Abstract
A Newcastle disease virus (NDV) outbreak in chickens was reported in the Dominican Republic in 2008. The complete genome of this isolate, chicken/DominicanRepublic(JuanLopez)/499-31/2008 (NDV-DR499-31/08), and the fusion proteins of three other related viruses from the Dominican Republic and Mexico were sequenced and phylogenetically analyzed. Genetically, these four isolates were highly distinct from all other currently known isolates of NDV, and together, they fulfill the newly established criteria for inclusion as a novel genotype of NDV (genotype XVI). The lack of any reported isolation of viruses related to this group since 1986 suggests that virulent viruses of this genotype may have evolved unnoticed for 22 years. The NDV-DR499-31/08 isolate had an intracerebral pathogenicity index (ICPI) score of 1.88, and sequencing of the fusion cleavage site identified multiple basic amino acids and a phenylalanine at position 117, indicating this isolate to be virulent. These results were further confirmed by a clinicopathological assessment in vivo. In 4-week-old chickens, NDV-DR499-31/08 behaved as a velogenic viscerotropic strain with systemic virus distribution and severe necrohemorrhagic lesions targeting mainly the intestine and the lymphoid organs. The clear phylogenetic relationship between the 2008, 1986, and 1947 ancestral viruses suggests that virulent NDV strains may have evolved in unknown reservoirs in the Caribbean and surrounding regions and underlines the importance of continued and improved epidemiological surveillance strategies to detect NDV in wild-bird species and commercial poultry.
INTRODUCTION
Newcastle disease (ND), caused by virulent isolates of Newcastle disease virus (NDV), infects poultry globally and results in significant economic losses and trade restrictions (1–3). Infections with these isolates have been shown to lead to fatality rates of up to 100% (3–6). NDV belongs to the order Mononegavirales, family Paramyxoviridae, subfamily Paramyxovirinae, and genus Avulavirus (7, 8). NDV is an enveloped virus that contains a negative-sense, single-stranded RNA genome of ∼15,200 nucleotides (nt) that contains six genes, which encode seven proteins, whose transcription occurs in the 3′-to-5′ orientation, resulting in decreasing amounts of protein with each ensuing gene (9, 10). The proteins encoded include nucleoprotein (NP), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin-neuraminidase (HN), and the RNA-dependent RNA polymerase (or large polymerase) (L) (10). The replication of the genome is controlled by the “rule of six,” which requires the genome length to be a multiple of 6 for the proper packaging of the RNA genome by NP into the virion (5, 11, 12).
The virulences of different NDV isolates vary remarkably, where clinical manifestations can range from a lack of clinical signs to severe malaise, respiratory distress, or neurological signs and death (3, 4). Due to such a protean presentation, different methods have been developed to classify the virulence of NDV isolates. Historically, NDV isolates are pathotyped into three groups based on their clinical presentation: the least virulent isolates are termed lentogenic and are usually asymptomatic, moderately virulent strains are termed mesogenic and typically present with respiratory or neurologic signs, and the most virulent strains are termed velogenic (viscerotropic or neurotropic) and are often fatal due to extensive necrosis and hemorrhaging (3, 4). Currently, the internationally recognized method for classifying the virulence of NDV strains is the intracerebral pathogenicity index (ICPI) score, which can be further supported by determining the cleavage site sequence of the F protein (13). Strains with ICPI scores of 0.7 to 1.5 are considered mesogenic, whereas those with ICPI scores of >1.5 are velogenic (3, 10); however, the United States designated all NDV strains with ICPI scores higher than 0.7 to be select agents that are virulent and reportable, to follow the World Organization for Animal Health (OIE) and European Union standards (13). The cleavage site of the F protein has been shown to be the main molecular determinant of NDV virulence, as strains with an F protein cleavage site with at least 3 arginine or lysine residues between positions 113 and 116 and a phenylalanine residue at position 117 are considered virulent (13).
NDV isolates all belong to one serotype but have been determined to contain significant genetic diversity and are therefore grouped into two distinct classes (class I or II), based on genome lengths and nucleotide sequences (14–18). Class I strains have genome sizes of 15,198 nt and are found worldwide, typically from waterfowl and shorebirds (15–17, 19). Class II strains have genome sizes of 15,186 nt, are also observed worldwide, and are typically found circulating within wild-bird and poultry species, and this class contains some of the more virulent genotypes (17). Furthermore, a third genome size has been recognized within class II strains with a genome length of 15,192 nt, and virus strains belonging to genotypes V to VIII, which represent only virulent strains, make up this group (15). The insertion of 6 nucleotides in the 5′ noncoding region (NCR) of the nucleoprotein was identified as the cause for the increase in the genome size (15, 20). Genotype VII strains are the predominant virulent strains circulating and have recently been isolated in broiler, layer, and breeder farms in Jordan, duck flocks in China, live-bird markets in Nigeria, pheasant farms in Spain, and poultry farms in Malaysia (21–24). NDV strains are constantly evolving, and numerous new genotypes and subgenotypes have been discovered in the last few decades (16). Recently, the genetic diversity of NDV was reassessed, and a system that separates the different NDV genotypes based on objective criteria was proposed (25). Under these criteria, class I viruses comprise a single genotype, and class II viruses are separated into 15 genetic groups, including the 10 previously established genotypes (genotypes I to IX and XI) and five new genotypes (genotypes X, XII, XIII, XIV, and XV) (25). As reviewed previously by Diel et al. (25), this classification system is based on the NDV genotyping system (26–29), and it uses the phylogenetic relationship and the evolutionary distances between genetic groups to classify NDV isolates into genotypes. The basis of this classification system allows researchers to easily determine whether they have identified a new genotype.
Together with this growing genetic diversity, there are increasing numbers of reports suggesting that traditional NDV vaccines may not be efficient in protecting against viruses belonging to genetically distant groups (30–32) or that new NDV isolates may have extended host ranges (33). ND is endemic to over 50% of the countries raising poultry but is considered exotic to the U.S. poultry industry (15, 23, 34). However, threats of exposure still occur through migration, through the illegal importation of birds from areas where ND is endemic, and possibly through apparently healthy wild-pigeon and cormorant populations that harbor virulent NDV (vNDV). Due to the extensive costs of containing vNDV outbreaks, the existence and maintenance of epidemiological surveillance strategies are critical. Here we report the complete genome and the clinicopathological characteristics of a novel NDV isolate [chicken/DominicanRepublic(JuanLopez)/499-31/2008] isolated in the Dominican Republic in 2008, referred to here as NDV-DR499-31/08. Phylogenetic analyses of this isolate, along with another isolate collected in the Dominican Republic in 2008 and two other ancestral viruses, suggest it to be a distinct genotype (genotype XVI), with features that classify this isolate as a viscerotropic velogenic isolate of NDV.
MATERIALS AND METHODS
Viruses.
In 1986 and 2008, the National Veterinary Services Laboratories (NVSL) (Ames, IA) isolated NDV from three separate specimens received from the Dominican Republic through the U.S. Embassy (USDA-APHIS-U.S. Embassy). Chicken/DominicanRepublic/28138-4/1986 (referred to here as NDV-DR28138-4/86) was isolated from allantoic fluid by the NVSL in 1986, and limited information is available regarding this isolate. Chicken/DominicanRepublic(JuanLopez)/499-31/2008 (referred to here as NDV-DR499-31/08) was isolated by the NVSL in 2008. Specimens were collected in February and March 2008 during routine surveillance of an apparently healthy flock of chickens after avian influenza virus was detected in the Dominican Republic in December 2007. Chicken/DominicanRepublic/867-2/2008 (referred to here as NDV-DR867-2/08) was also isolated by the NVSL in 2008. These specimens were received in May 2008 and were collected as a result of clinical signs in commercial hens from a flock of 80,000 that was experiencing an increase in the mortality rate (∼3%). Viruses were propagated by passage in 9- to 10-day-old embryonated chicken eggs inoculated by the chorioallantoic route. An egg passage virus stock was used for RNA extraction and for infectious inoculums in the animal experiments. Chicken/Mexico/Queretaro/452/1947 (referred to here as NDV-Mex452/47) was an NDV isolate from 1947 collected in Queretaro, Mexico, and was obtained from infected broilers. RNA of this isolate was a kind gift from Ruben Merino (University of Mexico, Mexico City, Mexico).
Eggs and chickens.
The sources of embryonating chicken eggs and chickens were the Charles River Avian Vaccine Services (formerly SPAFAS) at the NVSL and the Southeast Poultry Research Laboratory (SEPRL) specific-pathogen-free (SPF) White Leghorn flock. Birds for the ICPI and pathogenesis studies were housed in negative-pressure isolators under biosafety level 3 (BSL-3) enhanced (E) conditions at SEPRL and were provided food and water ad libitum.
ICPI test.
To characterize the virus, an ICPI test was performed according to standard protocols (13). Briefly, chickens were inoculated at 1 day of age with 0.05 ml of a 1:10 dilution of infective allantoic fluid. Chicks were monitored daily and scored as 0 for normal, 1 for sick or paralyzed, or 2 for dead, to compile a score for the 8-day observation period (13).
Clinicopathological assessment in chickens.
To determine the ability of NDV-DR499-31/08 to induce disease in chickens, two groups of 10 4-week-old SPF White Leghorn chickens (n = 20) were inoculated with 0.1 ml of an NDV-DR499-31/08 viral suspension (in brain heart infusion [BHI] broth), with half administered in the conjunctival sac and half in the choanal slit. Phosphate-buffered saline (PBS) was used for the uninfected control birds (n = 10). The target dose of the inoculum was 105.0 50% embryo infectious doses (EID50). The birds were clinically monitored every day, and two birds from each group were euthanized at 2, 5, 10, and 14 days postinoculation (dpi). Birds whose condition became critical were euthanized regardless of the scheduled sampling day.
Tissues (n = 25) (eyelid, spleen, bursa of Fabricius, thymus, Harderian gland, proventriculus, small intestine, cecal tonsils, large intestine, air sac, trachea, lung, heart, esophagus, pharynx, crop, brain, liver, pancreas, kidney, comb, head of left femur including bone marrow, and nasal turbinate) were collected and processed as previously described (8). All sampled tissues were routinely processed in paraffin, and 3-μm sections were cut for hematoxylin and eosin (HE) staining and immunohistochemistry (IHC).
Immunohistochemistry.
To determine the distribution of viral antigen in the organs of the infected chicken, IHC was carried out on the same organs collected for HE staining, as previously described (8).
RNA isolation and sequencing.
RNA isolation and sequencing were performed as previously described (35, 36). Briefly, viruses (NDV-DR499-31/08, NDV-DR867-2/08, and NDV-DR28138-4/86) were propagated in eggs, RNA was extracted from allantoic fluids, and F genes were then amplified by reverse transcription-PCR (RT-PCR) and sequenced. The full F gene primers of NDV-DR499-31/08, NDV-DR28138-4/86, and NDV-Mex452/47 are available upon request. Upon initially receiving the specimen, the full fusion gene of chicken/DominicanRepublic/867-2/2008 was sequenced and provided by the NVSL.
The complete genome sequence of NDV-DR499-31/08 was determined by using a shotgun RT-PCR/sequencing approach, as previously described (35).
Phylogenetic analysis.
The full F genes and complete NDV-DR499-31/08 genome were phylogenetically analyzed as previously described (35). Briefly, phylogenetic trees were constructed to localize NDV-DR499-31/08 among class II reference strains by using MEGA5 software (37). Evolutionary distances were inferred by using the maximum likelihood method, as implemented in MEGA5 software (38), and are shown as the number of base substitutions per site. Criteria to define new genotypes were based on those reported previously by Diel et al. (25).
Nucleotide sequence accession numbers.
The complete genome sequence of the NDV isolate chicken/DominicanRepublic(JuanLopez)/499-31/2008 is available in GenBank under accession no. JX119193 (39). The sequences of the full fusion genes of chicken/DominicanRepublic/867-2/2008, chicken/DominicanRepublic/28138-4/1986, and chicken/Mexico/Queretaro/452/1947 are available in GenBank under accession no. JX186997, JX915242, and JX915243, respectively.
RESULTS
NDV-DR499-31/08 belongs to the velogenic NDV pathotype.
The pathogenicity of isolate NDV-DR499-31/08 was assessed by sequencing of the F protein and by the ICPI test. As previously reported (39), the NDV F protein cleavage site has an amino acid sequence characterized by the presence of three basic residues at positions 113, 115, and 116 and a phenylalanine at position 117 (113RQKR*F117), which is consistent with a virulent NDV isolate (13). The ICPI score for NDV-DR499-31/08 was 1.88, classifying it as virulent and notifiable based upon OIE international standards. Sequencing and ICPI testing were also performed at the initial time of isolation by the NVSL, similarly determining NDV-DR499-31/08 to be a virulent isolate of NDV (Janice Pedersen, NVSL, personal communication), and the isolate was immediately reported to the OIE.
Phylogenetic analysis of NDV-DR499-31/08 among other known NDV genotypes classifies it as a novel class II genotype.
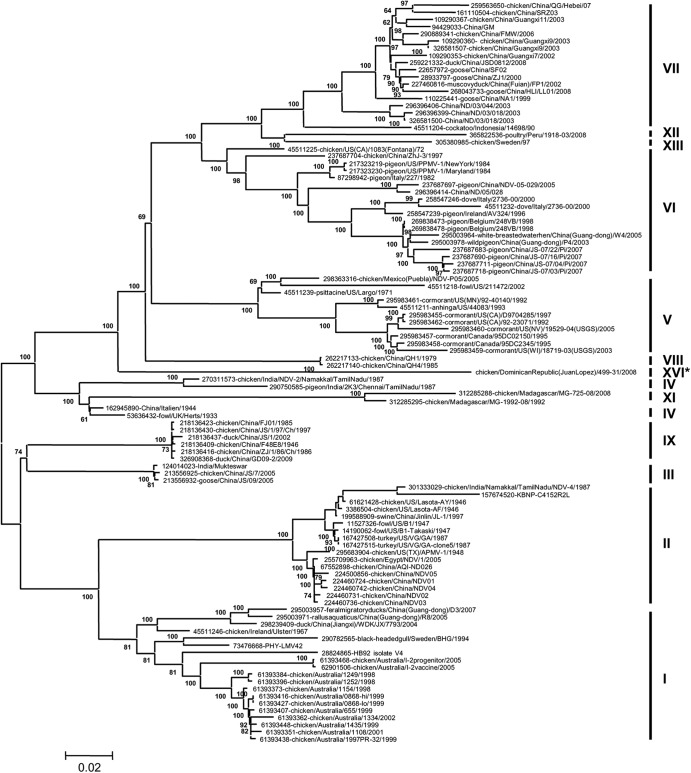
The complete genome of NDV-DR499-31/08 was sequenced (39) and phylogenetically analyzed with 103 representatives of other class II NDV genotype strains. This analysis identified NDV-DR499-31/08 to localize to a unique branch that is separate from all other currently known complete genomes of class II genotype strains (Fig. 1) by using the maximum likelihood method (40). The viruses that align on branches closest to this strain belong to genotypes IV and VIII. The possibility of a recombination event was analyzed by using RDP3 (41), but none were detected (data not shown). To further analyze NDV-DR499-31/08 among class II NDV genotype strains, the evolutionary distances between the complete genomes were analyzed by using MEGA5 and are shown as the number of base substitutions per site when averaged for each genotype. To test this, representative strains from 12 of the class II NDV genotypes were analyzed (genotype I, n = 19; genotype II, n = 17; genotype III, n = 3; genotype IV, n = 4; genotype V, n = 11; genotype VI, n = 18; genotype VII, n = 18; genotype VIII, n = 2; genotype IX, n = 6; genotype XI, n = 2; genotype XII, n = 1; genotype XIII, n = 1). These data showed that NDV-DR499-31/08 was significantly distant from all other class II reference strains (Table 1). NDV-DR499-31/08 has the shortest evolutionary distance with viruses in genotype VIII (0.185 substitutions per site), while the longest evolutionary distance is with viruses belonging to genotype XI (0.256 substitutions per site) (Table 1). These distances are all greater than 10%, suggesting that NDV-DR499-31/08 is significantly distinct.
Fig 1.
Molecular phylogenetic analysis of the complete NDV-DR499-31/08 genome sequence among 103 taxa available in GenBank. The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model (40). The tree with the highest log likelihood (−17,0944.0544) is shown. The percentages of trees in which the associated taxa clustered together are shown next to the branches. A discrete gamma distribution was used to model evolutionary rate differences among sites (4 categories [+G, parameter = 0.8193]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 35.2577% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 103 nucleotide sequences. Codon positions included were the first, second, third, and noncoding positions. All positions containing gaps and missing data were eliminated. There were a total of 15,073 positions in the final data set. Evolutionary analyses were conducted by using MEGA5 (37). Bootstrap values of less than 60 are not shown.
Table 1.
Estimates of evolutionary distances between the complete genomes of NDV-DR499-31/08 and NDV genotypes
| Strain or genotype | No. of base substitutions/site for strain or genotypea: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DR | I | II | III | IV | V | VI | VII | VIII | IX | XI | XII | XIII | |
| DR | (0.006) | (0.006) | (0.004) | (0.004) | (0.004) | (0.004) | (0.005) | (0.005) | (0.005) | (0.006) | (0.005) | (0.005) | |
| I | 0.237 | (0.003) | (0.003) | (0.003) | (0.003) | (0.004) | (0.003) | (0.003) | (0.003) | (0.005) | (0.004) | (0.004) | |
| II | 0.253 | 0.139 | (0.004) | (0.004) | (0.004) | (0.005) | (0.004) | (0.004) | (0.004) | (0.006) | (0.005) | (0.005) | |
| III | 0.212 | 0.131 | 0.158 | (0.003) | (0.004) | (0.003) | (0.002) | (0.004) | (0.002) | (0.005) | (0.004) | (0.004) | |
| IV | 0.197 | 0.148 | 0.172 | 0.116 | (0.003) | (0.003) | (0.003) | (0.004) | (0.002) | (0.004) | (0.004) | (0.004) | |
| V | 0.206 | 0.213 | 0.235 | 0.185 | 0.168 | (0.003) | (0.004) | (0.003) | (0.004) | (0.005) | (0.004) | (0.004) | |
| VI | 0.200 | 0.206 | 0.228 | 0.178 | 0.164 | 0.162 | (0.003) | (0.003) | (0.004) | (0.005) | (0.004) | (0.003) | |
| VII | 0.213 | 0.213 | 0.232 | 0.192 | 0.175 | 0.169 | 0.140 | (0.004) | (0.004) | (0.005) | (0.003) | (0.003) | |
| VIII | 0.185 | 0.190 | 0.217 | 0.159 | 0.149 | 0.144 | 0.143 | 0.154 | (0.004) | (0.006) | (0.003) | (0.004) | |
| IX | 0.212 | 0.138 | 0.160 | 0.102 | 0.123 | 0.181 | 0.181 | 0.190 | 0.163 | (0.005) | (0.004) | (0.004) | |
| XI | 0.256 | 0.222 | 0.245 | 0.192 | 0.153 | 0.228 | 0.226 | 0.234 | 0.210 | 0.186 | (0.006) | (0.006) | |
| XII | 0.229 | 0.228 | 0.250 | 0.205 | 0.194 | 0.186 | 0.152 | 0.127 | 0.167 | 0.207 | 0.249 | (0.004) | |
| XIII | 0.216 | 0.219 | 0.238 | 0.195 | 0.179 | 0.173 | 0.143 | 0.117 | 0.156 | 0.191 | 0.236 | 0.118 | |
The numbers of base substitutions per site from averaging over all sequence pairs between groups are shown for NDV-DR499-31/08 and genotypes of class II reference strains (n = 103). The numbers of sequences analyzed per group were as follows: 1 for NDV-DR499-31/08 (DR), 19 for genotype I, 17 for genotype II, 3 for genotype III, 4 for genotype IV, 11 for genotype V, 18 for genotype VI, 18 for genotype VII, 2 for genotype VIII, 6 for genotype IX, 2 for genotype XI, 1 for genotype XII, and 1 for genotype XIII. Analyses were conducted by using the maximum composite likelihood model in MEGA5. The codon positions included were the first, second, third, and noncoding positions. All positions containing gaps and missing data were eliminated. There were a total of 15,073 positions in the final data set. Values in parentheses are standard errors.
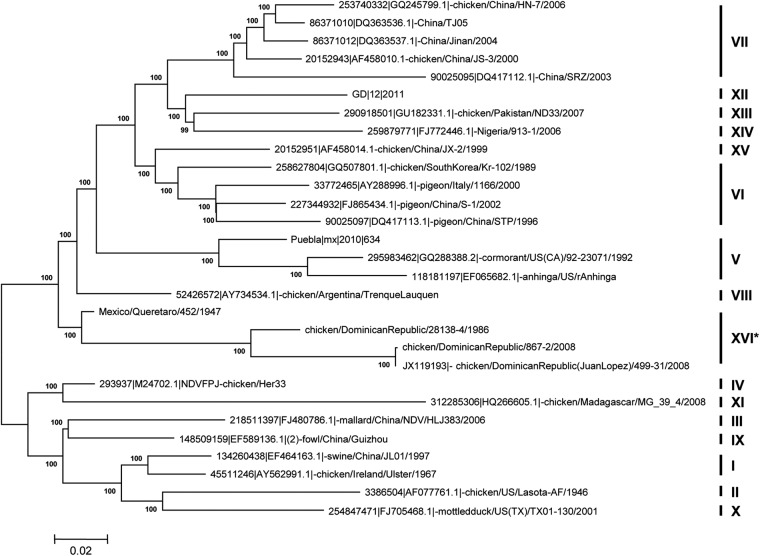
Because the complete genome analysis suggested that NDV-DR499-31/08 belongs to a previously unidentified NDV genotype, the full fusion gene sequences of novel viruses recovered from the surrounding geographical regions were also analyzed. These viruses included another isolate collected from a separate incident within the Dominican Republic in 2008 (chicken/DominicanRepublic/867-2/2008, referred to as NDV-DR867-2/08), an isolate from the Dominican Republic in 1986 (chicken/DominicanRepublic/28138-4/1986, referred to as NDV-DR28138-4/86), and an isolate collected in Mexico in 1947 (chicken/Mexico/Queretaro/452/1947, referred to as NDV-Mex452/47). When these isolates were compared with other full fusion gene sequences of class II reference strains, all four were located, and fully supported with a high bootstrap value, on a branch separate from the other reference strains (n = 29) (Fig. 2). Similarly, a separate tree analyzing over 600 NDV fusion nucleotide sequences also identified these four viruses as clustering together in a single clade (data not shown).
Fig 2.
Molecular phylogenetic analysis of the complete fusion (F) gene sequences of NDV-DR499-31/08, NDV-DR867-2/08, NDV-DR28138-4/86, and NDV-Mex452/47 among 25 other taxa available in GenBank. The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model (40). The tree with the highest log likelihood (−12,053.7125) is shown. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.5839]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 43.4250% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 29 nucleotide sequences. Codon positions included were the first, second, third, and noncoding positions. All positions containing gaps and missing data were eliminated. There were a total of 1,652 positions in the final data set. Evolutionary analyses were conducted with MEGA5 (37).
As before, the evolutionary distances between genotypes were determined by comparing the full fusion gene sequence of NDV-DR499-31/08 with those of other known NDV reference strains (n = 590). To test this, representative strains from each of the class II NDV genotypes were analyzed (genotype I, n = 66; genotype II, n = 102; genotype III, n = 9; genotype IV, n = 6; genotype V, n = 45; genotype VI, n = 57; genotype VII, n = 231; genotype VIII, n = 4; genotype IX, n = 19; genotype X, n = 18; genotype XI, n = 4; genotype XII, n = 6; genotype XIII, n = 8; genotype XIV, n = 6; genotype XV, n = 5; genotype XVI, n = 4), and the evolutionary distances were averaged for each genotype. The results showed that these four viruses are significantly distant (>10%) from all of the other class II genotype strains (Table 2). According to criteria reported previously by Diel et al. (25), who stated the requirements for the classification of a new NDV genotype, these strains can be classified as a novel class II NDV genotype, genotype XVI. Furthermore, these data suggest that NDV-DR28138-4/86 and NDV-Mex452/47 are ancestors of the two viruses isolated from the Dominican Republic in 2008 and that they may have been circulating and evolving in the region for many years unnoticed.
Table 2.
Estimates of evolutionary distances between the full fusion gene sequences of all NDV genotypes
| Genotype | No. of base substitutions/site for genotypea: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | IX | V | VI | VII | VIII | X | XI | XII | XIII | XIV | XV | XVI | |
| I | (0.008) | (0.008) | (0.007) | (0.008) | (0.010) | (0.009) | (0.010) | (0.009) | (0.006) | (0.009) | (0.010) | (0.009) | (0.009) | (0.007) | (0.009) | |
| II | 0.115 | (0.010) | (0.009) | (0.009) | (0.011) | (0.010) | (0.012) | (0.011) | (0.008) | (0.012) | (0.012) | (0.011) | (0.011) | (0.008) | (0.011) | |
| III | 0.106 | 0.131 | (0.006) | (0.008) | (0.009) | (0.009) | (0.010) | (0.009) | (0.008) | (0.010) | (0.009) | (0.010) | (0.010) | (0.008) | (0.010) | |
| IV | 0.093 | 0.119 | 0.080 | (0.007) | (0.008) | (0.007) | (0.008) | (0.007) | (0.007) | (0.008) | (0.009) | (0.008) | (0.008) | (0.006) | (0.008) | |
| IX | 0.098 | 0.118 | 0.090 | 0.077 | (0.009) | (0.009) | (0.010) | (0.009) | (0.008) | (0.010) | (0.010) | (0.009) | (0.010) | (0.006) | (0.010) | |
| V | 0.168 | 0.181 | 0.160 | 0.135 | 0.154 | (0.008) | (0.008) | (0.008) | (0.010) | (0.010) | (0.009) | (0.008) | (0.008) | (0.008) | (0.009) | |
| VI | 0.154 | 0.173 | 0.152 | 0.119 | 0.147 | 0.140 | (0.007) | (0.006) | (0.008) | (0.010) | (0.008) | (0.006) | (0.007) | (0.007) | (0.008) | |
| VII | 0.155 | 0.182 | 0.148 | 0.125 | 0.148 | 0.141 | 0.115 | (0.008) | (0.011) | (0.011) | (0.007) | (0.006) | (0.007) | (0.006) | (0.009) | |
| VIII | 0.130 | 0.150 | 0.124 | 0.095 | 0.119 | 0.123 | 0.108 | 0.115 | (0.009) | (0.009) | (0.008) | (0.007) | (0.007) | (0.007) | (0.007) | |
| X | 0.104 | 0.108 | 0.127 | 0.114 | 0.115 | 0.180 | 0.170 | 0.171 | 0.146 | (0.010) | (0.011) | (0.009) | (0.010) | (0.008) | (0.009) | |
| XI | 0.171 | 0.191 | 0.163 | 0.117 | 0.153 | 0.195 | 0.194 | 0.202 | 0.172 | 0.187 | (0.011) | (0.011) | (0.012) | (0.009) | (0.011) | |
| XII | 0.163 | 0.189 | 0.156 | 0.135 | 0.155 | 0.151 | 0.114 | 0.107 | 0.116 | 0.173 | 0.208 | (0.005) | (0.006) | (0.007) | (0.009) | |
| XIII | 0.154 | 0.185 | 0.153 | 0.127 | 0.146 | 0.148 | 0.119 | 0.104 | 0.117 | 0.171 | 0.196 | 0.100 | (0.005) | (0.006) | (0.008) | |
| XIV | 0.162 | 0.191 | 0.163 | 0.138 | 0.156 | 0.152 | 0.127 | 0.116 | 0.128 | 0.181 | 0.203 | 0.108 | 0.109 | (0.007) | (0.009) | |
| XV | 0.130 | 0.124 | 0.125 | 0.102 | 0.097 | 0.147 | 0.126 | 0.094 | 0.119 | 0.144 | 0.183 | 0.131 | 0.126 | 0.135 | (0.008) | |
| XVI | 0.151 | 0.175 | 0.148 | 0.117 | 0.144 | 0.152 | 0.141 | 0.150 | 0.120 | 0.163 | 0.197 | 0.148 | 0.148 | 0.159 | 0.150 | |
The numbers of base substitutions per site from averaging over all sequence pairs between groups are shown. Standard error estimates are shown above the diagonal (in parentheses). Analyses were conducted by using the maximum composite likelihood model (38). The differences in the composition bias among sequences were considered in evolutionary comparisons (45). The analysis involved 590 nucleotide sequences. The numbers of sequences analyzed per group were as follows: 66 for genotype I, 102 for genotype II, 9 for genotype III, 6 for genotype IV; 19 for genotype IX, 45 for genotype V, 57 for genotype VI, 231 for genotype VII, 4 for genotype VIII, 18 for genotype X, 4 for genotype XI, 6 for genotype XII, 8 for genotype XIII, 6 for genotype XIV, 5 for genotype XV, and 4 for genotype XVI. Codon positions included were the first, second, third, and noncoding positions. All positions containing gaps and missing data were eliminated. There were a total of 1,639 positions in the final data set. Evolutionary analyses were conducted with MEGA5 (37).
The fusion protein sequences of these novel genotype XVI viruses were compared to that of the LaSota strain to analyze their similarity to a common vaccine strain often used to protect against virulent NDV infections in the field. The results showed that the ancestral viruses NDV-Mex452/47 and NDV-DR/28138-4/86 had fusion protein sequences less divergent from that of LaSota (9.9% and 13%, respectively) than those of the NDV-DR499-31/08 and NDV-DR/867-2/08 strains, which were 14.6% divergent (Table 3). More specifically, this analysis shows that these genotype XVI viruses have actually evolved away from the LaSota fusion sequence over time as well as away from the ancestral viruses. Similarly, high levels of divergence between NDV-DR499-31/08 and LaSota were also observed for the HN (15.3%), M (13.2%), NP (9.6%), P (7.6%), and L (7.6%) proteins (data not shown).
Table 3.
Comparison of the full fusion protein of genotype XVI with that of the LaSota vaccine strain
| Strain | % similarity or divergence compared to strain: |
||||
|---|---|---|---|---|---|
| LaSota | NDV-DR867-2/08 | NDV-DR499-31/08 | NDV-DR28138-4/86 | NDV-Mex452/47 | |
| LaSota | 86.8a | 86.8a | 88.1a | 90.8a | |
| NDV-DR867-2/08 | 14.6b | 100a | 95.3a | 92.8a | |
| NDV-DR499-31/08 | 14.6b | 0b | 95.3a | 92.8a | |
| NDV-DR28138-4/86 | 13b | 4.9b | 4.9b | 93.9a | |
| NDV-Mex452/47 | 9.9b | 7.6b | 7.6b | 6.4b | |
Percent similarity.
Percent divergence.
Infection of chickens with NDV-DR499-31/08 results in characteristic viscerotropic velogenic Newcastle disease.
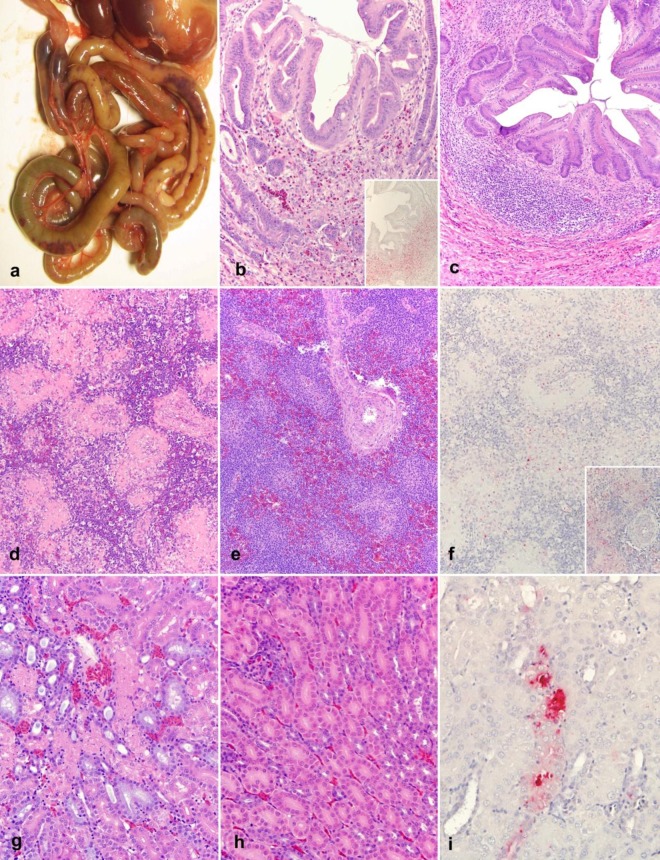
The clinicopathological characteristics and pathogenicity of NDV-DR499-31/08 were assessed by using 4-week-old SPF chickens. All birds were successfully infected with NDV-DR499-31/08, and clinical signs consisted of progressively worsening prostration, malaise, and mucous diarrhea, with all birds spontaneously dying or being euthanized by day 4 postinoculation (p.i.). Gross lesions were initially observed at day 2 p.i. and consisted of severe bilateral conjunctivitis, multifocal necrosis of the spleen, and multifocal necrosis and hemorrhages of the intestine (Fig. 3a) and cecal tonsils. Severe atrophy of the thymus was observed by day 3 p.i. and was associated with perithymic hemorrhages and edema. By day 4 p.i., infected birds had multiple ulcerations and necrosis of the proventricular mucosa. Control (PBS-treated) chickens did not exhibit any clinical symptoms or present any disease.
Fig 3.
Pathological changes and NDV nucleoprotein distribution in NDV-DR499-31/08-infected 4-week-old chickens. (a) Gross lesions of the intestine at 4 dpi. Multifocal areas of intestinal necrosis and hemorrhages are noticeable through the serosa. (b) Proventricular glands at 3 dpi (HE; magnification, ×20). In the submucosa, there is an extensive loss of lymphocytes, an accumulation of macrophages, scattered heterophils, and cellular debris. (Inset) Extensive immunolabeling (IHC; magnification, ×20) of the NDV nucleoprotein is observed in areas of necrosis in the same tissue as that shown panel b. (c) Proventricular glands of a bird experimentally inoculated with PBS (mock) at 3 dpi (HE; magnification, ×20). There was a normal accumulation of lymphocytes in the submucosa. (d) Spleen at 4 dpi (HE; magnification, ×20). Shown are diffuse lymphoid depletion and an accumulation of necrotic debris, fibrin, and macrophages surrounding the centrilobular arteries and within the splenic ellipsoid. (e) Spleen of a bird experimentally inoculated with PBS (mock) at 4 dpi (HE; magnification, ×20). The white pulp is characterized by a large number of lymphocytes surrounding the centrilobular arteries. (f) Spleen at 4 dpi. Scattered macrophages in the splenic ellipsoid are immunolabeled for the NDV nucleoprotein (IHC; magnification, ×20X). (Inset) Higher magnification (×40) of panel f showing immunolabeling between areas of necrosis and fibrin exudation. (g) Kidney at 4 dpi (HE; magnification, ×20). Shown are scattered tubules displaying a loss of tubular epithelial cells and an accumulation of cellular and karyorrhectic debris within the lumen. (h) Kidney of a bird experimentally inoculated with PBS (mock) at 4 dpi (HE; magnification, ×20). Shown is a normal arrangement of tubules. (i) Kidney at 4 dpi. Scattered tubular epithelial cells are immunolabeled for the NDV nucleoprotein (IHC; magnification, ×20).
Microscopic changes were observed for multiple organs, and the severity of lesions peaked at day 4 p.i. (end of the experiment). A summary of the severity and distribution of in these changes is reported Table 4. Overall, this virus targeted principally the eyelids (inoculation site); the mucosa-associated lymphoid tissues (MALT), such as the lymphoid patches surrounding the proventricular glands and the cecal tonsils (Fig. 3b); and the lymphoid organs, such as the thymus, bursa, and spleen (Fig. 3d). Lesions consisted of severe lymphoid depletion and necrosis that resulted in an accumulation of prominent macrophages, necrotic debris, and scattered heterophils. In the intestines, the destruction of the MALT resulted in necrosis and ulceration of the overlying epithelium. Other lesions consisted of multifocal necrosis of the tubular renal epithelium (Fig. 3g), multifocal necrosis of the lymphoid tissues surrounding the renal pelvis, and multifocal necrosis centered on the areas of extramedullary hematopoiesis situated in portal areas of the liver. In addition, infected animals showed multifocal areas of myocardial necrosis that were more prominent immediately beneath the epicardium and were characterized by multifocal myonecrosis with an accumulation of macrophages. In the respiratory tract, lesions involved the bronchial-associated lymphoid tissue (BALT), with the most severe lesions being observed in the laryngeal tonsils, characterized by necrosis of the submucosal lymphoid aggregates that resulted in multifocal epithelial ulceration and a loss of the overlying epithelium.
Table 4.
Clinicopathological findings for chickens infected with NDV-DR499-31/08
| Tissue and staining methoda | Result on day postinoculationb |
||
|---|---|---|---|
| 2 | 3 | 4 | |
| Eyelid | |||
| HE | + | + | ++ |
| IHC | ++ | +++ | ++++ |
| Spleen | |||
| HE | + | ++ | ++++ |
| IHC | + | ++ | +++ |
| Thymus | |||
| HE | − | ++ | +++ |
| IHC | + | +++ | ++++ |
| Bursa | |||
| HE | − | ++ | +++ |
| IHC | − | ++ | ++++ |
| Harderian gland | |||
| HE | − | − | − |
| IHC | − | − | + |
| Proventriculus | |||
| HE | + | ++ | ++ |
| IHC | − | ++++ | ++ |
| Pancreas | |||
| HE | + | ++ | |
| IHC | − | ++ | ++ |
| Small intestine | |||
| HE | − | + | + |
| IHC | − | + | ++ |
| Mekel's diverticulum | |||
| HE | + | − | +++ |
| IHC | + | − | ++ |
| Cecal tonsils | |||
| HE | + | ++ | +++ |
| IHC | ++ | ++ | + |
| Large intestine | |||
| HE | − | + | + |
| IHC | + | + | ++ |
| Air sac | |||
| HE | − | − | − |
| IHC | − | − | − |
| Trachea | |||
| HE | − | − | − |
| IHC | − | − | + |
| Lung | |||
| HE | − | + | + |
| IHC | − | + | + |
| Heart | |||
| HE | − | + | ++ |
| IHC | − | − | ++ |
| Esophagus | |||
| HE | − | − | − |
| IHC | − | + | + |
| Tongue | |||
| HE | − | − | − |
| IHC | − | − | − |
| Pharynx | |||
| HE | + | − | + |
| IHC | − | + | +++ |
| Crop | |||
| HE | − | − | − |
| IHC | − | − | ++ |
| Brain | |||
| HE | − | − | +/− |
| IHC | − | − | − |
| Liver | |||
| HE | − | − | + |
| IHC | − | − | ++ |
| Kidney | |||
| HE | − | + | + |
| IHC | − | + | ++ |
| Comb | |||
| HE | − | − | − |
| IHC | − | − | − |
| Femur | |||
| HE | − | + | ++ |
| IHC | − | − | ++ |
| Turbinates | |||
| HE | − | − | − |
| IHC | − | − | − |
HE, hematoxylin and eosin staining; IHC, immunohistochemistry staining for the NDV nucleoprotein.
For HE staining, the results are scored by tissue as follows: (i) for the spleen, + is moderate hyperplasia, ++ is mild (<20%) lymphocytic depletion, +++ is moderate (20 to 50%) lymphocyte depletion with macrophage accumulation and multifocal necrosis, and ++++ is severe (>50%) lymphocytic depletion, macrophage accumulation, and necrosis; (ii) for the thymus, cecal tonsil, gut-associated lymphoid tissue, and bursa, + is mild (<20%) lymphocytic depletion, ++ is moderate (20 to 50%) lymphocytic depletion with necrosis and macrophage accumulation, and +++ is severe (>50%) lymphocytic depletion, macrophage accumulation, and necrosis; (iii) for bone marrow, + is mild (<20%) bone marrow necrosis, ++ is moderate (20 to 50%) bone marrow necrosis, and +++ is severe (>50%) bone marrow necrosis; (iv) for the pancreas, + is mild (<3 areas) vacuolation and degeneration and ++ is moderate (>3 areas) vacuolation and degeneration; (v) for the brain, + is vascular reactivity, ++ is vascular reactivity and perivascular cuffing, and +++ is vascular reactivity, perivascular cuffing, and gliosis; and (vi) for other tissues, + is mild (<20%) necrosis, ++ is moderate (20 to 50%) necrosis, and +++ is severe (>50%) necrosis. For IHC staining, the results are scored as follows: −, no IHC signal present; +/−, very rare positive-signal cells observed; +, rare cells in the section are positive upon IHC staining; ++, positive-signal cells seen in <50% of all high-power fields; +++, positive-signal cells seen in 50 to 75% of high-power fields; ++++, abundant positive-signal cells in more than 75% of high-power fields.
In infected cells, immunolabeling by IHC for the NDV nucleoprotein showed intracytoplasmic staining that was finely to coarsely granular; however, extracellular signal (fine granular staining) was occasionally observed in necrotic areas. There was a widespread distribution of nucleoprotein staining observed, showing 20 positive tissues out of 25 analyzed. The strongest signal was found at day 4 p.i., when all the remaining birds died spontaneously or were euthanized in extremis. Organs with the strongest signal were the eyelids, the lymphoid organs, and the MALT in multiple tissues (Fig. 3b, inset). In the spleen, immunoreactivity was confined to the fixed macrophage-dependent areas around the penicillary arteries (Fig. 3f), while in the thymus and bursa, positive cells consisted mainly of lymphocytes and macrophages. In the respiratory system, the positive signal was confined to the pharyngeal tonsils and with scattered lymphoid aggregates closely associated with the secondary and tertiary bronchi (BALT). In the digestive tract, intense positivity for NDV was observed only within the submucosal lymphoid aggregates, with no staining being observed in the epithelial lining. Intense immunolabeling was observed in the tubular epithelial cells of the kidney and in scattered interstitial cells, often associated with areas of tubular necrosis (Fig. 3i). In the liver, immunolabeling was observed in the necrotic areas involving the areas of extramedullary hematopoiesis and in scattered Kupffer cells. Multifocal immunolabeling was also observed in the keratinocytes within the esophageal mucosa, associated mostly with areas of attenuation/erosion. In the heart, areas of positive signal were present in the epicardium. In the pancreas, there was intense staining in areas of cellular degeneration and necrosis.
DISCUSSION
Numerous outbreaks of ND throughout Central and South America are consistently reported to the World Organization for Animal Health (13), as ND is endemic to many of the countries located in these regions (http://web.oie.int/wahis/public.php?page=control&disease_type=Terrestrial&disease_id=16&selected_start_year=2011&selected_report_period=1). The isolate characterized here, NDV-DR499-31/08, was initially identified in apparently healthy chickens, whereas the NDV-DR867-2/08 isolate was isolated from chickens displaying clinical signs and an increase in the mortality rate (∼3%). The aim of this paper was to further characterize NDV-DR499-31/08 by full genomic sequencing, standard pathotyping, and detailed clinicopathological assessments of 4-week-old chickens. The ICPI score and the sequencing of the F protein cleavage site determined this isolate to be virulent and therefore notifiable (13). Clinicopathological characterization demonstrated that NDV-DR499-31/08 had the typical phenotype of a viscerotropic velogenic strain, similar to other virulent NDV strains currently circulating throughout the world (35, 36). Because the source of the outbreak remains unknown, further surveillance studies sampling local wild-bird species and healthy, in addition to clinically ill, poultry flocks throughout the region are highly recommended.
The NDV-DR499-31/08 isolate analyzed in the current study has a genome size of 15,192 nt, classifying it among the more evolutionarily recent strains of NDV. Phylogenetic analyses performed in this report identified it to be isolated within its own branch compared with 103 complete NDV reference genomes. Similarly, when the evolutionary distances were compared, none of the class II NDV genotypes were significantly related to NDV-DR499-31/08. These data suggest this isolate to be unique among the currently known and sequenced strains of NDV. Furthermore, other viruses isolated within the surrounding regions that were never previously sequenced were analyzed to determine if they were related. Phylogenetic analysis of the full fusion gene localized these viruses together in a single clade (bootstrap value, 100%). Recently, Diel et al. (25) reassessed the criteria for NDV genotype assignment, proposing that new genotypes (i) must be assigned based on a phylogenetic tree constructed by using the maximum likelihood method and the optimum nucleotide model with a bootstrap value greater than 60%, (ii) must have a mean evolutionary distance per site greater than 10% (maximum composite likelihood model), and (iii) must encompass at least four distinct isolates. The data presented in this paper fulfill these requirements and suggest that NDV-DR499-31/08, together with NDV-DR867-2/08, NDV-DR28138-4/86, and NDV-Mex452/47, belongs to a new genotype, referred to here as genotype XVI. Moreover, these data suggest that a virulent NDV isolate has been circulating and evolving throughout the Caribbean and surrounding regions unnoticed for over 2 decades and imply the possibility of an unknown reservoir for virulent NDV. It was previously reported that healthy white storks can harbor virulent NDV (genotype VII) (42), and isolates of genotype VII have been shown to cause severe clinical signs and increased mortality rates in this same species (43), thus implying the possibility of apparently healthy wild birds harboring the virus and then infecting other susceptible avian populations.
Clinicopathological assessments of NDV-DR499-31/08 determined it to be a classical viscerotropic velogenic strain of NDV. A similar tissue tropism has also been observed for other virulent strains (35, 36). Interestingly, NDV-DR499-31/08 showed marked tropism for the tubular epithelial cells of the kidney, often in association with tubular necrosis. The presence of NDV in the tubules of the kidney might be implicated in the transmission of the virus by shedding. For instance, the kidneys of double-crested cormorants have been shown to possess high titers of NDV, as observed by virus isolation (44).
Overall, the complete genome and evolutionary distance analyses of NDV-DR499-31/08 suggest that it is genetically distinct and, together with NDV-DR867-2/08, NDV-DR28138-4/86, and NDV-Mex452/47, belongs to a new genotype (genotype XVI) among class II NDV strains. Furthermore, epidemiological surveillance from the surrounding regions is required to determine whether this isolate is still circulating within wild-bird species and/or commercial poultry in an attempt to determine the currently unknown reservoir of this virus. Due to the extreme economic impact of virulent NDV outbreaks, extensively rigorous surveillance strategies need to be employed. Lastly, the analysis of the individual NDV-DR499-31/08 protein sequences here revealed it to be highly dissimilar to the commonly used LaSota vaccine strain, thereby suggesting that current vaccination programs may not protect from future outbreaks and further implying the necessity for improved control and diagnostic measures to prevent virulent NDV outbreaks from occurring.
ACKNOWLEDGMENTS
We thank Dawn Williams-Coplin and Tim Olivier for technical assistance, Roger Brock for help with animal experiments, and SEPRL sequencing facility personnel for nucleotide sequencing. We also acknowledge the generous support of the NVSL and the Dominican Republic Ministry of Agriculture for their invaluable collaboration and for sharing information on the outbreak and these viruses.
This work was supported by USDA funding (CRIS 6612-32000-064-00D).
Footnotes
Published ahead of print 28 November 2012
REFERENCES
- 1. Alexander D. 1988. Newcastle disease: methods of spread. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 2. Alexander D, Senne D. 2008. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, 12th ed Blackwell Publishing, Ames, IA [Google Scholar]
- 3. Alexander D, Senne D. 2008. Newcastle disease and other avian paramyxoviruses, p 135–141 In Dufour-Zavala L, Glisson JR, Jackwood MW, Pearson JE, Reed WM, Woolcock PR. (ed), A laboratory manual for the isolation, identification and characterization of avian pathogens, 4th ed American Association of Avian Pathologists, Athens, GA [Google Scholar]
- 4. Cattoli G, Susta L, Terregino C, Brown C. 2011. Newcastle disease: a review of field recognition and current methods of laboratory detection. J. Vet. Diagn. Invest. 23:637–656 [DOI] [PubMed] [Google Scholar]
- 5. Kim SH, Xiao S, Shive H, Collins PL, Samal SK. 2012. Replication, neurotropism, and pathogenicity of avian paramyxovirus serotypes 1-9 in chickens and ducks. PLoS One 7:e34927 doi:10.1371/journal.pone.0034927 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Pedersen JC, Senne DA, Woolcock PR, Kinde H, King DJ, Wise MG, Panigrahy B, Seal BS. 2004. Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002-2003 outbreak in California and other recent outbreaks in North America. J. Clin. Microbiol. 42:2329–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamb R, Kolakofsky CPLD, Melero JA, Nagai Y, Oldstone MBA, Pringle CR, Rima BK. 2005. The negative sense single stranded RNA viruses, p 607–738 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 8. Susta L, Miller PJ, Afonso CL, Estevez C, Yu Q, Zhang J, Brown CC. 2010. Pathogenicity evaluation of different Newcastle disease virus chimeras in 4-week-old chickens. Trop. Anim. Health Prod. 42:1785–1795 [DOI] [PubMed] [Google Scholar]
- 9. Collins PL, Hightower LE, Ball LA. 1980. Transcriptional map for Newcastle disease virus. J. Virol. 35:682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seal BS, King DJ, Sellers HS. 2000. The avian response to Newcastle disease virus. Dev. Comp. Immunol. 24:257–268 [DOI] [PubMed] [Google Scholar]
- 11. Calain P, Roux L. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peeters BP, Gruijthuijsen YK, de Leeuw OS, Gielkens AL. 2000. Genome replication of Newcastle disease virus: involvement of the rule-of-six. Arch. Virol. 145:1829–1845 [DOI] [PubMed] [Google Scholar]
- 13. World Organization for Animal Health 2012. Manual of diagnostic tests and vaccines for terrestrial animals. World Organization for Animal Health, Paris, France [Google Scholar]
- 14. Aldous EW, Mynn JK, Banks J, Alexander DJ. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239–256 [DOI] [PubMed] [Google Scholar]
- 15. Czegledi A, Ujvari D, Somogyi E, Wehmann E, Werner O, Lomniczi B. 2006. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 120:36–48 [DOI] [PubMed] [Google Scholar]
- 16. Miller PJ, Decanini EL, Afonso CL. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 10:26–35 [DOI] [PubMed] [Google Scholar]
- 17. Miller PJ, Kim LM, Ip HS, Afonso CL. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391:64–72 [DOI] [PubMed] [Google Scholar]
- 18. Wang Z, Liu H, Xu J, Bao J, Zheng D, Sun C, Wei R, Song C, Chen J. 2006. Genotyping of Newcastle disease viruses isolated from 2002 to 2004 in China. Ann. N. Y. Acad. Sci. 1081:228–239 [DOI] [PubMed] [Google Scholar]
- 19. Kim LM, King DJ, Curry PE, Suarez DL, Swayne DE, Stallknecht DE, Slemons RD, Pedersen JC, Senne DA, Winker K, Afonso CL. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81:12641–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Y, Wan HQ, Liu HQ, Wu YT, Liu XF. 2004. Genomic sequence of an isolate of Newcastle disease virus isolated from an outbreak in geese: a novel six nucleotide insertion in the non-coding region of the nucleoprotein gene. Brief report. Arch. Virol. 149:1445–1457 [DOI] [PubMed] [Google Scholar]
- 21. Ababneh MM, Dalab AE, Alsaad SR, Al-Zghoul MB, Al-Natour MQ. 2012. Molecular characterization of a recent Newcastle disease virus outbreak in Jordan. Res. Vet. Sci. 93:1512–1514 [DOI] [PubMed] [Google Scholar]
- 22. Solomon P, Abolnik C, Joannis TM, Bisschop S. 2012. Virulent Newcastle disease virus in Nigeria: identification of a new clade of sub-lineage 5f from livebird markets. Virus Genes 44:98–103 [DOI] [PubMed] [Google Scholar]
- 23. Tan SW, Ideris A, Omar AR, Yusoff K, Hair-Bejo M. 2010. Sequence and phylogenetic analysis of Newcastle disease virus genotypes isolated in Malaysia between 2004 and 2005. Arch. Virol. 155:63–70 [DOI] [PubMed] [Google Scholar]
- 24. Zhang S, Zhao L, Wang X, Zhang D, Zhao J, Zhang G. 2011. Serologic and virologic survey for evidence of infection with velogenic Newcastle disease virus in Chinese duck farms. Avian Dis. 55:476–479 [DOI] [PubMed] [Google Scholar]
- 25. Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL. 2012. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 12:1770–1779 [DOI] [PubMed] [Google Scholar]
- 26. Czegledi A, Herczeg J, Hadjiev G, Doumanova L, Wehmann E, Lomniczi B. 2002. The occurrence of five major Newcastle disease virus genotypes (II, IV, V, VI and VIIb) in Bulgaria between 1959 and 1996. Epidemiol. Infect. 129:679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herczeg J, Wehmann E, Bragg RR, Travassos Dias PM, Hadjiev G, Werner O, Lomniczi B. 1999. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch. Virol. 144:2087–2099 [DOI] [PubMed] [Google Scholar]
- 28. Ujvari D, Wehmann E, Kaleta EF, Werner O, Savic V, Nagy E, Czifra G, Lomniczi B. 2003. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. 96:63–73 [DOI] [PubMed] [Google Scholar]
- 29. Wehmann E, Czegledi A, Werner O, Kaleta EF, Lomniczi B. 2003. Occurrence of genotypes IV, V, VI and VIIa in Newcastle disease outbreaks in Germany between 1939 and 1995. Avian Pathol. 32:157–163 [DOI] [PubMed] [Google Scholar]
- 30. Cho SH, Kwon HJ, Kim TE, Kim JH, Yoo HS, Kim SJ. 2008. Variation of a Newcastle disease virus hemagglutinin-neuraminidase linear epitope. J. Clin. Microbiol. 46:1541–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dortmans JC, Peeters BP, Koch G. 2012. Newcastle disease virus outbreaks: vaccine mismatch or inadequate application? Vet. Microbiol. 160:17–22 [DOI] [PubMed] [Google Scholar]
- 32. Xiao S, Paldurai A, Nayak B, Samuel A, Bharoto EE, Prajitno TY, Collins PL, Samal SK. 2012. Complete genome sequences of Newcastle disease virus strains circulating in chicken populations of Indonesia. J. Virol. 86:5969–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wan H, Chen L, Wu L, Liu X. 2004. Newcastle disease in geese: natural occurrence and experimental infection. Avian Pathol. 33:216–221 [DOI] [PubMed] [Google Scholar]
- 34. Wakamatsu N, King DJ, Kapczynski DR, Seal BS, Brown CC. 2006. Experimental pathogenesis for chickens, turkeys, and pigeons of exotic Newcastle disease virus from an outbreak in California during 2002-2003. Vet. Pathol. 43:925–933 [DOI] [PubMed] [Google Scholar]
- 35. Diel DG, Susta L, Cardenas Garcia S, Killian ML, Brown CC, Miller PJ, Afonso CL. 2012. Complete genome and clinicopathological characterization of a virulent Newcastle disease virus isolate from South America. J. Clin. Microbiol. 50:378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Susta L, Miller PJ, Afonso CL, Brown CC. 2011. Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Vet. Pathol. 48:349–360 [DOI] [PubMed] [Google Scholar]
- 37. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Courtney SC, Gomez D, Susta L, Hines N, Pedersen JC, Miller PJ, Afonso CL. 2012. Complete genome sequencing of a novel Newcastle disease virus isolate circulating in layer chickens in the Dominican Republic. J. Virol. 86:9550 doi:10.1128/JVI.01491-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY [Google Scholar]
- 41. Martin DP. 2009. Recombination detection and analysis using RDP3. Methods Mol. Biol. 537:185–205 [DOI] [PubMed] [Google Scholar]
- 42. Kaleta EF, Kummerfeld N. 2012. Isolation of herpesvirus and Newcastle disease virus from white storks (Ciconia ciconia) maintained at four rehabilitation centres in northern Germany during 1983 to 2001 and failure to detect antibodies against avian influenza A viruses of subtypes H5 and H7 in these birds. Avian Pathol. 41:383–389 [DOI] [PubMed] [Google Scholar]
- 43. Kaleta EF, Kummerfeld N. 1983. Herpesviruses and Newcastle disease viruses in white storks (Ciconia ciconia). Avian Pathol. 12:347–352 [DOI] [PubMed] [Google Scholar]
- 44. Kuiken T, Wobeser G, Leighton FA, Haines DM, Chelack B, Bogdan J, Hassard L, Heckert RA, Riva J. 1999. Pathology of Newcastle disease in double-crested cormorants from Saskatchewan, with comparison of diagnostic methods. J. Wildl. Dis. 35:8–23 [DOI] [PubMed] [Google Scholar]
- 45. Tamura K, Kumar S. 2002. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 19:1727–1736 [DOI] [PubMed] [Google Scholar]