Abstract
We describe a novel, semiautomated Clostridium difficile typing platform that is based on PCR-ribotyping in conjunction with a semiautomated molecular typing system. The platform is reproducible with minimal intra- or interassay variability. This method exhibited a discriminatory index of 0.954 and is therefore comparable to more arduous typing systems, such as pulsed-field gel electrophoresis.
TEXT
Clostridium difficile, the etiological agent of C. difficile infection (CDI), is an important cause of both hospital- and community-acquired infectious diarrhea (1, 2). The emergence of hypervirulent C. difficile isolates and in particular the NAP1/BI/027 isolate has altered the epidemiology of C. difficile infections in many health care institutions, resulting in increased severity and duration of disease, with concomitant increases to the length and cost of hospitalization (1). Therefore, typing methods that can discriminate NAP1/BI/027 and other emerging hypervirulent C. difficile isolates may be important for understanding the transmission dynamics of the organism. In addition, it has recently been demonstrated that the analytical sensitivity and specificity of C. difficile diagnostic assays may be dependent on the C. difficile strain type (3). In addition, recent reports suggest that the relapse rate following treatment with certain novel antianaerobic and C. difficile-specific antimicrobials could correlate with C. difficile strain type (4, 5); thus, an appreciation of the isolate type may play a role in patient management in the future. As such, C. difficile typing may have the potential to improve the management of CDI beyond clinical surveillance, especially in the hospital setting, and it may be important for clinical laboratorians and infection control specialists to have a baseline understanding of the different C. difficile isolates circulating in their institutions.
Pulsed-field gel electrophoresis (PFGE) is the principal reference method employed for C. difficile typing in North America (6). While PFGE affords acceptable discriminatory power (6), it does suffer from some important limitations, in particular the labor intensity, technical expertise, turnaround time, and necessity for control strains to be processed alongside isolates of epidemiological interest. In Europe, the predominant method for C. difficile typing is PCR-ribotyping, which involves the PCR amplification of the intergenic space region between the 16S and 23S rRNA genes (7, 8, 9). For many years, epidemiologic studies for C. difficile have relied on PFGE and PCR-ribotyping to determine strain relatedness; multiple-locus variable-number tandem-repeat analysis (MLVA) for C. difficile typing has also been described for recent studies (6, 10, 11, 12). Although this method does appear to be reproducible and discriminatory, it requires access to a genetic analyzer/DNA sequencer, which is cost prohibitive to many laboratories.
The objective of this study was to develop and validate a semiautomated PCR-ribotyping platform that would further reduce labor intensity, allow analysis of isolates in real time, provide objective downstream data analysis, maintain discriminatory power, and be a feasible option for routine benchtop typing of C. difficile isolates in a clinical setting. As such, we evaluated the performance of PCR-ribotyping in conjunction with the DiversiLab system (bioMérieux, Durham, NC) and the 2100 Bioanalyzer instrument (Agilent; Santa Clara, CA, USA) for fragment separation and PCR-ribotype band pattern analysis. A significant advantage of this platform is that all resultant banding patterns are banked electronically so that they may be recalled and compared retrospectively.
(This work was presented in part at the 111th General Meeting of the American Society for Microbiology, New Orleans, LA, May 2011.)
Initially, we evaluated the intra- and interassay variability of the PCR-ribotyping/DiversiLab platform. Therefore, in a blinded experiment, prior to PCR-ribotype amplification, genomic DNA isolated from eight C. difficile isolates (labeled A to H) previously characterized as having unique PFGE types (6) was distributed into 24 tubes such that each tube contained DNA from only one isolate (C. difficile DNA was isolated using the BiOStic bacteremia DNA kit [MO BIO; Carlsbad, CA]). To perform the PCR-ribotyping reaction, a 25-μl PCR mixture that included 100 ng of C. difficile DNA, a Ready-To-Go randomly amplified polymorphic DNA (RAPD) analysis bead (GE Healthcare Life Sciences; Piscataway, NJ), and primers complementary to the 3′ end of the 16S rRNA gene (5′-CTGGGGTGAAGTCGTAACAAGG-3′) and the 5′ end of the 23S ribosomal gene (5′-GCGCCCTTTGTAGCTTGACC-3′) at a final concentration of 0.5 μM was assembled (13). The amplification conditions were 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The PCR products were loaded into two different DiversiLab DNA chips (bioMérieux; Durham, NC) according to the manufacturer's specifications and resolved using the 2100 Bioanalyzer instrument. The resultant banding patterns were analyzed using the DiversiLab Bacterial Barcodes software program, which uses curve-based Pearson coefficients for pairwise similarity scores (14). The data were organized into a dendrogram, and a similarity index (SI) for each pair was calculated (Fig. 1). By applying a SI of ≥95%, the PCR-ribotype band patterns were separated into eight clusters, with each cluster corresponding to an isolate. Upon examination of the resultant dendrograms, it is clear that independent PCR-ribotype patterns obtained for each C. difficile isolate cluster together when loaded onto either the same or different DNA chips and that this is both discriminatory and reproducible. Taken together, these data support our assessment of limited intra- and interassay variability.
Fig 1.
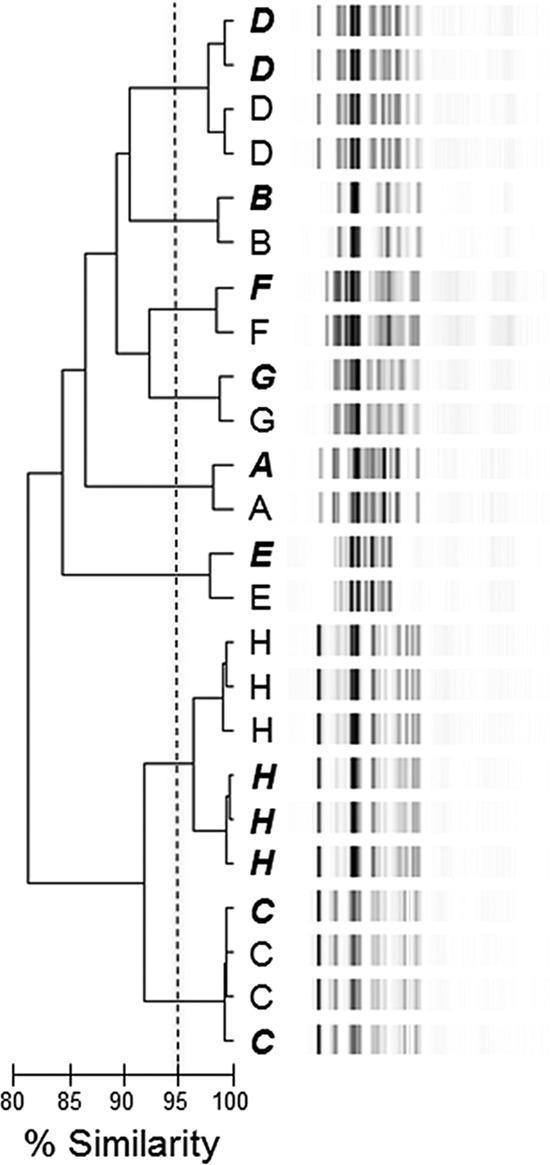
Intra- and interassay variability of the PCR-ribotyping/DiversiLab C. difficile typing platform. Dendrogram illustrating the gel electrophoresis profiles of 24 PCR-ribotyping reactions obtained for eight different C. difficile isolates (isolates A to H). Samples loaded onto the first Agilent chip are indicated by lightface capitalized letters, while PCR-ribotyping reactions loaded onto the second Agilent chip are indicated by letters in bold italics. The percent SI is indicated by the bar below the dendrogram; isolates with a SI of ≥95% are considered very related or identical.
To assess the effect of culture duration and subculture frequency on the reproducibility of the PCR-ribotype band patterns, a C. difficile isolate (ATCC 9689) was incubated anaerobically at 35°C on cycloserine-cefoxitin-fructose agar (CCFA) plates for 2, 5 or 14 days or consecutively passaged onto CCFA plates one, three or five times. Subsequently, the cells were collected, their DNA was extracted, and PCR-ribotyping was performed. The PCR-ribotype band patterns were analyzed using the DiversiLab Bacterial Barcodes software program, and dendrograms to illustrate strain relatedness were generated. From inspection of the PCR product electrophoresis profiles and the associated dendrograms, it was found that PCR-ribotype patterns are not altered significantly in response to either culture duration (SI > 98% across all conditions) or subculture frequency (SI > 99% across all conditions), indicating that the semiautomated typing platform can tolerate considerable variation in culture conditions; this has been demonstrated previously for other typing methods (15, 16).
In an effort to evaluate the performance of the semiautomated, PCR-ribotyping platform at typing C. difficile isolates at the local level, a collection of 50 C. difficile isolates cultured from the stools of patients with a positive toxin A/B assay in our institution and, as a calibrant, 24 C. difficile isolates whose PFGE pulsotypes had been previously determined (6) was analyzed (Fig. 2). To ensure a highly diverse calibrant collection, C. difficile isolates with the NAP1, NAP2, NAP4, NAP6, and NAP12 PFGE pulsotypes and isolates that did not map to previously characterized PFGE pulsotypes (UN for “unnamed” types) were included. The 74 isolates were separated into 17 clusters, with 15 unrelated isolates yielding a discrimination index (D) (17) of 0.954 with 95% confidence intervals for D of 0.931 to 0.977 (18), which is in very close agreement with findings of previous studies evaluating the discriminatory power of different C. difficile typing methods (6, 9). However, the discriminatory index of some of these prior studies could be somewhat skewed, since NAP1 strains are typically in abundance in contemporary epidemiological studies.
Fig 2.
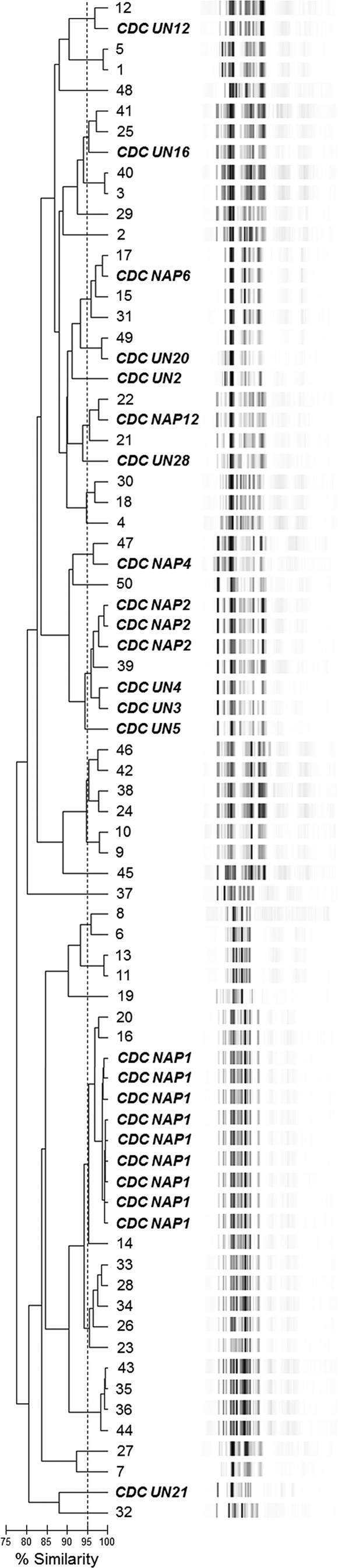
Evaluation of the performance of the PCR-ribotyping/DiversiLab C. difficile typing platform at the local level. The dendrogram illustrates the electrophoresis profiles of a collection of 74 C. difficile isolates obtained from both the CDC and our institution. Isolates obtained from the CDC are indicated by their PFGE pulsotype and are in bold italics, while patient isolates are indicated numerically. The collection was differentiated into 17 clusters with 15 unrelated strains, yielding a discrimination index of 0.954. The percent SI is indicated by the bar below the dendrogram; ≥95% are very related or identical.
A requirement of molecular typing methods is an ability to group related isolates and differentiate unrelated isolates. Our method can do both, grouping the PCR-ribotyping patterns obtained for isolates with identical PFGE pulsotypes and differentiating the PCR-ribotyping patterns attained for isolates with different PFGE pulsotypes. In an effort to further validate the discriminatory power of the semiautomated, PCR-ribotyping platform, a collection of patient isolates whose PCR-ribotyping patterns clustered with those obtained for isolates with known PFGE pulsotypes (i.e., calibrant isolates) was analyzed using PFGE. In all cases, the patient isolate yielded a PFGE pulsotype very similar, if not identical, to that of the calibrant isolate it had clustered with after PCR-ribotyping analysis (Table 1).
Table 1.
Agreement between PFGE and the semiautomated C. difficile PCR-ribotyping platform
| Patient isolate | Pulsotype inferred from PCR-ribotyping | Pulsotype determined by PFGE |
|---|---|---|
| 15 | NAP6 | NAP6 |
| 16 | NAP1 | NAP1 |
| 17 | NAP6 | NAP6 |
| 20 | NAP1 | NAP1 |
| 23 | NAP1 | NAP1 |
| 28 | NAP1 | NAP1 related |
| 35 | NAP1 | NAP1 |
| 39 | NAP2 | NAP2 |
| 47 | NAP4 | NAP4 |
The novel, semiautomated C. difficile typing platform described here has some advantages over PFGE and other more intensive molecular typing methodologies; in particular, it is less labor-intensive. Data analysis is automated, easy to perform, and can be done within a matter of minutes, while ensuring a high discriminatory power. In addition, the ability to bank isolate PCR-ribotype patterns for retrospective and prospective analysis is an advancement in C. difficile typing and could facilitate the rapid, streamlined examination of epidemiological trends.
At present, the primary utility of C. difficile typing is to assist epidemiological studies; however, it is possible that real-time C. difficile typing will become increasingly relevant in routine clinical management to identify the predominant C. difficile isolates in an institution prior to administration of expensive C. difficile-specific antimicrobials and to monitor diagnostic assay performance. After C. difficile is recovered in culture, the hands-on time per isolate to complete this typing method is approximately 1 h. After the initial investment of acquiring the hardware and software for this method, the cost of analyzing a single isolate is approximately $50. However, up to 12 samples can be analyzed in a single chip or run, and if 12 isolates are run in a batch, the cost per isolate is reduced to approximately $35. With the ability to bank banding patterns and compare isolates in real-time to previously typed isolates, it will be relatively easy for institutions to create their own C. difficile strain typing database.
Therefore, we believe that the rapid, facile, and discriminatory novel, semiautomated PCR-ribotyping platform described herein will be favorably useful to epidemiologists, clinicians, and laboratory medicine professionals involved in managing CDIs.
ACKNOWLEDGMENTS
Funding for this work was provided by DDRCC grant P30 DK52574 and NIH grant K23AI065806.
At the time this research was conducted, W. Michael Dunne, Jr., was a faculty member at the Washington University School of Medicine. W.M.D. is now an employee of bioMérieux.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 21 November 2012
REFERENCES
- 1. Dubberke ER, Wertherimer AI. 2009. Review of current literature on the economic burden of Clostridium difficile infection. Infect. Control Hosp. Epidemiol. 30:57–66 [DOI] [PubMed] [Google Scholar]
- 2. Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tenover FC, Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong E, Marlowe EM, Whitmore J, Pershing DH. 2010. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J. Clin. Microbiol. 48:3719–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK, OPT-80-003 Clinical Study Group 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364:422–431 [DOI] [PubMed] [Google Scholar]
- 5. Cornely CA, Crook DW, Esposito R, Poirier A, Somero MS, Weiss K, Shears P, Gorbach S, OPT-80-004 Clinical Study Group 2012. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomized controlled trial. Lancet Infect. Dis. 12:281–289 [DOI] [PubMed] [Google Scholar]
- 6. Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, Frost EH, Savelkoul P, Nicholson B, van den Berg RJ, Kato H, Sambol SP, Zukowski W, Woods C, Limbago B, Gerding DN, McDonald LC. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gürtler V. 1993. Typing of Clostridium difficile strains by PCR-amplification of variable length 16S-23S rDNA spacer regions. J. Gen. Microbiol. 139:3089–3097 [DOI] [PubMed] [Google Scholar]
- 8. O'Neill GL, Ogunsola FT, Brazier JS, Duerden BI. 1996. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe 2:205–209 [Google Scholar]
- 9. Bidet P, Lalande V, Salauze B, Burghoffer B, Avesani V, Delmée M, Rossier A, Barbut F, Petit JC. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38:2484–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hardy K, Manzoor S, Marriott C, Parsons H, Waddington C, Gossain S, Szczepura A, Stallard N, Hawkey PM. 2012. Utilizing rapid multiple-locus variable-number tandem-repeat analysis typing to aid control of hospital-acquired Clostridium difficile infection: a multicenter study. J. Clin. Microbiol. 50:3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marsh JW, O'Leary MM, Shutt KA, Pasculle W, Johnson S, Gerding DN, Muto CA, Harrison LH. 2006. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J. Clin. Microbiol. 44:2558–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van den Berg RJ, Schaap I, Templeton KE, Klaassen CHW, Kuijper EJ. 2007. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J. Clin. Microbiol. 45:1024–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stubbs SL, Brazier JS, O'Neill GL, Duerdden BI. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frye SR, Healy M. 2006. Molecular strain typing using repetitive sequence-based PCR, p 444–471 In Tang Y-W, Stratton CW. (ed), Advanced techniques in diagnostic microbiology, 1st ed Springer, New York, NY [Google Scholar]
- 15. Kang HP, Dunne WM. 2003. Stability of repetitive-sequence PCR patterns with respect to culture age and subculture frequency. J. Clin. Microbiol. 41:2694–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Higgins PG, Hujer AM, Hujer KM, Bonomo RA, Seifert H. 2012. Interlaboratory reproducibility of DiversiLab rep-PCR typing and clustering of Acinetobacter baumannii isolates. J. Med. Microbiol. 61:137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grundmann H, Hori S, Tanner G. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]


