Abstract
Molecular diagnostic tools have been used increasingly in the characterization of the transmission of cryptosporidiosis and microsporidiosis in developing countries. However, few studies have examined the distribution of Cryptosporidium species and Enterocytozoon bieneusi genotypes in AIDS patients receiving antiretroviral therapy. In the present study, 683 HIV-positive patients in the National Free Antiretroviral Therapy Program in China and 683 matched HIV-negative controls were enrolled. Cryptosporidium species and subtypes and Enterocytozoon bieneusi genotypes were detected and differentiated by PCR and DNA sequencing. The infection rates were 1.5% and 0.15% for Cryptosporidium and 5.7% and 4.2% for E. bieneusi in HIV-positive and HIV-negative participants, respectively. The majority (8/11) of Cryptosporidium cases were infections by zoonotic species, including Cryptosporidium meleagridis (5), Cryptosporidium parvum (2), and Cryptosporidium suis (1). Prevalent E. bieneusi genotypes detected, including EbpC (39), D (12), and type IV (7), were also potentially zoonotic. The common occurrence of EbpC was a feature of E. bieneusi transmission not seen in other areas. Contact with animals was a risk factor for both cryptosporidiosis and microsporidiosis. The results suggest that zoonotic transmission was significant in the epidemiology of both diseases in rural AIDS patients in China.
INTRODUCTION
Cryptosporidium spp. and microsporidia (especially Enterocytozoon bieneusi, the most common species infecting humans) are AIDS-defining pathogens (1). They induce chronic diarrhea, reduce life quality, and increase mortality in HIV-positive patients (2, 3). In developed countries, the morbidity and mortality caused by both pathogens have been reduced significantly by highly active antiretroviral therapy (HAART), introduced in 1996 (2, 3). However, HAART is still not widely available in developing countries. In most developing countries, including China, current HAART regimes do not include protease inhibitors, which appear to contribute significantly to the effect of HAART against cryptosporidiosis and microsporidiosis due to their effects on immune reconstitution (4). Thus, the effect of HAART on the occurrence of cryptosporidiosis and microsporidiosis in HIV-positive patients in developing countries remains unclear (5). Limited reports have shown that cryptosporidiosis and microsporidiosis are still common in HIV-positive patients receiving HAART in developing countries (6–8). Since there is no well-defined or reliable treatment for the diseases caused by Cryptosporidium spp. and E. bieneusi in immunocompromised persons, understanding their epidemiology is the key in the formulation of control strategies against cryptosporidiosis and microsporidiosis.
In recent years, molecular diagnostic tools have been widely used in the differentiation of Cryptosporidium species and subtypes and E. bieneusi genotypes and in the elucidation of transmission routes of cryptosporidiosis and microsporidiosis (9, 10). Data accumulated thus far suggest that anthroponotic transmission plays a more important role in the epidemiology of cryptosporidiosis in HIV-positive patients in developing countries and some industrialized nations such as the United States, but zoonotic transmission contributes significantly to the occurrence of cryptosporidiosis in HIV-positive patients in European countries (11, 12). In contrast, data on the distribution of E. bieneusi genotypes suggest that the transmission of E. bieneusi is mainly anthroponotic in developed countries (13–18) but both anthroponotic and zoonotic in developing countries (18–28). Differences in the transmission of the two diseases among countries and between rural and urban areas make studies of their epidemiology necessary.
Henan is the largest agricultural province in China and has been seriously affected by the HIV epidemic. Thousands of former plasma donors, almost all poor farmers who were paid to donate plasma by illegal plasma collectors in rural Henan, acquired HIV infection in the 1990s due to the unsanitary conditions during plasma collection. After the HIV outbreak, HIV began to spread via unprotected sexual contact and vertical transmission (29). By the end of 2006, 35,232 HIV-positive cases, with the majority (90.8%) of them being farmers, had been identified in 159 counties in Henan (29). In response to the large number of HIV-positive cases, the Chinese government initiated the National Free Antiretroviral Therapy Program (NFATP) in 2002, which provides free antiretroviral drugs to former plasma donors with HIV infection (30, 31). However, no data are available on the transmission of Cryptosporidium and E. bieneusi in HIV-positive persons in China, and few case-control studies have been conducted in developing countries to characterize the risk factors involved in the acquisition of the two opportunistic pathogens in HIV-positive patients (2, 3).
In this report, we describe the prevalence and genotype distribution of Cryptosporidium spp. and E. bieneusi in HIV-positive persons in NFATP in Henan, China, in a case-control study setting. Our data suggest that zoonotic transmission plays an important role in both cryptosporidiosis and microsporidiosis in HIV-positive patients on HAART.
MATERIALS AND METHODS
Study population.
In March and April 2007 and November 2010, 860 and 506 patients in hospitals in Henan were recruited to participate in the study, respectively. Among them, half were HIV-positive patients enrolled in the NFATP (cases), and the other half were HIV-negative patients with similar demographic and socioeconomic backgrounds (controls). The demographic data of the two groups are shown in Table 1. Among them, 641 (49.3%) were males and 660 (50.7%) were females. The patients were 1 to 82 years in age with a median age of 44 years. Most (91.2%) of the participants were farmers and lived in rural areas. Demographic data, presence of diarrhea, recent CD4+ cell counts, and potential risk factors related to food-borne, waterborne, person-to-person, and zoonotic transmission were collected from the participants by attending physicians using a structured questionnaire at the time of enrollment. Informed consent was obtained from all participants. No follow-up of participants was conducted. This study was approved by the institutional review board of the Henan Center for Disease Control and Prevention, China.
Table 1.
Demographics of the HIV-positive (case) and HIV-negative (control) groups in this study
| Demographic variable | Case | Controla |
|---|---|---|
| Gender, no. (%) | ||
| Male | 329 (48.9) | 312 (49.7) |
| Female | 344 (51.1) | 316 (50.3) |
| Age (yr) | 4–72 (mean, 44.2; median, 45) | 1–82 (mean, 41.3; median, 44) |
| Farming as occupation, no. (%) | 628 (93.0) | 384 (90.4) |
| Water supply source, no. (%) | ||
| Tap water | 37 (5.5) | 1 (0.2) |
| Hand pump water | 468 (69.2) | 355 (83.7) |
| Well water | 171 (25.3) | 68 (16.1) |
| Family size, no. (%) | ||
| ≤6 members | 640 (94.8) | 410 (96.5) |
| >6 members | 35 (5.2) | 15 (3.5) |
| Presence of diarrhea, no. (%) | ||
| Yes | 263 (44.5) | 127 (29.9) |
| No | 328 (55.5) | 298 (70.1) |
| Presence of animals in household, no. (%) | ||
| Yes | 425 (63.0) | 267 (62.8) |
| No | 250 (37.0) | 158 (37.2) |
| CD4+ cell count | 372 (mean), 340 (median) |
Only 425 participants provided information other than gender and age in this group.
Stool specimen collection and processing.
A single fecal specimen was collected from each case and control participant at the same time period and stored in 2.5% potassium dichromate solution at 4°C. Within several weeks of the two sampling rounds in 2007 and 2010, DNA was extracted from 200 μl of stool using the Fast DNA Spin kit for soil (Qbiogene, Irvine, CA), after the specimen was washed three times by centrifugation with deionized water. The DNA was extracted from the case and control pairs simultaneously and stored at −20°C before PCR analysis.
Cryptosporidium detection, genotyping, and subtyping.
An ∼830-bp fragment of the small-subunit (SSU) rRNA gene was amplified by nested PCR as previously described (32). Cryptosporidium species were differentiated by restriction fragment length polymorphism (RFLP) analysis of the secondary PCR products using restriction enzymes SspI and VspI. To identify Cryptosporidium hominis, Cryptosporidium parvum, and Cryptosporidium meleagridis subtypes, an ∼850-bp fragment of the 60-kDa glycoprotein (gp60) gene was amplified by nested PCR (33). The secondary PCR products were sequenced using the secondary PCR primers and an intermediary sequencing primer (5′-GAGATATATCTTGTTGCG-3′). The established subtype nomenclature was used to identify the gp60 subtypes (34).
Enterocytozoon bieneusi detection and genotyping.
To detect E. bieneusi, an ∼392-bp fragment of the rRNA gene, including the internal transcribed spacer (ITS), was amplified by nested PCR (28). Genotypes of E. bieneusi were determined by sequence analysis of the secondary PCR products and named according to the established nomenclature system (35).
DNA sequencing and data analysis.
After being purified using Montage PCR filters (Millipore, Bedford, MA), the secondary PCR products were sequenced directly in both directions using an ABI BigDye Terminator v.3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI3130 genetic analyzer (Applied Biosystems). The nucleotide sequences of Cryptosporidium spp. and E. bieneusi were aligned with reference sequences downloaded from GenBank using ClustalX (http://www.clustal.org/; last accessed in November 2012) to determine their genotype or subtype identity. The E. bieneusi genotypes identified in the study were compared with known E. bieneusi genotypes using a neighbor-joining analysis of the aligned E. bieneusi sequences implemented in the program Mega 5 (http://www.megasoftware.net/; last accessed in November 2012). Bootstrap analysis with 1,000 replicates was used to assess the robustness of clusters.
Statistical analysis.
A chi-square test or Fisher exact test was used to compare infection rates. The strength of association with risk factors was measured using the odds ratio (OR). Differences were considered significant at a P value of ≤0.05. All statistical analyses were performed using SPSS Statistics 17.0 (SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers.
Unique nucleotide sequences generated from the study were deposited in GenBank under accession numbers JF691560 to JF691566 and JQ029720 to JQ029736.
RESULTS
Infection rates of Cryptosporidium and E. bieneusi.
Cryptosporidium spp. were detected in 11 participants, including 10 (1.5%) HIV-positive patients and one (0.2%) HIV-negative patient. The difference in Cryptosporidium infection rates between HIV-positive and HIV-negative groups was significant (P < 0.01) (Table 2).
Table 2.
Risk factors for cryptosporidiosis and microsporidiosisc
| Risk factor | Specimen size, no. positive/total no.a |
Cryptosporidium infection |
E. bieneusi infection |
||||
|---|---|---|---|---|---|---|---|
| No. positive (%) | OR (95% CId) | P | No. positive (%) | OR (95% CId) | P | ||
| Age (yr) | |||||||
| 0–9 | 70/1,342 | 1 (1.43) | 6 (8.57) | ||||
| 10–19 | 48/1,342 | 0 | 6 (12.50) | ||||
| 20–29 | 30/1,342 | 1 (3.33) | 0 | ||||
| 30–39 | 257/1,342 | 0 | 0.36 | 10 (3.89) | 0.05 | ||
| 40–49 | 482/1,342 | 5 (1.04) | 18 (3.73) | ||||
| 50–59 | 364/1,342 | 4 (1.10) | 23 (6.32) | ||||
| ≥60 | 91/1,342 | 0 | 5 (5.49) | ||||
| Gender (male) | 641/1,301 | 3 (0.47) | 0.38 (0.10, 1.45) | 0.14 | 34 (5.30) | 1.03 (0.63, 1.68) | 0.90 |
| HIV infection | 683/1,366 | 10 (1.46) | 10.13 (1.29, 79.38) | <0.01 | 39 (5.71) | 1.37 (0.84, 2.24) | 0.21 |
| Diarrhea in patients | |||||||
| Case | 263/651 | 3 (1.14) | 0.74 (0.18, 2.96) | 0.93 | 23 (8.75) | 2.38 (4.66, 1.22) | 0.01 |
| Control | 127/425 | 7 (5.51) | 0.77 (0.32, 1.86) | 0.56 | |||
| CD4+ cells (<200)b | 158/590 | 3 (1.90) | 1.65 (0.39, 7.00) | 0.77 | 8 (5.06) | 1.16 (0.50, 2.70) | 0.73 |
| Contact with patients with diarrhea | 228/608 | 1 (0.44) | 0.55 (0.06, 5.35) | 1.00 | 14 (6.14) | 0.89 (0.46, 1.74) | 0.74 |
| Animal in household | |||||||
| Pig | 253/1,100 | 5 (1.98) | 3.40 (0.98, 11.82) | 0.10 | 14 (5.53) | 0.88 (0.48, 1.61) | 0.67 |
| Cattle | 39/1,100 | 1 (2.56) | 3.08 (0.38, 24.90) | 0.80 | 4 (10.26) | 1.81 (0.62, 5.25) | 0.44 |
| Dog | 475/1,100 | 5 (1.05) | 1.32 (0.38, 4.58) | 0.91 | 28 (5.89) | 0.94 (0.57, 1.55) | 0.81 |
| Cat | 189/1,100 | 3 (1.59) | 2.08 (0.53, 8.13) | 0.51 | 9 (4.76) | 0.74 (0.36, 1.51) | 0.40 |
| Sheep/goat | 180/1,100 | 5 (2.78) | 5.23 (1.50, 18.25) | 0.01 | 11 (6.11) | 1.00 (0.52, 1.96) | 0.99 |
| Chicken/duck | 253/1,100 | 3 (1.19) | 1.44 (0.37, 5.61) | 0.88 | 18 (7.11) | 1.25 (0.71, 2.18) | 0.44 |
| Water source | |||||||
| Tap water | 38/1,100 | 0 | 0.01 | 1 (2.63) | 0.42 | ||
| Hand pump water | 823/1,100 | 4 (0.49) | 48 (5.83) | ||||
| Well water | 239/1,100 | 6 (2.51) | 18 (7.53) | ||||
| Raw vegetable consumption | 988/1,101 | 10 (1.01) | 0.58 | 56 (5.67) | 0.56 (0.28, 1.10) | 0.09 | |
| Family of >6 members | 50/1,101 | 0 | 1.00 | 7 (14.00) | 2.64 (1.14, 6.12) | 0.04 | |
| Hand washing | 1,089/1,101 | 10 (0.92) | 1.00 | 64 (5.88) | 0.19 (0.05, 0.71) | 0.03 | |
| Alcohol consumption | 442/1,096 | 1 (0.23) | 0.16 (0.02, 1.29) | 0.10 | 23 (5.20) | 0.78 (0.46, 1.31) | 0.35 |
Sample sizes differ for various factors due to missing patient information.
Analysis conducted in the HIV-positive group only.
Bold type for values indicates statistical significance.
CI, confidence interval.
Enterocytozoon bieneusi was found in 68 study participants. Infection rates of E. bieneusi were 5.7% (39/683) and 4.2% (29/683) in HIV-positive and HIV-negative patients, respectively. The difference between the two groups was not significant (P = 0.21) (Table 2). Among the study patients, two were infected with both Cryptosporidium and E. bieneusi.
Species, genotypes, and subtypes of Cryptosporidium and E. bieneusi.
Four Cryptosporidium species were found, including C. meleagridis (5 cases), C. hominis (3 cases), C. parvum (2 cases), and C. suis (1 case). Subtypes of C. meleagridis (IIIbA26G1R1, IIIbA27G1R1, IIIbA29G1R1, and IIIeA26G2R1), C. hominis (IbA19G2 and IeA12G3T3), and C. parvum (IIdA19G1) were determined for all but one C. meleagridis patient (Table 3).
Table 3.
Cryptosporidium and E. bieneusi genotypes/subtypes in HIV-positive and HIV-negative participants in Henan, China
| Species | Subtype or genotype | No. of persons infected |
Major host(s)a | |
|---|---|---|---|---|
| HIV+ | HIV− | |||
| C. hominis | IbA19G2 | 2 | Humans | |
| IeA12G3T3 | 1 | Humans | ||
| C. parvum | IIdA19G1 | 2 | Humans, cattle, sheep, goats | |
| C. meleagridis | IIIbA27G1R1 | 1 | ||
| IIIbA29G1R1 | 1 | |||
| IIIbA26G1R1 | 1 | Chickens, turkeys, pet birds | ||
| IIIeA26G2R1 | 1 | |||
| Unknown | 1 | |||
| C. suis | 1 | Pigs | ||
| EbpC | 18 | 21 | Humans, pigs, wild mammals | |
| D | 7 | 5 | Humans, cattle, pigs, dogs, wild mammals | |
| IV | 6 | 1 | Humans, cats, cattle, dogs | |
| PigEBITS7 | 1 | Pigs | ||
| E. bieneusi | EbpD | 1 | Pigs | |
| Peru8 | 1 | Humans | ||
| Peru11 | 1 | Humans | ||
| Henan-I | 1 | Humans (this study) | ||
| Henan-II | 1 | Humans (this study) | ||
| Henan-III | 1 | Humans (this study) | ||
| Henan-IV | 1 | Humans (this study) | ||
| Henan-V | 1 | Humans (this study) | ||
| Unknown | 1 | |||
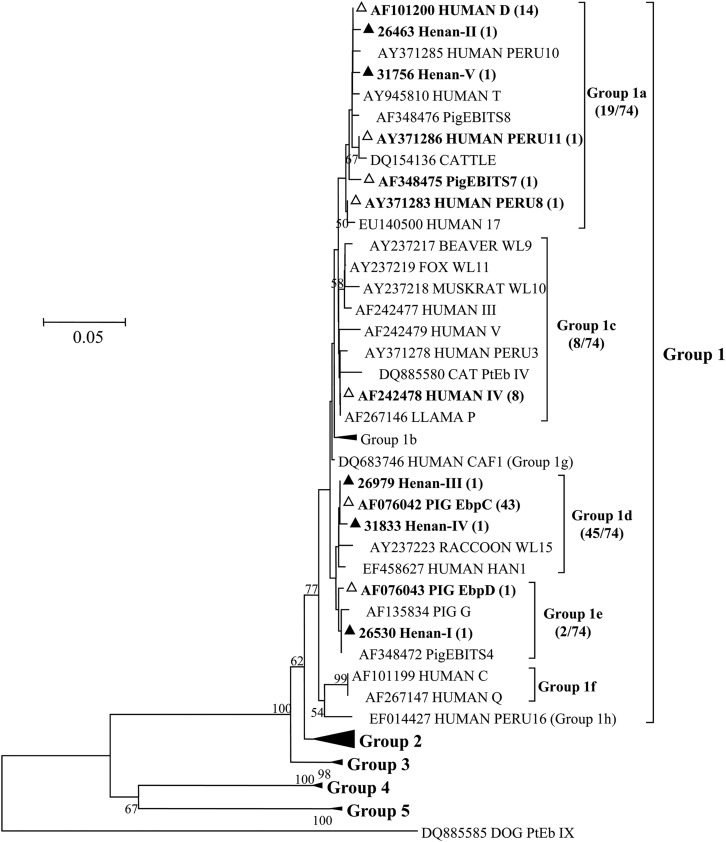
Twelve E. bieneusi genotypes were found. The most prevalent genotypes were EbpC (39 cases), D (12 cases), and type IV (7 cases). Others included Peru8, Peru11, PigEBITS7, EbpD, and five new genotypes named Henan-I to Henan-V and were found in one case each, mostly in the HIV-positive group (Table 3). These new genotypes were phylogenetically related to the genotype group 1, which contains most human-pathogenic E. bieneusi genotypes (Fig. 1). One patient with type IV and one with Peru8 were coinfected with Cryptosporidium meleagridis.
Fig 1.
Phylogenetic relationship of E. bieneusi genotypes identified in this study and other genotypes previously deposited in GenBank as inferred by a neighbor-joining analysis of ITS sequences, based on the p-distance model. Percent bootstrap values greater than 50% from 1,000 replicates are shown to the left of nodes. Novel and known genotypes identified in this study are indicated by ▲ and △, respectively.
Risk factors for cryptosporidiosis and microsporidiosis.
Cryptosporidium infection was significantly associated with HIV infection (10/683 in HIV-positive group versus 1/683 in HIV-negative group; P < 0.01) and use of well water as the drinking water supply (6/239 versus 4/861; P = 0.01). Patients with a history of animal contact in general had a higher occurrence of Cryptosporidium infection, but the association was significant only for sheep/goat keeping (P = 0.01) (Table 2).
A number of factors related to E. bieneusi infection were analyzed. An age of 10 to 19 years (P = 0.04) and living in a family with >6 members (P = 0.04) were associated with higher E. bieneusi prevalence, whereas hand washing before meals (P = 0.03) was a protective factor against E. bieneusi infection (Table 2).
A higher occurrence of diarrhea was observed in the HIV-positive group than in the HIV-negative group (263/651 versus 127/425; P < 0.01) (Table 1). In HIV-positive patients, patients with low CD4+ cell counts (<200 cells/ml) were more likely to have diarrhea (74/154 versus 148/411; P = 0.01). However, we did not identify a significant association between CD4+ cell count and Cryptosporidium or E. bieneusi infection (Table 2): the median CD4+ cell count was 295 in 8 Cryptosporidium-positive patients and 339 in 582 Cryptosporidium-negative patients, whereas the median was 357 in 27 E. bieneusi-positive patients and 337 in 563 E. bieneusi-negative patients. Among HIV-positive patients, infection with E. bieneusi was significantly associated with the occurrence of diarrhea (Table 2).
Risk factors were also analyzed for prevalent E. bieneusi genotypes (Table 4). The most common, EbpC, was associated with having pigs in the household (P = 0.01; Table 4). Genotype D was more prevalent in patients who were 0 to 19 years in age (6/12 versus 6/55; P < 0.01). The less common genotypes (those other than EbpC and D) were more often seen in the HIV-positive patients (P < 0.01) and patients with diarrhea (P = 0.04) (Table 4).
Table 4.
Significant genotype-specific risk factors in 67 E. bieneusi-positive patients
| Risk factor | Genotype | Sample sizea | No. positive (%)b | OR (95% CIc) | Pd |
|---|---|---|---|---|---|
| Age of 0–19 yr | D | 12 | 6 (50.00) | 8.17 (1.99, 33.58) | <0.01 |
| Female gender | Not EbpC or D | 33 | 11 (33.33) | 2.90 (0.88, 9.57) | 0.07 |
| HIV infection | Not EbpC or D | 39 | 14 (35.90) | 7.28 (1.50, 35.35) | <0.01 |
| Diarrhea | Not EbpC or D | 30 | 11 (36.67) | 3.47 (1.04, 11.57) | 0.04 |
| Animal in household | EbpC | ||||
| Pig | 14 | 12 (85.71) | 6.23 (1.27, 30.58) | 0.01 | |
| Cattle | 4 | 2 (50.00) | 0.75 (0.10, 5.67) | 1.00 | |
| Dog | 27 | 13 (48.15) | 0.56 (0.21, 1.50) | 0.24 | |
| Cat | 9 | 4 (44.44) | 0.57 (0.14, 2.32) | 0.66 | |
| Sheep/goat | 11 | 7 (63.64) | 1.41 (0.37, 5.37) | 0.86 | |
| Chicken/duck | 18 | 12 (66.67) | 1.77 (0.57, 5.47) | 0.32 |
Number out of 67 patients (65 patients for diarrhea) with the risk factor.
Number of patients with the genotype.
CI, confidence interval.
Bold type for values indicates statistical significance.
DISCUSSION
The low infection rates of Cryptosporidium (1.5%) and E. bieneusi (5.7%) in HIV-positive patients in this study could be mostly attributed to the NFATP. The infection rates were lower than those in studies conducted in other developing countries, where infection rates of 13.3 to 73.6% and 9.5 to 76.9% were reported for Cryptosporidium and microsporidia/E. bieneusi, respectively (7, 19, 22, 24, 26, 36–39). The infection rate of Cryptosporidium in this study was similar to that in HAART-receiving HIV-positive patients in Taiwan (1.2% in 332 patients) (40) and Denmark (1.0% in 96 patients) (41), as well as in Brazilian patients in the HAART era (0.3%) (42). A previous study also showed that infection rates of Cryptosporidium decreased significantly in patients after the initiation of HAART in Brazil: 8.1% in 482 pretreated patients and none in 100 posttreated patients (43). In a large cohort study in 10 European countries and Australia, the relative risk for contracting cryptosporidiosis was reduced by 96% in the HAART era (44). Therefore, even though protease inhibitors were not used, HAART in the NFATP was probably able to reduce the transmission of cryptosporidiosis. Previously, it was shown that NFATP was effective in reducing mortality in HIV-positive former plasma donors in China (31).
In agreement with the gradual decrease of E. bieneusi prevalence in HIV-positive patients receiving HAART in developed countries (17, 45), a significant decrease in E. bieneusi occurrence was observed in HIV-positive patients enrolled in 2010 compared with those enrolled in 2007 (8/253 versus 31/430; P = 0.03) in this study, suggesting that the NFATP might have played a role in this reduction, especially in cryptosporidiosis. In developing countries, the relationship between HAART and E. bieneusi occurrence in HIV-positive patients has seldom been examined. A gradual decrease in the prevalence of E. bieneusi was seen in a Thai orphanage (including 77 HIV-positive patients on HAART and 463 HIV-negative patients), although it was attributed to increased sanitation rather than antiretroviral therapy (25).
Zoonotic contact appeared to be an important risk factor in the acquisition of cryptosporidiosis in HIV-positive farmers in this study. Three of the four Cryptosporidium species detected, including C. parvum, C. meleagridis, and C. suis, were known parasites of animals. Although C. meleagridis was reported in six pediatric patients in China in one recent study (46), C. parvum and C. suis were reported in humans in China for the first time. The dominance of zoonotic Cryptosporidium spp. observed is different from most observations on cryptosporidiosis in humans in developing countries, where anthroponotic transmission plays a more important role in the epidemiology of cryptosporidiosis in HIV-positive patients (11, 12). Previously, it was thought that cryptosporidiosis transmission in China was also largely anthroponotic in nature (33, 46, 47). Differences in study areas (rural versus urban) could be responsible for the discrepancy in major transmission routes between current and earlier studies.
The occurrence of zoonotic cryptosporidiosis was supported by C. parvum subtyping. Both C. parvum cases were caused by the IIdA19G1 subtype, which was detected recently in calves in Henan (48). Although thus far they have not been found in sheep and goats in Henan (49), C. parvum IId subtypes are known to preferentially infect these animals rather than cattle (50, 51), a fact in agreement with the identification of sheep/goat keeping as a risk factor for cryptosporidiosis in this study. Another related C. parvum IId subtype, IIdA15G1, is common in rodents in Henan (52).
Consistent with the relatively high occurrence of zoonotic Cryptosporidium species, 90% of E. bieneusi infections in the farmers were caused by genotypes commonly found in animals, including all three dominant genotypes (EbpC, D, and type IV), PigEBITS7, and EbpD. Only Peru8 and Peru11, which were found in one HIV-positive and HIV-negative patient each, have thus far been reported only in humans (35). The common occurrence of the EbpC genotype in HIV-positive patients in Henan differs from findings in other areas. Thus far, only 10 cases of infection by EbpC strains have been reported in HIV-positive patients in developing countries and none have been reported in industrialized nations. Previously, the zoonotic genotypes D (90 cases) and type IV (87 cases) and the anthroponotic genotype A (142 cases) were the most common genotypes of E. bieneusi in HIV-positive patients in developing countries (18–23, 25–28). In contrast, the anthroponotic genotype B was the dominant genotype in HIV-positive patients in Europe and Australia, infecting at least 164 HIV-positive patients, compared to only two patients infected by the zoonotic genotype D and 14 patients infected by type IV (13–18). Interestingly, the most common anthroponotic genotype in HIV-positive patients in developing countries, genotype A, was found in only three HIV-positive patients in Europe (15), and similarly, the dominant anthroponotic genotype in developed countries, genotype B, was found in only one HIV-positive patient in Africa (24).
The likely occurrence of zoonotic cryptosporidiosis and microsporidiosis in this study was also supported by risk factor analysis. In this study, contact with animals, especially sheep and goats, was associated with cryptosporidiosis. Likewise, for E. bieneusi, contact with pigs was strongly associated with the most prevalent genotype EbpC, which was initially identified in pigs and is a common genotype in this species of animals (35). The zoonotic microsporidiosis transmission was likely caused by direct contact with infected animals or contaminated premises, as neither water nor eating raw vegetables was a risk factor. Correspondingly, hand washing before meals was a protective factor against microsporidiosis. Poor hygiene in general was probably a risk factor for microsporidiosis, as indicated also by the association of microsporidiosis with big families.
In conclusion, zoonotic Cryptosporidium species and E. bieneusi genotypes are still present in some HAART-receiving patients in rural China. Minimizing contact with animals and maintaining good hygiene practices should be advocated to reduce the transmission of Cryptosporidium spp. and E. bieneusi in HIV-positive rural patients in addition to the adherence to HAART regimes.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 31110103901, 31229005, and 41001316); the National Basic Research Program of China (973 Project) (2011CB200903); Project of Medical Science and Technique of Henan, China (No. 201003142); Special Funding of the Henan Health Science and Technology Innovation Talent Project; Fundamental Research Funds for the Central Universities, China; and Open Funding Projects of the State Key Laboratory of Veterinary Etiological Biology at the Lanzhou Veterinary Research Institute and State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology.
We thank the attending physicians for assistance in patient enrollment.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Sharpstone D, Phelan M, Gazzard B. 2000. Differential metabolic response in AIDS-related chronic protozoal diarrhoea. HIV Med. 1:102–106 [DOI] [PubMed] [Google Scholar]
- 2. Didier ES. 2005. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 94:61–76 [DOI] [PubMed] [Google Scholar]
- 3. Hunter PR, Nichols G. 2002. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin. Microbiol. Rev. 15:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pozio E, Morales MA. 2005. The impact of HIV-protease inhibitors on opportunistic parasites. Trends Parasitol. 21:58–63 [DOI] [PubMed] [Google Scholar]
- 5. Ajjampur SS, Sankaran P, Kang G. 2008. Cryptosporidium species in HIV-infected individuals in India: an overview. Natl. Med. J. India 21:178–184 [PubMed] [Google Scholar]
- 6. Certad G, Arenas-Pinto A, Pocaterra L, Ferrara G, Castro J, Bello A, Nunez L. 2005. Cryptosporidiosis in HIV-infected Venezuelan adults is strongly associated with acute or chronic diarrhea. Am. J. Trop. Med. Hyg. 73:54–57 [PubMed] [Google Scholar]
- 7. Tuli L, Gulati AK, Sundar S, Mohapatra TM. 2008. Correlation between CD4 counts of HIV patients and enteric protozoan in different seasons—an experience of a tertiary care hospital in Varanasi (India). BMC Gastroenterol. 8:36 doi:10.1186/1471-230X-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Werneck-Silva AL, Prado IB. 2009. Gastroduodenal opportunistic infections and dyspepsia in HIV-infected patients in the era of highly active antiretroviral therapy. J. Gastroenterol. Hepatol. 24:135–139 [DOI] [PubMed] [Google Scholar]
- 9. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]
- 10. Santin M, Fayer R. 2011. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 90:363–371 [DOI] [PubMed] [Google Scholar]
- 11. Xiao L, Feng Y. 2008. Zoonotic cryptosporidiosis. FEMS Immunol. Med. Microbiol. 52:309–323 [DOI] [PubMed] [Google Scholar]
- 12. Xiao L, Ryan UM. 2008. Molecular epidemiology, p 119–172 In Fayer R, Xiao L. (ed), Cryptosporidium and cryptosporidiosis, 2nd ed CRC Press, New York, NY [Google Scholar]
- 13. Liguory O, David F, Sarfati C, Derouin F, Molina JM. 1998. Determination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosis. J. Clin. Microbiol. 36:1882–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liguory O, Sarfati C, Derouin F, Molina JM. 2001. Evidence of different Enterocytozoon bieneusi genotypes in patients with and without human immunodeficiency virus infection. J. Clin. Microbiol. 39:2672–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rinder H, Katzwinkel-Wladarsch S, Loscher T. 1997. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol. Res. 83:670–672 [DOI] [PubMed] [Google Scholar]
- 16. Sadler F, Peake N, Borrow R, Rowl PL, Wilkins EG, Curry A. 2002. Genotyping of Enterocytozoon bieneusi in AIDS patients from the north west of England. J. Infect. 44:39–42 [DOI] [PubMed] [Google Scholar]
- 17. Stark D, van Hal S, Barratt J, Ellis J, Marriott D, Harkness J. 2009. Limited genetic diversity among genotypes of Enterocytozoon bieneusi strains isolated from HIV-infected patients from Sydney, Australia. J. Med. Microbiol. 58:355–357 [DOI] [PubMed] [Google Scholar]
- 18. ten Hove RJ, Van Lieshout L, Beadsworth MB, Perez MA, Spee K, Claas EC, Verweij JJ. 2009. Characterization of genotypes of Enterocytozoon bieneusi in immunosuppressed and immunocompetent patient groups. J. Eukaryot. Microbiol. 56:388–393 [DOI] [PubMed] [Google Scholar]
- 19. Akinbo FO, Okaka CE, Omoregie R, Dearen T, Leon ET, Xiao L. 2012. Molecular epidemiologic characterization of Enterocytozoon bieneusi in HIV-infected persons in Benin City, Nigeria. Am. J. Trop. Med. Hyg. 86:441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bern C, Kawai V, Vargas D, Rabke-Verani J, Williamson J, Chavez-Valdez R, Xiao L, Sulaiman I, Vivar A, Ticona E, Navincopa M, Cama V, Moura H, Secor WE, Visvesvara G, Gilman RH. 2005. The epidemiology of intestinal microsporidiosis in patients with HIV/AIDS in Lima, Peru. J. Infect. Dis. 191:1658–1664 [DOI] [PubMed] [Google Scholar]
- 21. Breton J, Bart-Delabesse E, Biligui S, Carbone A, Seiller X, Okome-Nkoumou M, Nzamba C, Kombila M, Accoceberry I, Thellier M. 2007. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J. Clin. Microbiol. 45:2580–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Espern A, Morio F, Miegeville M, Illa H, Abdoulaye M, Meyssonnier V, Adehossi E, Lejeune A, Cam PD, Besse B, Gay-Andrieu F. 2007. Molecular study of microsporidiosis due to Enterocytozoon bieneusi and Encephalitozoon intestinalis among human immunodeficiency virus-infected patients from two geographical areas: Niamey, Niger, and Hanoi, Vietnam. J. Clin. Microbiol. 45:2999–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leelayoova S, Subrungruang I, Suputtamongkol Y, Worapong J, Petmitr PC, Mungthin M. 2006. Identification of genotypes of Enterocytozoon bieneusi from stool samples from human immunodeficiency virus-infected patients in Thailand. J. Clin. Microbiol. 44:3001–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ojuromi OT, Izquierdo F, Fenoy S, Fagbenro-Beyioku A, Oyibo W, Akanmu A, Odunukwe N, Henriques-Gil N, del Aguila C. 2012. Identification and characterization of microsporidia from fecal samples of HIV-positive patients from Lagos, Nigeria. PLoS One 7:e35239 doi:10.1371/journal.pone.0035239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagornrat W, Leelayoova S, Rangsin R, Tan-Ariya P, Naaglor T, Mungthin M. 2009. Carriage rate of Enterocytozoon bieneusi in an orphanage in Bangkok, Thailand. J. Clin. Microbiol. 47:3739–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saksirisampant W, Prownebon J, Saksirisampant P, Mungthin M, Siripatanapipong S, Leelayoova S. 2009. Intestinal parasitic infections: prevalences in HIV/AIDS patients in a Thai AIDS-care centre. Ann. Trop. Med. Parasitol. 103:573–581 [DOI] [PubMed] [Google Scholar]
- 27. Sarfati C, Bourgeois A, Menotti J, Liegeois F, Moyou-Somo R, Delaporte E, Derouin F, Ngole EM, Molina JM. 2006. Prevalence of intestinal parasites including microsporidia in human immunodeficiency virus-infected adults in Cameroon: a cross-sectional study. Am. J. Trop. Med. Hyg. 74:162–164 [PubMed] [Google Scholar]
- 28. Sulaiman IM, Bern C, Gilman R, Cama V, Kawai V, Vargas D, Ticona E, Vivar A, Xiao L. 2003. A molecular biologic study of Enterocytozoon bieneusi in HIV-infected patients in Lima, Peru. J. Eukaryot. Microbiol. 50(Suppl):591–596 [DOI] [PubMed] [Google Scholar]
- 29. Li N, Wang Z, Sun D, Zhu Q, Sun G, Yang W, Wang Q, Nie Y, Wu Z. 2010. HIV among plasma donors and other high-risk groups in Henan, China. J. Acquir. Immune Defic. Syndr. 53(Suppl 1):S41–S47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang F, Dou Z, Ma Y, Zhang Y, Zhao Y, Zhao D, Zhou S, Bulterys M, Zhu H, Chen RY. 2011. Effect of earlier initiation of antiretroviral treatment and increased treatment coverage on HIV-related mortality in China: a national observational cohort study. Lancet Infect. Dis. 11:516–524 [DOI] [PubMed] [Google Scholar]
- 31. Zhang F, Dou Z, Yu L, Xu J, Jiao JH, Wang N, Ma Y, Zhao Y, Zhao H, Chen RY. 2008. The effect of highly active antiretroviral therapy on mortality among HIV-infected former plasma donors in China. Clin. Infect. Dis. 47:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feng Y, Zhao X, Chen J, Jin W, Zhou X, Li N, Wang L, Xiao L. 2011. Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in Shanghai, China. Appl. Environ. Microbiol. 77:3609–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Y, Li N, Duan L, Xiao L. 2009. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J. Clin. Microbiol. 47:153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santin M, Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 56:34–38 [DOI] [PubMed] [Google Scholar]
- 36. Cama VA, Bern C, Sulaiman IM, Gilman RH, Ticona E, Vivar A, Kawai V, Vargas D, Zhou L, Xiao L. 2003. Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J. Eukaryot. Microbiol. 50(Suppl):531–533 [DOI] [PubMed] [Google Scholar]
- 37. Gumbo T, Sarbah S, Gangaidzo IT, Ortega Y, Sterling CR, Carville A, Tzipori S, Wiest PM. 1999. Intestinal parasites in patients with diarrhea and human immunodeficiency virus infection in Zimbabwe. AIDS 13:819–821 [DOI] [PubMed] [Google Scholar]
- 38. Houpt ER, Bushen OY, Sam NE, Kohli A, Asgharpour A, Ng CT, Calfee DP, Guerrant RL, Maro V, Ole-Nguyaine S, Shao JF. 2005. Short report: asymptomatic Cryptosporidium hominis infection among human immunodeficiency virus-infected patients in Tanzania. Am. J. Trop. Med. Hyg. 73:520–522 [PubMed] [Google Scholar]
- 39. Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, Akiyoshi DE, Tzipori S. 2005. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am. J. Trop. Med. Hyg. 73:921–925 [PubMed] [Google Scholar]
- 40. Hung CC, Tsaihong JC, Lee YT, Deng HY, Hsiao WH, Chang SY, Chang SC, Su KE. 2007. Prevalence of intestinal infection due to Cryptosporidium species among Taiwanese patients with human immunodeficiency virus infection. J. Formos. Med. Assoc. 106:31–35 [DOI] [PubMed] [Google Scholar]
- 41. Stensvold CR, Nielsen SD, Badsberg JH, Engberg J, Friis-Moller N, Nielsen SS, Nielsen HV, Friis-Moller A. 2011. The prevalence and clinical significance of intestinal parasites in HIV-infected patients in Denmark. Scand. J. Infect. Dis. 43:129–135 [DOI] [PubMed] [Google Scholar]
- 42. Cardoso LV, Galisteu KJ, Schiesari A, Junior, Chahla LA, Canille RM, Belloto MV, Franco C, Maia IL, Rossit AR, Machado RL. 2011. Enteric parasites in HIV-1/AIDS-infected patients from a Northwestern Sao Paulo reference unit in the highly active antiretroviral therapy era. Rev. Soc. Bras. Med. Trop. 44:665–669 [DOI] [PubMed] [Google Scholar]
- 43. Bachur TP, Vale JM, Coelho IC, Queiroz TR, de Souza Chaves C. 2008. Enteric parasitic infections in HIV/AIDS patients before and after the highly active antiretroviral therapy. Braz. J. Infect. Dis. 12:115–122 [DOI] [PubMed] [Google Scholar]
- 44. Pozio E. 2004. Highly active antiretroviral therapy and opportunistic protozoan infections. Parassitologia 46:89–93 [PubMed] [Google Scholar]
- 45. Conteas CN, Berlin OG, Lariviere MJ, Pandhumas SS, Speck CE, Porschen R, Nakaya T. 1998. Examination of the prevalence and seasonal variation of intestinal microsporidiosis in the stools of persons with chronic diarrhea and human immunodeficiency virus infection. Am. J. Trop. Med. Hyg. 58:559–561 [DOI] [PubMed] [Google Scholar]
- 46. Feng Y, Wang L, Duan L, Gomez-Puerta A, Zhang L, Zhao X, Hu J, Zhang N, Xiao L. 2012. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg. Infect. Dis. 18:312–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang R, Zhang X, Zhu H, Zhang L, Feng Y, Jian F, Ning C, Qi M, Zhou Y, Fu K, Wang Y, Sun Y, Wang Q, Xiao L. 2011. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp. Parasitol. 127:42–45 [DOI] [PubMed] [Google Scholar]
- 48. Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, Ning C, Xiao L. 2011. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J. Clin. Microbiol. 49:1077–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y, Feng Y, Cui B, Jian F, Ning C, Wang R, Zhang L, Xiao L. 2010. Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol. Res. 106:341–347 [DOI] [PubMed] [Google Scholar]
- 50. Quilez J, Torres E, Chalmers RM, Robinson G, Del Cacho E, Sanchez-Acedo C. 2008. Cryptosporidium species and subtype analysis from dairy calves in Spain. Parasitology 135:1613–1620 [DOI] [PubMed] [Google Scholar]
- 51. Quilez J, Torres E, Chalmers RM, Hadfield SJ, Del Cacho E, Sanchez-Acedo C. 2008. Cryptosporidium genotypes and subtypes in lambs and goat kids in Spain. Appl. Environ. Microbiol. 74:6026–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lv C, Zhang L, Wang R, Jian F, Zhang S, Ning C, Wang H, Feng C, Wang X, Ren X, Qi M, Xiao L. 2009. Cryptosporidium spp. in wild, laboratory, and pet rodents in China: prevalence and molecular characterization. Appl. Environ. Microbiol. 75:7692–7699 [DOI] [PMC free article] [PubMed] [Google Scholar]