixture was loaded into each port of the card and centrifuged twice at 1,200 rpm for 1 min. The card was sealed, the loading ports were excised, and then the card was inserted into a ViiA7 instrument (Life Technologies) and run under the same cycling conditions as above. Upon receipt, this lot of cards was tested with a no-template control that confirmed no amplification.
Table 1.
Assay primer and probe sequencesa
| Organism | Target | Sequence | Reference |
|---|---|---|---|
| Adenovirus | Hexonb | F, GCCACGGTGGGGTTTCTAAACTT | 7 |
| R, GCCCCAGTGGTCTTACATGCACATC | |||
| P, TGCACCAGACCCGGGCTCAG | |||
| Astrovirus | Capsid | F, CAGTTGCTTGCTGCGTTCA | 4 |
| R, CTTGCTAGCCATCACACTTCT | |||
| P, CACAGAAGAGCAACTCCATCGC | |||
| Norovirus GII | ORF1-ORF2 | F, CARGARBCNATGTTYAGR TGGATGAG | 8 |
| R, TCGACGCCATCTTCATTCACA | |||
| P, TGGGAGGGCGATCGCAATCT | |||
| Rotavirus | NSP3 | F, ACCATCTWCACRTRACCCTCTATGAG | 9 |
| R, GGTCACATAACGCCCCTATAGC | |||
| P, AGTTAAAAGCTAACACTGTCAAA | |||
| Sapovirus | RdRp | F, GAYCAGGCTCTCGCYACCTAC | 10 (modified) |
| F, TTTGAACAAGCTGTGGCATGCTAC | |||
| R, CCCTCCATYTCAAACACTA | |||
| P, CYTGGTTCATAGGTGGTRCAG | |||
| P, CAGCTGGTACATTGGTGGCAC | |||
| EAEC | aaiC | F, ATTGTCCTCAGGCATTTCAC | 11 (modified) |
| R, ACGACACCCCTGATAAACAA | |||
| P, TAGTGCATACTCATCATTTAAG | |||
| aatA | F, CTGGCGAAAGACTGTATCAT | 11 | |
| R, TTTTGCTTCATAAGCCGATAGA | |||
| P, TGGTTCTCATCTATTACAGACAGC | |||
| STEC | stx1 | F, ACTTCTCGACTGCAAAGACGTATG | 12 (modified) |
| R, ACAAATTATCCCCTGWGCCACTATC | |||
| P, CTCTGCAATAGGTACTCCA | |||
| stx2 | F, CCACATCGGTGTCTGTTATTAACC | 12 | |
| R, GGTCAAAACGCGCCTGATAG | |||
| P, TTGCTGTGGATATACGAGG | |||
| EPEC | eae | F, CATTGATCAGGATTTTTCTGGTGATA | 13 (modified) |
| R, CTCATGCGGAAATAGCCGTTA | |||
| P, ATACTGGCGAGACTATTTCAA | |||
| bfpA | F, TGGTGCTTGCGCTTGCT | 14 (modified) | |
| R, CGTTGCGCTCATTACTTCTG | |||
| P, CAGTCTGCGTCTGATTCCAA | |||
| ETEC | LT | F, TTCCCACCGGATCACCAA | 12 |
| R, CAACCTTGTGGTGCATGATGA | |||
| P, CTTGGAGAGAAGAACCCT | |||
| ST | Fh, GCTAAACCAGYAGRGTCTTCAAAA | 15 (modified) | |
| Fp, TGAATCACTTGACTCTTCAAAA | |||
| Rh, CCCGGTACARGCAGGATTACAACA | |||
| Rp, GGCAGGATTACAACAAAGTT | |||
| Ph, TGGTCCTGAAAGCATGAA | |||
| Pp, TGAACAACACATTTTACTGCT | |||
| EIEC/Shigella | ipaHc | F, CCTTTTCCGCGTTCCTTGA | 16 |
| R, CGGAATCCGGAGGTATTGC | |||
| P, CGCCTTTCCGATACCGTCTCTGCA | |||
| Campylobacter jejuni/C. coli | cadF | F, CTGCTAAACCATAGAAATAAAATTTCTCAC | 17 |
| R, CTTTGAAGGTAATTTAGATATGGATAATCG | |||
| P, CATTTTGACGATTTTTGGCTTGA | |||
| Salmonella | invA | F, TCGGGCAATTCGTTATTGG | 18 (modified) |
| R, GATAAACTGGACCACGGTGACA | |||
| P, AAGACAACAAAACCCACCGC | |||
| V. cholerae | toxR | F, GTTTGGCGAGAGCAAGGTTT | 18 (modified) |
| R, TCTCTTCTTCAACCGTTTCCA | |||
| P, CGCAGAGTCGAAATGGCTTGG | |||
| C. difficile | tcdB | F, GGTATTACCTAATGCTCCAAATAG | 19 (modified) |
| R, TTTGTGCCATCATTTTCTAAGC | |||
| P, CCTGGTGTCCATCCTGTTTC | |||
| Cryptosporidium | 18S | F, GGGTTGTATTTATTAGATAAAGAACCA | 20 (modified) |
| R, AGGCCAATACCCTACCGTCT | |||
| P, TGACATATCATTCAAGTTTCTGAC | |||
| Giardia | 18S | F, GACGGCTCAGGACAACGGTT | 21 |
| R, TTGCCAGCGGTGTCCG | |||
| P, CCCGCGGCGGTCCCTGCTAG | |||
| E. histolytica | 18S | F, ATTGTCGTGGCATCCTAACTCA | 21 |
| R, GCGGACGGCTCATTATAACA | |||
| P, TCATTGAATGAATTGGCCATTT | |||
| Ascaris | ITS1 | F, GTAATAGCAGTCGGCGGTTTCTT | 22 |
| R, GCCCAACATGCCACCTATTC | |||
| P, TTGGCGGACAATTGCATGCGAT | |||
| Trichuris | 18S | F, TTGAAACGACTTGCTCATCAACTT | This study |
| R, CTGATTCTCCGTTAACCGTTGTC | |||
| P, CGATGGTACGCTACGTGCTTACCATGG | |||
| PhHV | gB | F, GGGCGAATCACAGATTGAATC | 23 (modified) |
| R, GCGGTTCCAAACGTACCAA | |||
| P, TATGTGTCCGCCACCATCT | |||
| MS2 | MS2g1 | F, TGGCACTACCCCTCTCCGTATTCAC | 24 |
| R, GTACGGGCGACCCCACGATGAC | |||
| P, CACATCGATAGATCAAGGTGCCTACAAGC | |||
| Bacterial 16S rRNA gene | F, TCCTACGGGAGGCAGCA | 25 (modified) | |
| R, GGACTACCAGGGTATCTAATCCTG | |||
| P, CGTATTACCGCGGCTGCT |
F, forward primer; R, reverse primer; P, probe, labeled with FAM (6-carboxyfluorescein) at 5′ and MGB at 3′; h, STh; p, STp.
The assay detected all adenovirus serotypes.
The ipaH gene was targeted for detection of both Shigella and EIEC.
Fig 1.
Enteric TaqMan Array Card layout. Format 48 was used.
Combined positive controls.
Two combined positive controls, one for DNA targets and one for RNA targets, were designed according to the method of Kodani and Winchell (26). Plasmids were synthesized by GeneWiz (South Plainfield, NJ) (see the supplemental material for the sequences). For DNA targets, the plasmid was directly utilized, while the RNA template was generated by amplification of the insert portion with M13 primers and in vitro transcription with the AmpliScribe T7 High Yield Transcription kit (Epicentre, Madison, WI) (4).
PCR-Luminex.
PCR-Luminex panels, including viral, bacterial, E. coli, protozoan, and helminth panels, were run according to previously established protocols (4, 6, 18, 27). Results were reported in units of median fluorescence intensity (MFI). To compare real-time PCR cycle threshold (CT) values versus PCR-Luminex MFI values, relative fluorescence intensity was calculated as the percentile versus the highest fluorescence intensity observed for each target, i.e., the MFI cutoff for positivity was set as 0 and the maximal MFI as 100%.
Analytical performance.
Linearity was tested with 10-fold serial dilutions of combined positive controls. For limit of detection (LOD) and precision (repeatability and reproducibility), positive materials (either whole organisms or the DNA/RNA templates as indicated above) were spiked into healthy donor stool, extracted, and then amplified. Repeatability was tested with eight repeats of two samples respectively spiked with a high and a low concentration of each target. Reproducibility was tested with 10 identically spiked samples for each concentration (two concentrations, high and low, were interrogated) that were extracted and assayed over 5 days. LOD was defined as the lowest concentration at which the target could be detected in all 10 spiked samples. Matrix inhibition was tested with combined positive controls spiked into three different lots of stool from healthy donors. Analytical accuracy was evaluated with reference samples. Nineteen pathogens were divided into three groups: A, consisting of adenovirus, astrovirus, norovirus GII, rotavirus, sapovirus, Ascaris, and Trichuris; B, consisting of Campylobacter, Salmonella, Shigella, V. cholerae, and C. difficile; and C, consisting of EAEC, STEC, EPEC, ETEC, Cryptosporidium, Giardia, and E. histolytica. Combinations of three groups at different concentrations (negative, low, and high) were prepared and spiked into healthy donor stool. A total of 30 samples were prepared and assayed.
Statistics.
Correlation was tested by regression analysis using the analysis of variance (ANOVA) test. CT values were compared by the t test using IBM SPSS software. All P values were two-tailed, and values of <0.05 were considered statistically significant.
RESULTS
Analytical performance.
We compared CT values between single template and pooled templates of all 19 targets, and no significant differences were observed (data not shown). Therefore, for development purposes the spiking materials for all 19 pathogens were mixed together and assayed as one sample. Under the universal conditions on the microfluidic card, all assays showed good linearity (R2 = 0.988 to 1, P < 0.05) and 90 to 105% amplification efficiency within the tested range from 10 to 106 organisms per reaction mixture (Table 2). The limit of detection of all the assays, defined as 100% detection among 10 distinct samples, ranged from 103 to 107 copies of organisms or nucleic acid template per gram of stool, equivalent to 0.1 to 500 copies (prior to extraction) per 1 μl of reaction mixture (Table 2). This LOD was within 10-fold of the cognate assays tested on plates (data not shown). The CT values of 23 pathogen assays had within-run variance from 0.7% to 3.6% (repeatability, n = 8) and between-run variance from 2.5% to 7.9% (reproducibility, n = 10) (Table 2). Stool is a difficult substrate to use for extraction and amplification because of the presence of a variety of inhibitors, which can vary between samples. We tested matrix inhibition using three lots of stool from healthy donors spiked with combined positive controls and extracted and amplified in duplicate. As shown in Fig. S1 in the supplemental material, no significant difference was observed among three lots for any of the assays.
Table 2.
Analytical performance of enteric TaqMan Array Carda
| Organism | Linearityb R2 (efficiency, %) | LOD (equiv. no. of copies)c | Low- and high-concn CV (%) ford: |
Accuracy (%)d |
||
|---|---|---|---|---|---|---|
| Repeatability | Reproducibility | Sensitivitye | Specificity | |||
| Adenovirus | 0.997 (99.7) | 106 (100) | 1.1, 1.7 | 2.6, 4.8 | 100 | 100 |
| Astrovirus | 0.999 (93.0) | 5 × 106 (500) | 2.2, 2.6 | 3.2, 5.3 | 95 | 100 |
| Norovirus GII | 0.998 (99.2) | 5 × 106 (500) | 1.9, 1.1 | 3.5, 5.3 | 97 | 100 |
| Rotavirus | 0.993 (98.4) | 5 × 106 (500) | 1.5, 1.8 | 4.2, 7.5 | 100 | 100 |
| Sapovirus | 0.994 (94.9) | 5 × 106 (500) | 1.8, 2.9 | 5.8, 7.9 | 97 | 100 |
| EAEC | ||||||
| aaiC | 0.995 (99.7) | 105 (10) | 1.4, 2.8 | 4.2, 6.6 | 100 | 100 |
| aatA | 0.998 (95.5) | 105 (10) | 2.0, 3.0 | 6.3, 7.9 | 98 | 100 |
| STEC | ||||||
| stx1 | 0.999 (96.3) | 105 (10) | 0.9, 2.9 | 3.5, 5.8 | 100 | 100 |
| stx2 | 0.999 (94.6) | 105 (10) | 0.9, 1.8 | 4.2, 5.5 | 100 | 100 |
| EPEC | ||||||
| eae | 0.998 (100.2) | 105 (10) | 1.2, 1.5 | 3.6, 4.9 | 100 | 100 |
| bfpA | 0.998 (96.7) | 105 (10) | 0.9, 1.3 | 4.2, 5.2 | 100 | 100 |
| ETEC | ||||||
| LT | 0.998 (95.8) | 105 (10) | 0.7, 1.3 | 2.5, 3.6 | 100 | 100 |
| ST | 0.998 (95.5) | 105 (10) | 1.0, 1.1 | 3.1, 6.2 | 100 | 100 |
| EIEC/Shigella | 0.997 (104.2) | 105 (10) | 1.2, 1.3 | 3.6, 5.2 | 100 | 100 |
| Campylobacter | 0.997 (95.9) | 106 (100) | 0.9, 2.4 | 4.9, 7.2 | 100 | 100 |
| Salmonella | 0.997 (100.9) | 106 (100) | 2.3, 2.1 | 7.2, 7.5 | 98 | 100 |
| V. cholerae | 0.997 (102.1) | 106 (100) | 1.1, 1.7 | 7.6, 8.4 | 100 | 100 |
| C. difficile | 1.000 (99.4) | 106 (100) | 2.2, 1.9 | 4.2, 5.1 | 97 | 100 |
| Cryptosporidium | 0.998 (100.4) | 103 (0.1) | 2.0, 2.9 | 3.6, 6.3 | 100 | 100 |
| Giardia | 0.998 (101.4) | 103 (0.1) | 1.7, 3.0 | 5.2, 4.7 | 100 | 100 |
| E. histolytica | 0.988 (93.0) | 103 (0.1) | 2.0, 3.6 | 6.8, 6.7 | 100 | 100 |
| Ascaris | 0.993 (90.6) | 106 (100) | 1.3, 2.5 | 4.5, 7.3 | 100 | 100 |
| Trichuris | 0.999 (94.6) | 106 (100) | 0.7, 0.7 | 3.6, 5.2 | 100 | 100 |
| PhHV | 0.998 (98.3) | 3.1, 1.8 | 4.2, 3.5 | |||
| MS2 | 0.999 (96.8) | 1.4, 2.1 | 7.2, 8.4 | |||
| Bacterial 16S rRNA gene | 0.995 (91.0) | 4.9, 4.2 | 5.1, 4.7 | |||
Linearity was tested with combined positive controls. LOD and precision (repeatability and reproducibility) were tested with pooled positive materials spiked into healthy donor stool. LOD is the number of copies per gram of stool that were 100% detectable with 10 distinct extractions/amplifications.
The linearity range was 10 to 106 copy numbers per reaction for all the targets.
LOD, copy number of an organism or artificial template per gram of stool; equiv. no. of copies, equivalent copy numbers per 1-μl volume of reaction mixture prior to extraction.
Coefficients of variance (CVs) at both low (LOD) and high spiked concentrations are shown. High-concentration spikes were 1,000-fold higher for viral targets and 100-fold higher for bacterial and parasitic targets than LOD. The same concentrations were applied to accuracy tests.
Sensitivity was calculated on the total number of PCRs for each target.
Analytical accuracy.
Combinations of pathogens or nucleic acid templates at different concentrations were spiked into stool samples from a healthy donor and assayed with TAC. Among a total of 1,260 reactions, all 23 targets spiked at high concentration were detected in all replicates. Twelve of 23 targets spiked at low concentration were detected in all replicates, 10 of 23 targets spiked at low concentration were detected in only one of the replicates, and the C. difficile low-concentration spike was not detected. Detection sensitivity and specificity for each target are listed in Table 2. Overall, this revealed 98.7% sensitivity and 100% specificity on these analytical specimens.
Clinical performance.
Clinical samples from Tanzania and Bangladesh were tested with TAC and then compared with previously obtained results from conventional methods as well as our laboratory-developed PCR-Luminex assays (Table 3). Conventional tests included immunoassay (adenovirus, astrovirus, rotavirus, Campylobacter, Cryptosporidium, Giardia, and E. histolytica), culture (Salmonella, V. cholerae, and Shigella), culture with PCR of 5 picked colonies (ETEC, EIEC, EPEC, EAEC, and STEC), and microscopy (Ascaris and Trichuris). All samples that were positive by conventional methods for adenovirus, Salmonella, V. cholerae, EIEC, both ST and LT gene targets of ETEC, eae and bfpA of EPEC, aatA of EAEC, Ascaris, and Trichuris were identified with TAC, while some samples that tested positive by conventional methods for astrovirus, rotavirus, Campylobacter, Cryptosporidium, Giardia, E. histolytica, and the aaiC target of EAEC did not amplify in TAC (5/11, 2/8, 13/35, 4/21, 3/20, 5/13, and 1/18, respectively). However, all these TAC-negative results were confirmed with PCR-Luminex. Overall, sensitivity and specificity of TAC versus conventional results were 85% and 77%, respectively, while versus PCR-Luminex they were 98% and 96%, respectively. To confirm specific amplification among the TAC-positive/PCR-Luminex-positive/conventional-assay-negative specimens, amplicons from 81 of these PCRs were sequenced and all were confirmed to be the proper sequence (n = 18 targets).
Table 3.
Comparison of TAC results with conventional methods and PCR-Luminex results on clinical samples from Tanzania and Bangladesha
| Target | Conventional assay positive |
Conventional assay negative |
PCR-Luminex positive |
PCR-Luminex negative |
||||
|---|---|---|---|---|---|---|---|---|
| TAC+ | TAC− | TAC+ | TAC− | TAC+ | TAC− | TAC+ | TAC− | |
| Adenovirus | 5 | 0 | 34 | 32 | 53 | 0 | 11 | 45 |
| Astrovirus | 6 | 5 | 3 | 58 | 14 | 0 | 1 | 94 |
| Rotavirus | 6 | 2 | 23 | 50 | 30 | 0 | 4 | 75 |
| Campylobacter | 22 | 13 | 5 | 28 | 44 | 2 | 5 | 58 |
| Cryptosporidium | 17 | 4 | 4 | 55 | 24 | 1 | 2 | 82 |
| Giardia | 17 | 3 | 11 | 49 | 32 | 0 | 8 | 69 |
| E. histolytica | 8 | 5 | 9 | 66 | 20 | 2 | 1 | 86 |
| Salmonella | 6 | 0 | 0 | 72 | 9 | 0 | 0 | 100 |
| V. cholerae | 8 | 0 | 0 | 72 | 9 | 0 | 0 | 100 |
| Shigella/EIEC | 1 | 0 | 12 | 59 | 37 | 0 | 0 | 72 |
| ETEC | ||||||||
| ST | 2 | 0 | 20 | 46 | 38 | 1 | 7 | 63 |
| LT | 8 | 0 | 33 | 27 | 63 | 2 | 4 | 40 |
| EPEC | ||||||||
| eae | 12 | 0 | 44 | 12 | 76 | 0 | 8 | 25 |
| bfpA | 7 | 0 | 22 | 39 | 34 | 0 | 9 | 66 |
| EAEC | ||||||||
| aaiC | 17 | 1 | 22 | 28 | 57 | 2 | 5 | 45 |
| aatA | 26 | 0 | 28 | 14 | 77 | 4 | 5 | 23 |
| STEC | ||||||||
| stx1 | 0 | 0 | 6 | 62 | 11 | 0 | 0 | 98 |
| stx2 | 0 | 0 | 9 | 59 | 10 | 0 | 1 | 98 |
| Ascaris | 8 | 0 | 1 | 79 | 9 | 0 | 0 | 100 |
| Trichuris | 8 | 0 | 1 | 79 | 16 | 0 | 0 | 93 |
| Norovirus GII | ND | ND | ND | ND | 31 | 0 | 3 | 75 |
| Sapovirus | ND | ND | ND | ND | 18 | 0 | 1 | 90 |
| Total | Sensitivity = 85% | Specificity = 77% | Sensitivity = 98% | Specificity = 96% | ||||
Values indicate the number of samples. TAC+, TAC-positive samples; TAC−, TAC-negative samples; ND, not done (including C. difficile).
We examined the correlation of CT values from TAC with those from the conventional assay results. Statistically, CT values of conventional-assay-positive and TAC-positive samples were significantly lower than those of conventional-assay-negative and TAC-positive samples for most targets (Fig. 2). Likewise, we examined the correlation of CT values from TAC with the PCR-Luminex MFI values. Samples that were discrepant for PCR-Luminex and TAC were generally lower-burden infections toward the lower detection limit (Fig. 3). Five samples were detected as C. difficile positive by TAC, all of which were confirmed with a secondary real-time PCR assay (28). Ultimately, use of this TAC led to detection of a greater number of infections in the clinical samples than did conventional methods, with an average of 5.9 pathogens per sample by TAC and 2.5 by conventional methods (Fig. 4, P < 0.001).
Fig 2.
Quantitative comparison of pathogen burdens in clinical samples that were positive or negative with conventional methods. Box plots with medians were generated with IBM SPSS software. Asterisks (*) indicate that CT was lower for conventional positive than for conventional negative samples (P < 0.05), including adenovirus (P = 0.02), rotavirus (P = 0.01), Cryptosporidium (P < 0.0001), Giardia (P < 0.001), E. histolytica (P < 0.001), LT of ETEC (P = 0.04), eae of EPEC (P = 0.02), and both aaiC and aatA of EAEC (P < 0.001 for both). A trend was observed for astrovirus (P = 0.06), ST of ETEC (P = 0.08), and bfpA of EPEC (P = 0.14), whereas correlation was poor between TAC and the Campylobacter enzyme immunoassay (EIA) (P = 0.59). Statistics were not applicable to stx1 and stx2 of STEC, Shigella/EIEC, Salmonella, V. cholerae, Ascaris, and Trichuris.
Fig 3.
Analysis of TAC and PCR-Luminex assays. CT values from TAC are shown for both PCR-Luminex-positive and -negative samples. Likewise, relative fluorescence intensities from PCR-Luminex are shown for both TAC-positive and -negative samples. To adjust for different Luminex fluorescence intensity scales, the relative fluorescence intensity of a PCR-Luminex-positive sample is shown as the percentile versus the highest fluorescence intensity observed. Asterisks (*) indicate that CT values were lower for PCR-Luminex-positive than for PCR-Luminex-negative samples and Luminex fluorescence intensity (FI) values were higher for TAC-positive than for TAC-negative samples (P < 0.05).
Fig 4.

Numbers of mixed infections detected with TAC versus conventional methods for 16 pathogens on samples from Tanzania (medians; P < 0.001).
DISCUSSION
This is the first described TaqMan Array Card for enteropathogens. We designed this assay to capture a wide range of enteropathogens relevant in developing country settings, where childhood diarrhea burden is highest. We developed the TaqMan Array Card as a singleplex real-time PCR platform, allowing for multitarget detection through spatial distribution. The singleplex format means a simpler assay design and better quantification and sensitivity than multiplex real-time PCR; however, we did need to adapt most of the primers and probes, usually by modifying their length, so that they performed well under the universal conditions on the card.
We had to handle a broad range of enteropathogen types including both DNA and RNA genomes. Therefore, an RT-PCR protocol was applied to enable amplification of all targets. Since the targeted genes for parasites were rRNA genes, rRNA was far more abundant than genomic DNA, and this boosted the sensitivity of the assay by at least 2 log compared to regular PCR (data not shown). Panspecies or pangenus assays were usually chosen, and multicopy targets were utilized whenever possible to ensure sensitivity. We originally had some concern that the small sample volume might limit the lower-level detection of pathogens in clinical samples (3), so we maximized the sample volume (theoretically 0.4 μl) in each reaction and observed a <10-fold decrease of sensitivity compared with cognate assays performed on plates where 2 μl of sample was added. Our LODs, as shown in Table 2, were close to the theoretical detection limit of one single copy of template per reaction. This translates to an LOD adequate for most clinical cases, where the pathogen loads were reported as 103 to 109 CFU/g stool for bacteria, 103 to 105 CFU/g for protozoa, and 104 to 1011 CFU/g for viruses.
Eighty-five percent of the positive samples by conventional detection were identified as positive with TAC; however, TAC detected an additional ∼160% more positives, and this was particularly prevalent with the bacteria. We can envision several possible reasons for this. First, culture in stool is challenging, and bacteria can be in a viable but nonculturable state. Second, for E. coli, as is common, only five colonies were picked for typing, whereas the TAC assay examines stool in its entirety. Third, TAC could be detecting only nonviable nucleic acids. Another explanation for discrepancies in the case of Campylobacter is that the Campylobacter ELISA may detect a range of species (18) whereas we designed the TAC assay to be specific for C. jejuni/C. coli. In contrast to the discrepancies seen in comparison with conventional methods, the correlation between the molecular platforms of TAC and PCR-Luminex surpassed 96%, with only rare discrepancies, which were seen exclusively with low-burden samples. In general, TAC detected slightly more pathogens than PCR-Luminex, which we speculate reflects true low-burden infections picked up with the singleplex versus multiplex amplification methods.
With such sensitive and comprehensive molecular diagnostics, we were not surprised to find a high rate of mixed infections. Albert et al. found up to 5 pathogens in a Bangladeshi population using conventional methods (29). Lindsay et al. used a combination of culture, immunologic, and conventional PCR to detect 26 pathogens in India and found as many as six pathogens in a single specimen (30). Our findings with conventional methods were similar to the ones reported by those studies. We think, with the advent of highly sensitive molecular assays to interrogate multiple targets in stool, that we will detect even more pathogen nucleic acids in low socioeconomic settings (29–31). Understanding which infections are dominant or likely contributors and which ones are less likely playing a role will take a significant amount of work, ideally through prospective studies and probably interventional trials targeted at key pathogens. In our view, quantitative assays using a platform such as TAC, which minimizes target-to-target, panel-to-panel, and plate-to-plate variations, can serve as an important tool in this effort.
Quantitation of organisms in stool samples is challenging. We introduced two extrinsic controls to monitor the extraction and amplification inhibition and efficiency. We observed CT values for PhHV that ranged from 30 to 35 and for MS2 from 31 to 38, which represented up to 40- to 150-fold differences in DNA and RNA extraction/amplification efficiency, respectively. Furthermore, we included bacterial 16S rRNA gene (16S) assays as an indicator of total bacterial content in each sample. Our results showed that the CT values of the 16S assay ranged from 15 to 24, equivalent to a 500-fold difference. Normalization to 16S may be useful for accurate quantitation in the cases where it is difficult to measure the true sample mass, e.g., in watery stool. For best quantitation, should one be so inclined, we would propose first calculating the extraction/amplification efficiency by comparing the CT values of PhHV and MS2 in a given sample with the CT values of pure PhHV and MS2 at equivalent spiking concentrations. Next, take the CT value of a certain pathogen in that sample to calculate the copy number via a standard curve, and divide the copy number by the efficiency to yield the pathogen load per gram of stool. Finally, one could further normalize this to the copy number of 16S. An important caveat, however, is that for targets such as Shigella, Cryptosporidium, and Giardia, the quantitation may be less accurate due to the uncertainty of target copy numbers per cell (either plasmid or rRNA).
We would emphasize how molecular assays such as TAC greatly simplify the diagnostic process and procurement versus conventional methods. To detect 16 pathogens with conventional methods, we had to procure seven ELISA kits from two different companies, three types of bacterial culture media, biochemical identification reagents, a PCR gel electrophoresis system, and a microscope. At current prices, conventional reagent costs alone can run $200 per sample versus approximately $60 for TAC. The turnaround time of TAC is also significantly shorter than that of conventional methods.
There are limitations to the TAC method, however. Repeats are expensive, since one must run all the targets on the card even if only one or two require further investigation. Therefore, ideally one wants to have backup assays in plate format. We have found utility in spotting the TAC in duplicates, because in our work in developing countries there are often low-level infections at the lower limit of detection, and duplicate results allow greater confidence. Finally, the quantitative PCR (qPCR) platform is costly. In sum, however, the TAC offers a sensitive, broad-range screen for enteropathogens that is useful for surveillance or clinical purposes at equipped sites. The assay is modular, so once the assay performance is established under the universal cycling conditions, different pathogen combinations can be spotted for the specific purpose of the end user.
Supplementary Material
ACKNOWLEDGMENTS
This project was undertaken in collaboration with the MAL-ED Network. The Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development Project (MAL-ED) is carried out as a collaborative project supported by the Bill & Melinda Gates Foundation, the Foundation for the NIH, and the National Institutes of Health/Fogarty International Center.
We thank the staff and participants of the MAL-ED Network Project for their important contributions. We also thank Martin Schutten, Erasmus MC, Department of Virology, Rotterdam, The Netherlands, for kindly providing PhHV.
Footnotes
Published ahead of print 21 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02658-12.
REFERENCES
- 1. Boschi-Pinto C, Velebit L, Shibuya K. 2008. Estimating child mortality due to diarrhoea in developing countries. Bull. World Health Organ. 86:710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, Schrag SJ, Taylor TH, Jr, Beall BW, Breiman RF, Feikin DR, Njenga MK, Mayer LW, Oberste MS, Tondella ML, Winchell JM, Lindstrom SL, Erdman DD, Fields BS. 2011. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J. Clin. Microbiol. 49:2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rachwal PA, Rose HL, Cox V, Lukaszewski RA, Murch AL, Weller SA. 2012. The potential of TaqMan Array Cards for detection of multiple biological agents by real-time PCR. PLoS One 7:e35971 doi:10.1371/journal.pone.0035971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J, Kibiki G, Maro V, Maro A, Kumburu H, Swai N, Taniuchi M, Gratz J, Toney D, Kang G, Houpt E. 2011. Multiplex reverse transcription PCR Luminex assay for detection and quantitation of viral agents of gastroenteritis. J. Clin. Virol. 50:308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mondal D, Minak J, Alam M, Liu Y, Dai J, Korpe P, Liu L, Haque R, Petri WA., Jr 2012. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin. Infect. Dis. 54:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taniuchi M, Walters CC, Gratz J, Maro A, Kumburu H, Serichantalergs O, Sethabutr O, Bodhidatta L, Kibiki G, Toney DM, Berkeley L, Nataro JP, Houpt ER. 2012. Development of a multiplex polymerase chain reaction assay for diarrheagenic Escherichia coli and Shigella spp. and its evaluation on colonies, culture broths, and stool. Diagn. Microbiol. Infect. Dis. 73:121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228–239 [DOI] [PubMed] [Google Scholar]
- 8. Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng SQ, Halkosalo A, Salminen M, Szakal ED, Puustinen L, Vesikari T. 2008. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Methods 153:238–240 [DOI] [PubMed] [Google Scholar]
- 10. Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu FT, White PA, Takeda N. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 78:1347–1353 [DOI] [PubMed] [Google Scholar]
- 11. Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. 2008. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect. Immun. 76:3281–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hidaka A, Hokyo T, Arikawa K, Fujihara S, Ogasawara J, Hase A, Hara-Kudo Y, Nishikawa Y. 2009. Multiplex real-time PCR for exhaustive detection of diarrhoeagenic Escherichia coli. J. Appl. Microbiol. 106:410–420 [DOI] [PubMed] [Google Scholar]
- 13. Bugarel M, Martin A, Fach P, Beutin L. 2011. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 11:142 doi:10.1186/1471-2180-11-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen EM, Andersen MT. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41:2884–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. 2005. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J. Clin. Microbiol. 43:755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vu DT, Sethabutr O, Von Seidlein L, Tran VT, Do GC, Bui TC, Le HT, Lee H, Houng HS, Hale TL, Clemens JD, Mason C, Dang DT. 2004. Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J. Clin. Microbiol. 42:2031–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. 2010. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J. Clin. Microbiol. 48:2929–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J, Gratz J, Maro A, Kumburu H, Kibiki G, Taniuchi M, Howlader AM, Sobuz SU, Haque R, Talukder KA, Qureshi S, Zaidi A, Haverstick DM, Houpt ER. 2012. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-luminex assay. J. Clin. Microbiol. 50:98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houser BA, Hattel AL, Jayarao BM. 2010. Real-time multiplex polymerase chain reaction assay for rapid detection of Clostridium difficile toxin-encoding strains. Foodborne Pathog. Dis. 7:719–726 [DOI] [PubMed] [Google Scholar]
- 20. Stroup SE, Roy S, McHele J, Maro V, Ntabaguzi S, Siddique A, Kang G, Guerrant RL, Kirkpatrick BD, Fayer R, Herbein J, Ward H, Haque R, Houpt ER. 2006. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J. Med. Microbiol. 55:1217–1222 [DOI] [PubMed] [Google Scholar]
- 21. Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 42:1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiria AE, Prasetyani MA, Hamid F, Wammes LJ, Lell B, Ariawan I, Uh HW, Wibowo H, Djuardi Y, Wahyuni S, Sutanto I, May L, Luty AJ, Verweij JJ, Sartono E, Yazdanbakhsh M, Supali T. 2010. Does treatment of intestinal helminth infections influence malaria? Background and methodology of a longitudinal study of clinical, parasitological and immunological parameters in Nangapanda, Flores, Indonesia (ImmunoSPIN Study). BMC Infect. Dis. 10:77 doi:10.1186/1471-2334-10-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niesters HG. 2002. Clinical virology in real time. J. Clin. Virol. 25(Suppl. 3):S3–S12 [DOI] [PubMed] [Google Scholar]
- 24. Rolfe KJ, Parmar S, Mururi D, Wreghitt TG, Jalal H, Zhang H, Curran MD. 2007. An internally controlled, one-step, real-time RT-PCR assay for norovirus detection and genogrouping. J. Clin. Virol. 39:318–321 [DOI] [PubMed] [Google Scholar]
- 25. Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 26. Kodani M, Winchell JM. 2012. Engineered combined-positive-control template for real-time reverse transcription-PCR in multiple-pathogen-detection assays. J. Clin. Microbiol. 50:1057–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taniuchi M, Verweij JJ, Noor Z, Sobuz SU, Lieshout L, Petri WA, Jr, Haque R, Houpt ER. 2011. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am. J. Trop. Med. Hyg. 84:332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sloan LM, Duresko BJ, Gustafson DR, Rosenblatt JE. 2008. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 46:1996–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D. 1999. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J. Clin. Microbiol. 37:3458–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindsay B, Ramamurthy T, Sen Gupta S, Takeda Y, Rajendran K, Nair GB, Stine OC. 2011. Diarrheagenic pathogens in polymicrobial infections. Emerg. Infect. Dis. 17:606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sinha A, Sengupta S, Guin S, Dutta S, Ghosh S, Mukherjee P, Mukhopadhyay AK, Ramamurthy T, Takeda Y, Kurakawa T, Nomoto K, Nair GB, Nandy RK. 2011. Culture-independent real-time PCR reveals extensive polymicrobial infections in hospitalized diarrhoea cases in Kolkata, India. Clin. Microbiol. Infect. doi:1111/j.1469-0691.2011.03746x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.