Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been evaluated for the identification of multidrug-resistant Acinetobacter baumannii nosocomial outbreaks in comparison with the repetitive sequence-based PCR DiversiLab system. The results suggest that MALDI-TOF MS can be used for real-time detection of Acinetobacter outbreaks before results from DNA-based systems are available.
TEXT
Acinetobacter baumannii has become a major nosocomial threat, causing infections with high rates of morbidity and mortality (1–4).
Acinetobacter baumannii hospital outbreaks have been assessed with various DNA typing methods (5–8). Very few typing methods assess outbreaks in real time. Recently, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been developed for the rapid identification of pathogens (9). There are a growing number of reports showing the utility of MALDI-TOF MS to type different bacterial species for easy and early detection of nosocomial outbreaks (10–16).
The aim of this study was to assess the potential utility of MALDI-TOF MS to detect the nosocomial spread of A. baumannii isolates. For this purpose, 35 multidrug-resistant strains of A. baumannii, isolated from colonized or infected patients hospitalized in a general hospital in Italy in 2010, were studied using the repetitive sequence-based PCR (rep-PCR) DiversiLab system (DL; bioMérieux, Marcy l'Etoile, France) and MALDI-TOF MS system (Bruker Daltonics, Bremen, Germany), and the results were compared. After storage at −70°C, the isolates were thawed out and cultured simultaneously on MacConkey agar.
DiversiLab rep-PCR (bioMérieux) was performed according to the manufacturer's instructions (5, 17). Briefly, DNA was extracted from a bacterial culture, amplified using rep-PCR, loaded in LabChip, and run using an Agilent 2100 Bioanalyzer (Agilent Technologies). The results were analyzed by DiversiLab with the Pearson correlation (PC) coefficient to emphasize peak intensities more than peak presence or absence. The strains were considered indistinguishable, similar, or different if the similarity was ≥97.5% without differences in the fingerprinting pattern, >95% and <97.5%, or ≤95%, respectively.
For MALDI-TOF MS, bacteria were cultured for 24 h at 37°C on MacConkey agar and extracts were prepared as described previously (18, 19). For isolate identification, the row spectra were compared with those in the Biotyper database and a log(score) of ≥2.3 was considered to represent a secure species identification. Spectra were acquired with a microflex LT mass spectrometer (Bruker Daltonics) and recorded in the positive linear mode at a laser frequency of 20 Hz, ion source 1 voltage of 20 kV, ion source 2 voltage of 8.5 kV, and mass range from 2,000 to 20,000 kDa, as described elsewhere (18). Reference spectra of newly created main spectra were added to the original Biotyper database. To evaluate the spectrum variation within each strain and to estimate the mass spectral variance of biological replicates, a principal component analysis (PCA) was performed; two biological replicates from six randomly selected Acinetobacter strains were tested, as described above, and spectra for each selected strain were acquired in the same experiment on the mass spectrometer. Technical mass-spectral variance was also evaluated, analyzing five technical replicates from each Acinetobacter independent culture on the same MALDI target plate. For main spectral projection construction, a total of 5,000 shots were acquired from 10 technical replicates, each spectrum was visually inspected by Flex analysis 3.0 software (Bruker Daltonics), and peak profiles with variable occurrence among strains were removed. Reference spectra of newly created main spectra were added to the original Biotyper database. A log(score) value higher than 2.3 was obtained for all spectra from biological and technical replicates of the six different Acinetobacter strains matching their own newly created reference main spectra. PCA performed on spectra from biological and technical replicates within each Acinetobacter strain selected revealed discrete clusters, indicating little variation between replicates and supporting the reproducibility of the method described. Hierarchical clustering of the spectra was performed by applying the average linkage algorithm to a distance matrix containing the Hamming distance between all pairs of peak profiles.
The discriminatory power of each typing method was assessed using Simpson's index of diversity (SID), calculating the probability that two unrelated strains sampled from the test population will be placed into different typing groups (20), and the 95% confidence intervals (CI) of the SID values were calculated as described previously (21). The quantitative concordance between typing methods was analyzed by using adjusted Rand and Wallace coefficients (22).
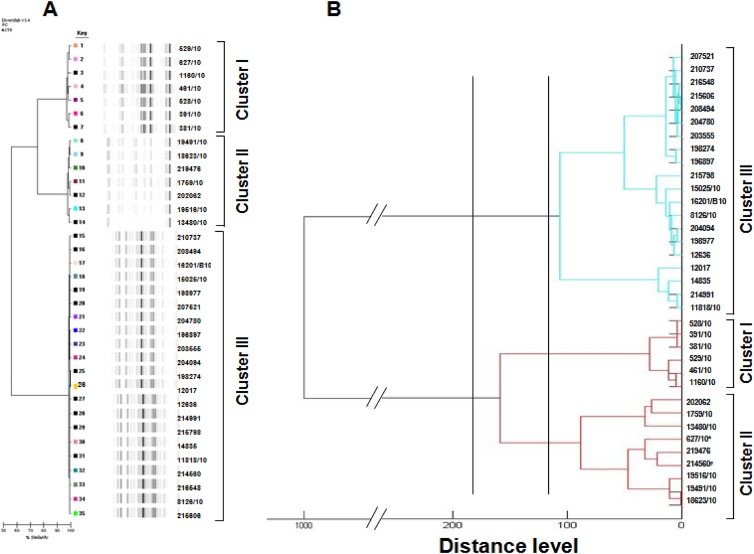
The rep-PCR system identified three different clusters of A. baumannii isolates (Fig. 1A): cluster I, 7 indistinguishable isolates, with an average genomic similarity ratio of >98.5%; cluster II, 7 indistinguishable isolates, with an average genomic similarity ratio of >98.5%; and cluster III, 21 indistinguishable isolates, with an average genomic similarity ratio of >98.5%. The three clusters were significantly different from each other in that the percentage of similarity between clusters I and II was around 75% and was around 55% between clusters I and III and clusters II and III (Fig. 1A).
Fig 1.
Hierarchical clustering of Acinetobacter baumannii isolates by rep-PCR and MALDI-TOF MS. (A) Dendrogram describing percentage similarities and banding patterns of A. baumannii by repetitive PCR, interpreted by the Pearson correlation coefficient. (B) Dendrogram representation of hierarchical cluster analysis provided by MALDI-TOF MS. The vertical lines indicate the similarity cutoff values around 200 and 100 used arbitrarily for MALDI group definition. Isolate assigned to cluster I by the DiversiLab system is indicated by an asterisk. Isolate assigned to cluster III by the DiversiLab system is indicated by a number sign.
All isolates were identified as A. baumannii by MALDI-TOF with a log(score) of ≥2.3. The hierarchical clustering of MALDI-TOF peak profiles identified three different clusters of A. baumannii isolates, substantially interchangeable with those obtained with the rep-PCR system (Fig. 1B), except for isolates 627/10 and 214560, which were assigned to different clusters (clusters I and III) with rep-PCR and included in the same cluster (cluster II) with MALDI-TOF.
The statistical analysis of the data showed that the rep-PCR system (Simpson's index, 0.595; 95% CI, 0.477 to 0.713) and MALDI-TOF MS system (Simpson's index, 0.576; 95% CI, 0.444 to 0.709) provided similar results, with a good concordance between the two methods (adjusted Rand's coefficient, 0.858) and a high probability of MALDI-TOF MS to predict rep-PCR results (Wallace coefficient, 0.892). Arbitrarily chosen cutoff values around 200 or 100 for the MALDI-TOF MS distance level seem to match with the 55% or 75% similarity obtained by rep-PCR (Fig. 1B).
A rapid and accurate detection of nosocomial outbreaks of A. baumannii is essential for the appropriate management of and timely intervention in infection control. Nowadays, sequence-based methods available for typing of Acinetobacter spp., such as pulsed-field electrophoresis (8, 23) and multilocus sequence typing (MLST) (17), are rather time-consuming, laborious, and expensive and require a long period of time before generating a typing result, useful for outbreak investigation and control (24, 25). The use of DL rep-PCR has been intensively investigated for detecting outbreaks of nosocomial infections. This method is considered comparable with the above-described sequence-based methods for different bacterial species, including A. baumannii (5, 17, 23). Moreover, it is an affordable method for laboratories of clinical microbiology.
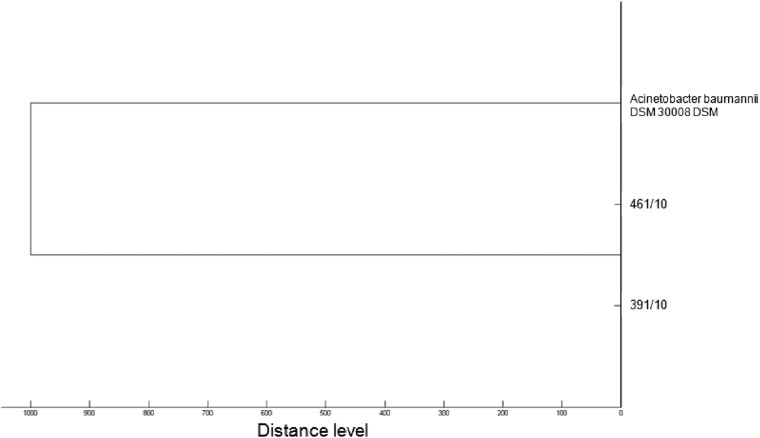
To verify if the relatedness between isolates observed with MALDI-TOF MS and DL rep-PCR was evidenced also by a reference method such as MLST, two A. baumannii strains (461/10 and 391/10), classified in cluster I by both MALDI-TOF MS and rep-PCR, were assessed using an MLST scheme based on fragments of the seven housekeeping genes gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD (http://pubmlst.org/abaumannii/). Acinetobacter baumannii reference strain DSM 30008 DSM was added as an unrelated strain. MLST analysis was performed as described previously (26), and amplicons were sequenced using an ABI Prism 3130xl sequence analyzer (Applied Biosystem, Foster City, CA). Sequences were analyzed using Chromas Lite 2.1 software (Technelysium) and compared with the A. baumannii database at the MLST website (http://pubmlst.org/abaumannii/). The two isolates belonged to sequence type 128, while the reference strain number DSM 30008 DSM belonged to sequence type 13 and was classified by MALDI-TOF MS as a unique isolate (Fig. 2). This finding could be particularly relevant, as often the clinical question is to know if a new isolate is an outbreak strain or a unique isolate.
Fig 2.
Dendrogram representation of hierarchical cluster analysis of A. baumannii isolates 461/10 and 391/10 and unrelated strain DSM 30008 DSM.
The accuracy and speed of data acquisition which can be achieved using MALDI-TOF MS make this a potentially important tool for a rapid and sensitive identification of nosocomial outbreaks caused by various alert organisms. In line with recent studies (5, 12, 17), the results of the present work suggest that MALDI-TOF MS can be used for a rapid and optimal detection of nosocomial A. baumannii outbreaks, as described for other microorganisms (10, 11, 13–16).
In this study, MALDI-TOF MS classified two isolates (isolates 627/10 and 214560) in different clusters from the ones they were classified in with rep-PCR. These results were out from three separate experiments, in which the two isolates were analyzed together with all the other isolates. A possible explanation for this result could be that the quality of the spectra obtained for these isolates was not clear enough to allow an exact typing. Indeed, different technical or biological causes, such as the quality of the spectra (27, 28) and the incubation time of cultures, can affect the MALDI-TOF results (29). Bacteria adapt rapidly to environmental changes, modifying the production of proteins or other cellular processes when they are stored, handled, or cultured over different time periods. This makes it important to carefully control all the above-described variables in experiments aimed at bacterial typing with mass spectrometry.
In conclusion, the present study suggests that MALDI-TOF MS can be successfully used in routine clinical microbiology for real-time identification of nosocomial outbreaks from A. baumannii isolates before results from DNA-based systems are available.
Footnotes
Published ahead of print 21 November 2012
REFERENCES
- 1. Bergogne-Berezin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giamarellou H, Antoniadou A, Kanellakopoulou K. 2008. Acinetobacter baumannii: a universal threat to public health? Int. J. Antimicrob. Agents 32:106–119 [DOI] [PubMed] [Google Scholar]
- 3. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Towner KJ. 2009. Acinetobacter: an old friend, but a new enemy. J. Hosp. Infect. 73:355–363 [DOI] [PubMed] [Google Scholar]
- 5. Higgins PG, Hujer AM, Hujer KM, Bonomo RA, Seifert H. 2011. Interlaboratory reproducibility of DiversiLab rep-PCR typing and clustering of Acinetobacter baumannii isolates. J. Med. Microbiol. 61:137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koeleman JG, Stoof J, Biesmans DJ, Savelkoul PH, Vandenbroucke-Grauls CM. 1998. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J. Clin. Microbiol. 36:2522–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu PY, Wu WL. 1997. Use of different PCR-based DNA fingerprinting techniques and pulsed-field gel electrophoresis to investigate the epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Diagn. Microbiol. Infect. Dis. 29:19–28 [DOI] [PubMed] [Google Scholar]
- 8. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lay JO., Jr 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172–194 [DOI] [PubMed] [Google Scholar]
- 10. Eddabra R, Prévost G, Scheftel JM. 2012. Rapid discrimination of environmental Vibrio by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol. Res. 167:226–230 [DOI] [PubMed] [Google Scholar]
- 11. Fujinami Y, Kikkawa HS, Kurosaki Y, Sakurada K, Yoshino MJ, Yasuda J. 2011. Rapid discrimination of Legionella by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol. Res. 166:77–86 [DOI] [PubMed] [Google Scholar]
- 12. Huang YF, Wang JR, Gu HT, Shang XR, Cao JJ, Wang M, Fan YY, Sui WJ, Lu XX. 2012. Correlation factor analysis of pan-drug resistant Acinetobacter baumannii strains in acquired infections. Zhonghua Yi Xue Za Zhi 91:2525–2529 [PubMed] [Google Scholar]
- 13. Siegrist TJ, Anderson PD, Huen WH, Kleinheinz GT, McDermott CM, Sandrin TR. 2007. Discrimination and characterization of environmental strains of Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). J. Microbiol. Methods 68:554–562 [DOI] [PubMed] [Google Scholar]
- 14. Treviño M, Areses P, Peñalver MD, Cortizo S, Pardo F, del Molino ML, García-Riestra C, Hernández M, Llovo J, Regueiro BJ. 2012. Susceptibility trends of Bacteroides fragilis group and characterization of carbapenemase-producing strains by automated REP-PCR and MALDI TOF. Anaerobe 18:37–43 [DOI] [PubMed] [Google Scholar]
- 15. Treviño M, Navarro D, Barbeito G, García-Riestra C, Crespo C, Regueiro BJ. 2011. Molecular and epidemiological analysis of nosocomial carbapenem-resistant Klebsiella spp. using repetitive extragenic palindromic-polymerase chain reaction and matrix-assisted laser desorption/ionization-time of flight. Microb. Drug Resist. 17:433–442 [DOI] [PubMed] [Google Scholar]
- 16. Wolters M, Rohde H, Maier T, Belmar-Campos C, Franke G, Scherpe S, Aepfelbacher M, Christner M. 2011. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int. J. Med. Microbiol. 301:64–68 [DOI] [PubMed] [Google Scholar]
- 17. Higgins PG, Janßen K, Fresen MM, Wisplinghoff H, Seifert H. 2012. Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J. Clin. Microbiol. 50:3493–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, Dunn J, Hall G, Wilson D, Lasala P, Kostrzewa M, Harmsen D. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spanu T, De Carolis E, Fiori B, Sanguinetti M, D'Inzeo T, Fadda G, Posteraro B. 2011. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to rpoB gene sequencing for species identification of bloodstream infection staphylococcal isolates. Clin. Microbiol. Infect. 17:44–49 [DOI] [PubMed] [Google Scholar]
- 20. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grundmann H, Hori S, Tanner G. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fluit AC, Terlingen AM, Andriessen L, Ikawaty R, van Mansfeld R, Top J, Cohen Stuart JW, Leverstein-van Hall MA, Boel CH. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J. Clin. Microbiol. 48:3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diep BA, Perdreau-Remington F, Sensabaugh GF. 2003. Clonal characterization of Staphylococcus aureus by multilocus restriction fragment typing, a rapid screening approach for molecular epidemiology. J. Clin. Microbiol. 41:4559–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zaidi N, Konstantinou K, Zervos M. 2003. The role of molecular biology and nucleic acid technology in the study of human infection and epidemiology. Arch. Pathol. Lab Med. 127:1098–1105 [DOI] [PubMed] [Google Scholar]
- 26. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carbonnelle E, Mesquita C, Bille E, Day N, Dauphin B, Beretti JL, Ferroni A, Gutmann L, Nassif X. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 28. Williams TL, Andrzejewski D, Lay JO, Musser SM. 2003. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 14:342–351 [DOI] [PubMed] [Google Scholar]
- 29. Keys CJ, Dare DJ, Sutton H, Wells G, Lunt M, McKenna T, McDowall M, Shah HN. 2004. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 4:221–242 [DOI] [PubMed] [Google Scholar]