Abstract
Experimental techniques for the investigation of three-dimensional (3D) genome organization are being developed at a fast pace. Currently, the associated computational methods are mostly specific to the individual experimental approach. Here we present a general statistical framework that is widely applicable to the analysis of genomic contact maps, irrespective of the data acquisition and normalization processes. Within this framework DNA–DNA contact data are represented as a complex network, for which a broad number of directly applicable methods already exist. In such a network representation, DNA segments and contacts between them are denoted as nodes and edges, respectively. Furthermore, we present a robust method for generating randomized contact networks that explicitly take into account the inherent 3D nature of the genome and serve as realistic null-models for unbiased statistical analyses. By integrating a variety of large-scale genome-wide datasets we demonstrate that meiotic crossover sites display enriched genomic contacts and that cohesin-bound genes are significantly colocalized in the yeast nucleus. We anticipate that the complex network framework in conjunction with the randomization of DNA–DNA contact networks will become a widely used tool in the study of nuclear architecture.
INTRODUCTION
The development of chromatin conformation capture (3C) provided us with the first in situ method for studying the three-dimensional (3D) organization of DNA at genomic loci of interest (1). Since its invention, interest in the application and improvement of the 3C approach has been ever increasing. New derivative high-resolution, high-throughput methods that enable us to study DNA–DNA contacts at a much larger scale are rapidly being developed (2). The number of available datasets employing the 4C (3,4), 5C (5), GCC (6) and particularly the Hi-C (7,8) and TCC (9) approaches is steadily on the rise. In the past 2 years alone, researchers have obtained information on the global 3D organization of DNA in human lymphoblastoid cells (8–10), the budding yeast (11,6) and fission yeast (12) nuclei, Drosophila embryonic cells (13), and, most recently, Arabidopsis thaliana (14). These studies provide us with maps of DNA–DNA contacts, and accumulate evidence on both a local and a genome-wide scale that 3D genome architecture is highly non-random.
Typically, the analysis of DNA–DNA contact data is intrinsically linked to the individual experimental approach of each study, as is the case for the Hi-C-specific probabilistic background model of Yaffe and Tanay (15). This powerful approach allows us to minimize experimental and computational biases, and to decide whether the observed contacts could have arisen from chance. Since their model is so tightly linked to Hi-C data, however, it is not easily transferrable to non-Hi-C-based approaches. To accommodate and to be able to integrate data from diverse existing and upcoming experimental approaches there is a need for a unifying framework that can be used to study DNA–DNA contact data in general, irrespective of the underlying data acquisition and normalization processes. Within this framework each dataset would undergo (i) an experimental approach dependent pre-processing, in which false-positive contacts are filtered; and (ii) experimental approach independent representation and analysis, which is based on the inherent 3D nature of the data (Figure 1).
Figure 1.
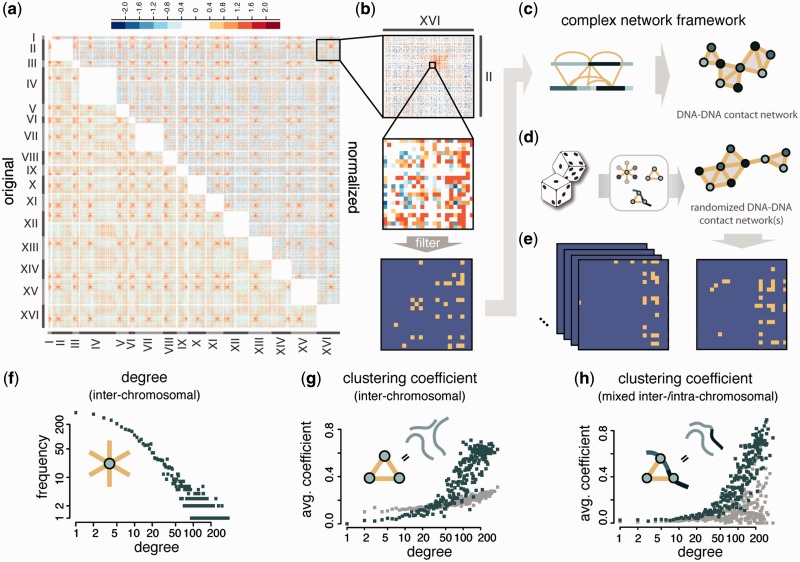
Data normalization and filtering, complex network framework and topological network properties (a) Log2 contact enrichment of yeast inter-chromosomal DNA–DNA contact data (11) before and after normalization using the probabilistic model of Yaffe and Tanay (15). (b) Filtering of contacts. (c) The high-confidence map of filtered DNA–DNA contacts is represented as a complex network. (d) Randomized networks that maintain essential properties of the original. (e) A large number of random networks is generated using the procedure suggested in this study. (f–h) Topological analysis of the yeast inter-chromosomal segment contact network (SCN). Green denotes yeast SCN nodes, grey denotes random networks of the same size and degree sequence as the yeast SCN (‘rewired networks’, not to be confused with the randomization approach). (f) Degree distribution. (g) Average clustering coefficient distribution. (h) Average clustering coefficient distribution for mixed triangles (where two nodes are neighbours on the same chromosome).
Here we adapt the complex network framework, which has proven successful for many types of genomic data in systems biology (16–19) and offers a large number of existing analysis methods (19–22), to the study of DNA–DNA contact data. These methods can be used to study a number of important biological questions regarding the 3D arrangement of DNA in a network context: for instance, is it possible to identify enriched patterns such as cliques or motifs in these networks (23–25), and what is their biological relevance? Or can we identify subnetworks with a specific local functional enrichment?
We demonstrate the framework on a recently published Hi-C dataset for Saccharomyces cerevisiae (11) by investigating the following questions: (i) How can we apply existing methods for Hi-C data normalization to construct a high-confidence network of DNA–DNA contacts?; (ii) Which topological properties characterize the resulting network of contacts?; (iii) Can we design a network randomization procedure to obtain an unbiased ‘null-model’ network that mimics the 3D behaviour of the underlying data?; and (iv) How can the null-model network be used in statistical enrichment tests, and how does the method compare to existing approaches? Specifically, we choose the assessment of contact enrichment between genomic loci as one particular example of application for our approach: we exploit several a large-scale datasets, including 136 reported meiotic recombination hotspots (26) and the experimentally verified binding sites of over 200 chromatin regulatory proteins (27), to investigate colocalization between the corresponding genomic loci.
MATERIALS AND METHODS
Saccharomyces cerevisiae inter-chromosomal DNA SCN
To construct the yeast inter-chromosomal DNA segment-contact network (SCN), we first generated a filtered list of DNA–DNA contacts from the inter-chromosomal HindIII libraries of a recently published Hi-C experiment in S. cerevisiae (11). We use a q-value-based method described in the Supplementary Material of Duan et al. (11) to filter unspecific contacts, with the important difference that we do not assume uniform contact probabilities of all DNA segments. Instead, we applied the procedure described in Yaffe and Tanay (15) to obtain contact probabilities for each fragment-pair that take into account known biases of Hi-C experiments (see Supplementary Materials and Methods).
S. cerevisiae inter-chromosomal GCN
We obtained coordinates of open reading frames (ORFs) and transposable-element genes from the SGD (28). These genes form the set of nodes in the yeast gene–gene contact network (GCN). A gene is assigned to the DNA segment that contains the largest part of its ORF, ensuring a unique segment assignment for each gene. Genes inherit the contacts of their ‘parent’ segments, i.e. every gene–gene combination of an interacting segment pair will be a new edge in the GCN.
Random contact data, artificial SCNs and GCNs
We generated 50 sets of random contact data in similar fashion to Witten and Noble (29): 500 nodes were randomly distributed uniformly in a unit cube. Euclidean distances between all pairs of nodes were measured and edges drawn between nodes with a distance among the smallest 2% of all distances, leading to an average node degree of 10. For each of these 50 artificial SCNs 1000 corresponding random networks were generated according to the protocol described below (Network randomization procedure) and in Figure 5. Networks are intentionally kept small to decrease to computational workload of randomization. For simulations of larger networks please refer to the Supplementary Materials and Methods, and Control Calculations. From these 50 artificial SCNs we generated corresponding artificial GCNs by randomly assigning 1–3 ‘genes’ to each node and splitting nodes into their constituent genes as described above.
Figure 5.

Unbiased randomization procedure for DNA–DNA contact networks. Our randomization approach for DNA–DNA contact networks aims at maintaining the defining network properties of the original network, thereby creating an appropriate ‘null-model’ for comparison. The toy example illustrates the effects of each randomization step at the network and the DNA level (in a 2D representation). Step 1: This ‘rewiring’ part of the randomization procedure shuffles the contacts between DNA segments by selecting pairs of edges and swapping their targets. While this maintains the exact degrees of every node, the number of triangles is significantly reduced, essentially disrupting the strong clustering behaviour. Thus, no compact representation exists for the rewired network. Step 2: To correct the long-range clustering behaviour, triangles are introduced into the rewired network by an established Markov-chain procedure (31), until the transitivity T (the ratio of observed triangles to possible triangles in the network) of the random network matches that of the original network Torig. Step 3: Since the number of ‘mixed triangles’ (where two of the participating nodes are not connected by an edge, but are neighbours on the same chromosome) is also decreased in the rewired networks, this step increases the corresponding mixed transitivity T′ until it matches the original mixed transitivity T′orig in the same fashion as Step 2. Step 3 is optional, especially in colocalization assessment, because it will only have an effect if genes or DNA segments of interest are actually chromosomal neighbours. In Steps 2 and 3, the overall clustering behaviour is restored, however, individual nodes are allowed to vary in their clustering behaviour. See Supplementary Figure S9 for comparison of rewired and fully randomized networks.
Clustering coefficient
Here we use the definition of clustering coefficient of a node provided by Soffer and Vasquez (30). The Soffer–Vasquez clustering coefficient explicitly considers joint degree constraints, i.e. it directly takes i account the degree of neighbouring nodes when calculating the clustering coefficient of individual nodes: it calculates the clustering coefficient of a node as the number of triangles formed with its neighbours divided by the maximum possible number of triangles given the neighbours degree sequence. This is important in the network randomization procedure (see below), as global transitivities might otherwise not be attainable with traditional clustering measures in rewired networks due to degree constraints (31). We formulate the clustering coefficient for ‘mixed triangles’, which we define as a triplet of nodes s, t, u where u and s are direct ‘chromosomal neighbours’, i.e. they lie within c base-pairs (here c = 2500 bp) of each other on the same chromosome, and the edges (u, t) and (s, t) exist. For details see Supplementary Materials and Methods.
Network randomization procedure
We designed a custom randomization procedure for DNA–DNA contact networks, which is based on work by Bansal et al. (31) and explained in Figure 5. For the exact details of the algorithm, please refer to the Supplementary Materials and Methods.
Randomization approach for colocalization assessment
The basics of the randomization approach for colocalization assessment are explained schematically in Figure 2, more detail can be found in the Supplementary Materials and Methods.
Figure 2.
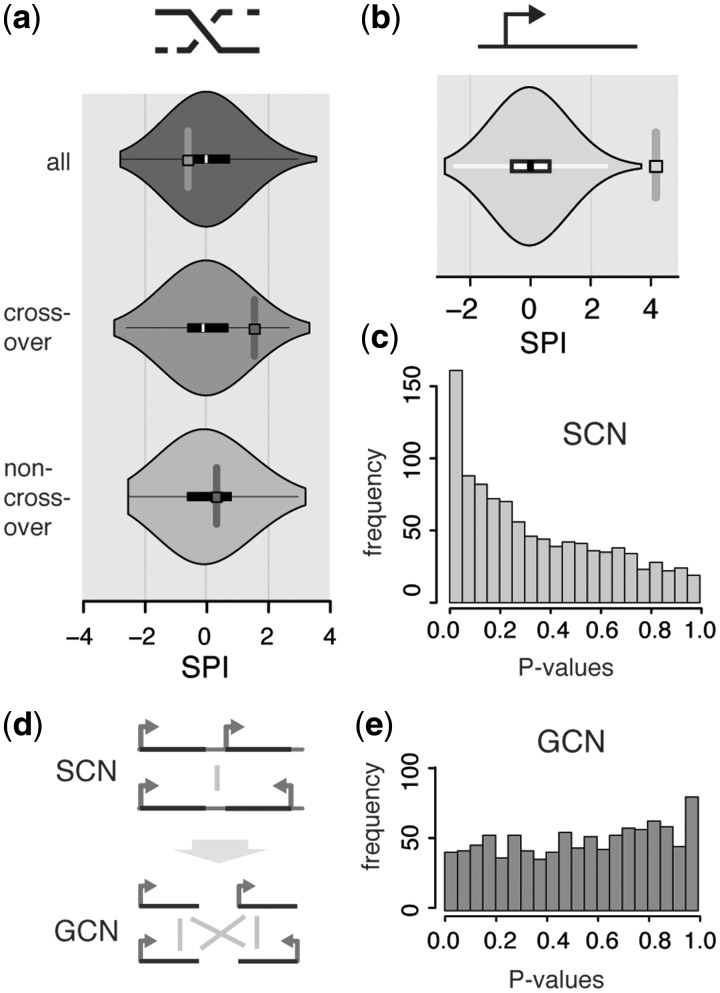
Colocalization assessment in DNA–DNA contact networks. (a) Colocalization assessment using our randomization approach, in direct comparison to the resampling approach (Supplementary Materials and Methods). Green nodes are the nodes investigated for colocalization. Rectangular nodes correspond to the nodes investigated in each of the random null-models. (b) Colocalization assessment tests for 250 random sets of 100 nodes in each of 50 artificial SCNs (Materials and Methods section). Grey (background): rewired random networks (no correction of clustering), excess of low P-values (arrow). Green (foreground): randomization approach. (c, d) Boxplots of node degrees in the above-mentioned sets binned by colocalization P-value for the resampling approach (light-grey) and the randomization approach (green), respectively. P-values obtained by Wilcoxon rank-sum test. Outliers have been omitted for clarity. (e) Boxplots showing that the degree of nodes in the SCN is dependent on the corresponding segment-lengths. Segments have been binned according to their degree into four categories, where 0 is unconnected, while low, med and high are formed by the first, second + third and fourth quartile of nodes, respectively. P-value calculated using the Wilcoxon rank-sum test (f) Schematic of colocalized nodes with low versus high degrees.
Randomly sampled node sets
Segment and gene sets were generated by sampling from the respective networks uniformly at random without replacement. For the yeast SCN, we generated 1000 sets of nodes consisting of 200 segments each; for the yeast GCN we generated 1000 sets of nodes consisting of 250 genes each; for each of the artificial SCNs we generated node sets of varying sizes (250 × 100 nodes, 250 × 150 nodes).
Meiotic recombination hotspot dataset
We obtained a dataset of 136 meiotic recombination hotspots and categorized subsets of hotspots into crossover (CO) and non-crossover (NCO) sites as described by the authors. Hotspots were mapped to DNA segments by their midpoint.
Gene and chromatin regulatory protein binding dataset
We obtained microarray data of gene and chromatin regulatory protein binding to genes in the budding yeast genome from Venters et al. (27). The data for each of the 202 proteins are divided into three groups, based on the location of the microarray probe: upstream activating sequence (UAS), transcription start site (TSS) and ORF. For each protein and group we construct a gene set that is bound by the protein at the corresponding position (UAS, TSS, ORF): binding is defined according to the false-discovery rate (FDR) cutoff provided by the authors. Note that data are not available for every protein at every microarray probe location, so that the size of each dataset for TSS, UAS and ORF can vary.
Correction for multiple testing
Due to the size of the dataset and the lower limit of obtainable P-values (here 10−3), a strict multiple testing correction (such as the Bonferroni correction) might obscure some of the truly colocalized gene sets. Although the randomization process can be slow for large networks, the actual enrichment test is comparatively fast once the random networks have been obtained. Thus, the method lends itself well to a permutation-based P-value correction method (32). To obtain the permutation-based P-value, the chromatin regulatory protein binding data are randomly shuffled among genes. The adjusted P-value is then the fraction of permuted datasets where the maximum SPI is larger than the original SPI of the investigated protein. A similar approach is very popular, for example, in eQTL studies (33). Also see Supplementary Materials and Methods.
RESULTS
DNA–DNA contact data lend itself well to the representation as a complex network, where DNA loci form the network nodes, and 3D contacts between them are represented as edges (34,35). As an example, we focus in this work on the inter-chromosomal DNA–DNA contacts in S. cerevisiae, whose compact, haploid genome allows for an unambiguous mapping of most Hi-C reads to each chromosome, and which has a large number of comprehensive, genome-wide datasets available, which can be studied in the context of DNA–DNA contacts. We only study inter-chromosomal contact data in S. cerevisiae, because the dependency of the intra-chromosomal contact probability of two DNA loci is hard to control for (11,13,15). However, most of the introduced methodology can be applied to intra-chromosomal contacts with minor adaptations.
Building a DNA SCN that accounts for experimental biases
The first part of the analysis of 3D genome architecture in a complex network framework is formed by the experimental approach dependent data acquisition and filtering to obtain a high-confidence list of DNA–DNA contacts. In the special case of Hi-C sequencing data (11) the current standard for data normalization is provided by Yaffe and Tanay (15). Their probabilistic method explicitly accounts for specific experimental and computational biases affecting contact probabilities between DNA segments, specifically GC content, length and computational mappability. Unspecific contacts are filtered according to both their probability of occurrence and frequency of observation (Figure 1b and Materials and Methods section, details in Supplementary Materials and Methods, Supplementary Table ST1 and Supplementary Figure S1). Filtering strictness has been chosen to maximize the network’s information content while minimizing the influence of random contacts (Supplementary Control Calculations, Supplementary Table T2 and Supplementary Figure S2). The filtered contact list is then converted into a DNA SCN, consisting of 44 720 edges (contacts) between 4454 nodes (DNA segments) (Figure 1c).
Inter-chromosomal SCNs are defined by their connectivity and clustering behaviour
To distinguish contacts in SCNs that have arisen by chance from biologically significant associations it is necessary to design a robust null-model that can be used in unbiased statistical comparisons. To this end it is important to understand the inherent structure of the original network. In other words, what are the topological properties that describe and define the yeast inter-chromosomal SCN?
The connectivity (degree) of nodes in the network ranges from unconnected to several hundred contacts, as evident from the network's degree distribution (Figure 1f). The centromeric regions in yeast remain clustered during interphase. DNA segments that lie within this crowded environment have a higher likelihood of making contacts and, consequently, tend to be among the segments with the highest degree.
Perhaps the most important characteristic of DNA–DNA contact networks is their inherently strong clustering behaviour. Edges in SCNs describe dynamic spatial relationships, which we expect to see reflected in the general structure of the network. As an example consider three DNA segments X, Y and Z. If X is spatially close to both Y and Z it is very likely that Y and Z are also in close proximity. The corresponding nodes in the SCN form a triangle (schematic in Figure 1g). The increased likelihood of contacts in the centromeric regions discussed above also leads to a higher occurrence of triangles in that region of space. Consequently, the average clustering coefficient increases for segments with a high degree in the yeast SCN (Figure 1g and Materials and Methods section). The same 3D relationship holds for DNA segments that do not share an edge between them in the SCN, but lie within a short linear distance of each other on the same chromosome. Therefore, in ‘mixed triangles’, where two of the participating DNA segments are ‘chromosomal neighbours’ (schematic in Figure 1h), we can observe the same clustering trend (Figure 1h and Materials and Methods section).
Custom randomization procedure for DNA–DNA contact networks maintains important 3D network properties
Working with the right null-model is crucial for making biologically relevant interpretations of large amounts of data. A tried and tested approach to obtain a suitable null-model for statistical comparisons is that of network randomization, in which contacts between DNA segments are randomly reassigned (36). The challenge for this type of approach lies in reproducing the original network’s properties in the random network to avoid the introduction of artificial biases (24,37,38).
Using the knowledge on topological properties of the yeast SCN gained in the previous section, we can design a null-model network that reproduces the original network’s wide range of segment connectivities and inherently strong clustering behaviour. The most common randomization method, often called rewiring, is insufficient for DNA–DNA contact networks, as it disrupts the network’s global clustering behaviour (Figure 5). We suggest a tailored randomization approach, which addresses this issue by first rewiring contacts in the network (Step 1), and then, unlike previous approaches, performing additional steps to correct the random network’s long-range clustering behaviour (Step 2) and [optionally] increase the network's short-range clustering behaviour (including chromosomal neighbours; Step 3). This ensures well-shuffled random networks, which maintain both the degree distribution and the clustering behaviour of the original network (Figure 5 and Supplementary Materials and Methods).
Randomized SCNs can be used to assess DNA segment colocalization
We focus on one particular application for the generated null-model networks: the assessment of colocalization between genomic loci. In other words, which genomic features display a contact enrichment in the 3D genome? Currently, there are three main approaches used to assess contact enrichment: the Hi-C background model (15), comparing observed contacts to expected contacts, the resampling approach (12,29), comparing observed contact counts to contacts in randomly selected regions, and the hypergeometric test (11,39), based on contingency tables of genomic contacts. The latter could previously shown to be heavily biased towards low P-values (29). Here, we demonstrate that our randomization approach is well-suited for assessing the colocalization between genomic regions and can provide substantial advantages over the previously existing approaches (see Table 1 for summary).
Table 1.
Comparison of approaches for colocalization assessment in DNA–DNA contact data
| Feature/Property | Kruse et al. (randomization approach) | Yaffe and Tanay (Hi-C background model) | Witten and Noble (resampling approach) | Dai et al. (hypergeometric test) |
|---|---|---|---|---|
| Quantification of colocalization | ✓ | ✓ | ✓ | ◯ |
| Uniform P-values on random data | ✓ | NA | ✓ | ✗ |
| Unaffected by experimental biases | ✓ | ✓ | ✗ | ✗ |
| Experimental approach independent | ✓ | ✗ | ✓ | ✗ |
| Widely applicable to other questions | ✓ | ✗ | ✗ | ✗ |
Circle in the Dai et al. column signifies that colocalization is indirectly quantified by the P-value. ‘NA’ in the Yaffe and Tanay column signifies that the method does not produce P-values.
Figure 2a shows a comparison of our randomization approach to the resampling approach. In both methods, contact counts among DNA segments of interest are compared to contact counts in corresponding null-models. The approaches differ primarily in the choice of this null-model: the resampling approach maintains the original network’s structure and contacts, but randomly selects a new set of segments from the network (re-labelling); the randomization approach keeps the segment set of interest identical, but shuffles the network contacts according to the above-described procedure. In each case, we can obtain a global measure of colocalization, here called the segment-proximity index (SPI), and a corresponding P-value from the comparison of observed to random contacts (Supplementary Materials and Methods).
To confirm the validity of the colocalization assessment using our randomization approach, we tested it on a large number of random segment sets in several artificially created contact networks [Materials and Methods section, as previously described (29)]. If the approach is valid, the distribution of P-values obtained from assessing colocalization on these artificial networks must be uniform. If the original network is merely rewired, we can observe a clear over-representation of low P-values on the random data (Figure 2b, background histogram), demonstrating the importance of clustering in the network. In random networks that maintain the clustering behaviour of the original, this effect disappears (Figure 2b, foreground histogram). The slight skew in the distribution towards high P-values is an effect of sample size, which can also be observed in the resampling approach (Supplementary Control Calculations, Supplementary Figure S3). A more exhaustive evaluation of the effects of network size and sample size on the uniformity of P-values can be found in the Supplementary Control Calculations and Supplementary Figure S4.
The colocalization results on artificial networks and random segments in real-world yeast Hi-C data are in very good agreement with the enrichment test of Yaffe and Tanay (15), demonstrating that our method is consistent with the contact enrichment in the Hi-C background model. It can reproduce results from experimental approach-specific analyses, while additionally providing P-values for the assessment of statistical significance (Supplementary Control Calculations and Supplementary Figures S5 and S6). We also ensured independently that the approach using random networks accurately reflects colocalization of DNA segments. For each node in the artificial contact networks we selected its 100 closest neighbours in space and performed a colocalization assessment between them. All resulting P-values were very low (P < 10−3).
The randomization-based colocalization assessment is not biased by node degree
We have shown that the colocalization assessment using random networks produces uniform P-values, but why is it more accurate than the existing resampling approach? Since the resampling approach does not explicitly take into account the network's degree sequence when assessing colocalization, it will, on average, favour sets of nodes with high degrees. We demonstrate this effect by assessing the colocalization of a large number of random DNA segment sets in the yeast genome, and plotting the degrees of segments in each set in dependence of the resulting colocalization P-value (Figure 2c, and Materials and Methods section). The random networks generated according to our protocol preserve the original network's degree distribution. In addition, the randomization approach never changes the set of investigated nodes in the entire process (Figure 2a). Consequently, it does not suffer from the same degree bias (Figure 2d).
Why is this degree bias problematic? While the probabilistic normalization approach developed by Yaffe and Tanay (15) removes individual unspecific contacts on a pairwise basis, the total number of contacts (degree) made by a particular DNA segment can still be affected by these biases. For example, a long DNA segment can potentially occupy a larger nuclear volume, and thus make more contacts than a short one (shown in Figure 2e). Stricter contact filtering cannot abolish this effect. To avoid these biases, the network’s degree sequence has to be explicitly taken into account when testing for contact enrichment between DNA segments. In addition, we argue that degree-compensating approach is more likely to be able to discover interpretable patterns of contacts among colocalized nodes, since it is difficult to assign meaning to DNA segments with highly promiscuous contacts (example in Figure 2f). In yeast, this is especially crucial, since the degree of DNA segments in the centromeric region outweighs degrees of segments elsewhere in the genome.
Meiotic CO site association and confirmation of previous colocalization results
We applied the randomization approach to various genome-wide datasets, investigating colocalization between genomic features of interest. First we verified results from previous analyses (11,29) by confirming tRNA clustering and colocalization of early firing replication origins in S. cerevisiae (Supplementary Control Calculations and Supplementary Figures S7 and S8). We then mapped a genome-wide, high-resolution dataset of meiotic recombination hotspots (26) onto the yeast SCN to study the relationship between meiotic COs and 3D genome architecture. The data, which were obtained by studying the products of 56 yeast meiosis using a dense set of genetic markers, provides information on the locations of 136 meiotic recombination hotspots, 72 of which are enriched in COs, and 49 of which are enriched in NCO type recombination. We are unable to detect a significant colocalization between meiotic recombination sites, but show that the subset of CO sites is enriched in 3D contacts in the yeast nucleus (Figure 3a, randomization approach, SPI 1.48, respectively, P = 0.098).
Figure 3.
Colocalization assessment results of randomization approach in the yeast SCN and GCN. (a) Meiotic recombination hotspot contact enrichment. Figure shows the distribution of contact counts in the randomized networks in comparison to the contact count between recombination hotspots in the original network (violin plot = combined density- and boxplot), measured by the SPI. (b) ORF colocalization (c, e) P-value distribution from the colocalization assessment of 1000 gene subsets of 250 genes each. (c) Yeast SCN. Skew towards low P-values is clearly visible. (d) Schematic showing the SCN to GCN (gene contact network) conversion (e) Yeast GCN. Skew towards low P-values is no longer visible.
GCNs can resolve colocalization biases of gene subsets
Using the randomization approach, we find that ORFs in the yeast genome are significantly colocalized (SPI = 4.171, P < 10−3, Figure 3b). In a network context this means that the randomized networks on average display fewer contacts among genes than the original network. This inherent colocalization of the full set of genes will bias colocalization assessments of gene subsets in the same direction. The extent of this bias is visible when we select random gene subsets in the yeast SCN and plot a histogram of their colocalization P-values (Figure 3c).
So how can we separate the effect of a global gene colocalization from the colocalization of gene subsets? A flexible solution is to break down individual DNA segments into their constituent nodes, rather than restricting the contact partners of individual DNA segments. The genes then form the nodes of the new network, which each inherit the connections to genes their parent segments make (Materials and Methods section, Figure 3d). In this manner, we can convert the original SCN to a GCN. Upon randomization the network's degree sequence, and with it the number of gene–gene contacts, will be preserved. The corresponding distribution of P-values of random gene subsets is now much more uniform (Figure 3e); we do not expect a completely uniform distribution, as we performed the calculations on the real-world yeast GCN. We also ensured that the GCN conversion still produces valid P-values in artificial networks (Supplementary Control Calculations and Supplementary Figure S3). An analogous network conversion will have to be performed when analysing subsets of data that display a similar (or inverse) dependency as the set of genes.
Genes bound by the cohesin subunit IRR1p are significantly colocalized
We apply the colocalization assessment using randomized yeast GCNs to a real-world example: can we identify colocalized sets of genes that are bound by a particular transcription or chromatin regulator? We exploit a large-scale dataset which documents the DNA binding of over 200 gene or chromatin regulatory proteins in yeast (27) to study the colocalization of each protein’s target genes. Analyses and permutation-based multiple testing corrections were performed separately for each dataset, which correspond to the approximate binding site of the protein, UAS, TSS and ORF (Figure 4a and Materials and Methods section).
Figure 4.
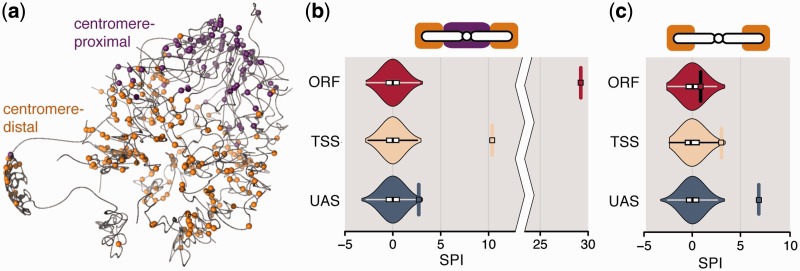
Cohesin subunit IRR1p colocalization (a) Cohesin binding sites at the UAS overlaid on a 3D representation of the budding yeast genome (11). (b, c) Violinplots of SPIs for IRR1p-bound genes in random networks. SPI of original contact count indicated by rectangle and horizontal line. One plot for each of the three datasets (ORF, TSS and UAS). (b) SPIs of full dataset—note the broken axis between SPI 10 and 25. (c) SPIs of centromere-distal IRR1p-bound genes.
The results of the ORF and TSS datasets show only one protein with significantly colocalized target genes after P-value adjustment (Figure 4b): IRR1p (ORF: SPI = 29.23, P < 10−3; TSS: SPI = 10.38, P < 10−3). Contacts between IRR1p-bound genes in the UAS dataset are enriched, but not significantly (SPI = 3.55, P = 0.116). IRR1p is a subunit of the yeast cohesin complex (the only cohesin subunit examined in the dataset), known to be involved in mediating sister chromatid cohesion during cell division, but also implicated in DNA repair and gene regulation. Full colocalization results for all chromatin regulatory proteins can be found in the Supplementary Datasets 1–3, a description of the file structure can be found in the Supplementary Control Calculations.
Since cohesin is known to display enriched binding in centromeric regions (40,41), we investigated if the observed colocalization can be explained entirely by the known centromeric clustering in yeast. For each dataset, we split the cohesin binding sites on each chromosome into two groups of comparable size: centromere-proximal (within 100 kb from the centromere) and centromere-distal (example for UAS in Figure 4a). As expected, the SPI of centromere-proximal genes increases for the TSS and ORF datasets. Interestingly, the SPI of centromere-proximal IRR1p-bound genes in the UAS dataset is also strongly increased (SPI = 10.908, P < 10−3). In the centromere-distal groups genes no longer display colocalization in the ORF and TSS datasets (Figure 4c, ORF: SPI = 0.825, P = 1.0; TSS: SPI = 3.021, P = 0.218). In contrast, the centromere-distal group of IRR1p-bound genes in the UAS dataset now displays significant colocalization (UAS: SPI = 7.024, P < 10−3). A possible explanation for this observation is that target genes of IRR1p, which are bound at the UAS, are not globally colocalized, but form two or more relatively independent clusters of colocalized genes. The above results have been obtained by only performing Steps 1 and 2 in the randomization procedure (Figure 5), Step 3 has no significant influence.
DISCUSSION
The study of genome-wide DNA–DNA contacts is starting to reveal the close relationship between the DNA's 3D architecture and diverse biological processes. However, the computational analysis of genome-wide DNA–DNA contact data is still in its early stages. Most of the currently available methods are specific to the experimental approach, or subject to various experimental and computational biases (15). In this work, we presented a unifying, complex network framework for the computational and statistical analysis of DNA–DNA contact data that builds upon the experiment-specific normalization and filtering procedures, but provides a robust approach for their analysis independent of the experimental method used. The randomization approach we developed explicitly takes into account the inherent 3D structure of DNA–DNA contact data, allowing rigorous and unbiased statistical enrichment tests.
The colocalization assessment using the randomization approach confirmed the global colocalization of genes observed in other organisms (11,12,13,15), as well as previous findings in S. cerevisiae, such as tRNA and replication origin clustering. The latter has been implicated in human cancer based on somatic copy-number alterations (SCNAs), where it was possible to predict a significant proportion of SCNAs using long-range DNA–DNA contacts and replication timing data alone (42). In similar fashion, we show that meiotic CO sites have a tendency to colocalize genome-wide in the yeast nucleus. While we cannot show a significant colocalization, this is consistent with the finding that genomic areas with a strong ‘intermingling’ of DNA are associated with a higher frequency of double-strand breaks in these regions (43). The 3D association of CO sites could therefore also be associated with SCNAs in cancer, as discussed by Fudenberg et al. (44).
It has recently been shown that the higher-order genome organization in yeast can be accurately modelled by merely considering the physical tethering of heterochromatic regions to nuclear landmarks and volume exclusion as major driving forces of genomic organization (45). This random encounter model satisfactorily explains many of the experimental observations made in the 3D S. cerevisiae nuclear architecture. These results suggest that genome organization works largely independently of biochemically mediated DNA–DNA interactions, and that the linear arrangement of DNA features on the chromosomes can serve to position them in 3D space. This notion is in good agreement with results from a previous study which demonstrates that transcription factor target genes tend to be encoded within the same area on one chromosome, effectively increasing their spatial proximity (21).
In this context, the results obtained in this analysis on cohesin suggest that it could serve as global organizer that can tether DNA loci in a seemingly directed fashion. Evidence for this can be found in the strong contact enrichment of IRR1p-bound target genes, both at and away from the centromere. This alone is not sufficient to identify cohesin as a cause of the observed colocalization. There is, however, strong evidence on a local scale that cohesin mediates chromatin looping and tethering in different organisms (46). This is supported by the structure of the cohesin complex itself, which forms a ring that, at least in theory, could entrap up to two strands of DNA simultaneously (47), providing a mechanism of how the tethering could be achieved.
It is unclear why we observe differences in colocalization of cohesin-bound genes based on the location of the binding site. A straightforward assumption would be that the role of ORF- and, in part, TSS-bound cohesin lies primarily in mediating centromeric clustering, whereas UAS binding confers a different functionality, such as gene regulation (46). It has been suggested, for example, that cohesin mediates both gene-enhancer and boundary-insulator contacts in humans (48).
In summary, our complex network framework provides a versatile tool for enrichment analyses in DNA–DNA contact networks. With the steady increase in new experimental methods that accelerate the acquisition of genome-wide contact datasets in diverse organisms, cell types and conditions, we expect a complex network approach to become a central tool in the elucidation of nuclear architecture and its relationship to diverse biological processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables T1 and T2, Supplementary Figures 1–9, Supplementary Materials and Methods, Supplementary Control Calculations, Supplementary Datasets 1–3 and Supplementary References [49, 50].
FUNDING
Medical Research Council [U105185859]; Human Frontier Science Program [RGY0073/2010 to S.S.]; LMB International PhD Programme (to K.K.); LMB, EMBO Young Investigator Programme, Trinity College, ERASysBio+ and HFSP [RGY0073/2010 to M.M.B.]. Funding for open access charge: Medical Research Council [U105185859].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Charles Ravarani, Sreenivas Chavali, David Wright and Sarah Teichmann for their comments on this work.
REFERENCES
- 1.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 2.Osborne CS, Ewels PA, Young ANC. Meet the neighbours: tools to dissect nuclear structure and function. Brief. Funct. Genom. 2011;10:11–17. doi: 10.1093/bfgp/elq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 4.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 5.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodley CDM, Bertels F, Jones B, O’Sullivan JM. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet. Biol. FG & B. 2009;46:879–886. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 7.van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. JoVE. 2010;39:1–7. doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJG, Zhu Y, Kaaij LJT, van Ijcken W, Gribnau J, Heard E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K-I. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 16.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 17.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 18.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 19.Jothi R, Balaji S, Wuster A, Grochow JA, Gsponer J, Przytycka TM, Aravind L, Babu MM. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol. Syst. Biol. 2009;5:294. doi: 10.1038/msb.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 21.Janga SC, Collado-Vides J, Babu MM. Transcriptional regulation constrains the organization of genes on eukaryotic chromosomes. Proc. Natl Acad. Sci. USA. 2008;105:15761–15766. doi: 10.1073/pnas.0806317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 23.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 24.Itzkovitz S, Milo R, Kashtan N, Ziv G, Alon U. Subgraphs in random networks. Phys. Rev. E. 2003;68:1–8. doi: 10.1103/PhysRevE.68.026127. [DOI] [PubMed] [Google Scholar]
- 25.Itzkovitz S, Alon U. Subgraphs and network motifs in geometric networks. Phys. Rev. E. 2005;71:1–9. doi: 10.1103/PhysRevE.71.026117. [DOI] [PubMed] [Google Scholar]
- 26.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witten DM, Noble WS. On the assessment of statistical significance of three-dimensional colocalization of sets of genomic elements. Nucleic Acids Res. 2012:1–7. doi: 10.1093/nar/gks012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soffer S, Vázquez A. Network clustering coefficient without degree-correlation biases. Phys. Rev. E. 2005;71:2–5. doi: 10.1103/PhysRevE.71.057101. [DOI] [PubMed] [Google Scholar]
- 31.Bansal S, Khandelwal S, Meyers LA. Exploring biological network structure with clustered random networks. BMC Bioinform. 2009;10:405. doi: 10.1186/1471-2105-10-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westfall PH, Young SS. Resampling-Based Multiple Testing. John Wiley & Sons Hoboken, NJ 07030-5774, United States of America; 1993. [Google Scholar]
- 33.Kendziorski C, Wang P. A review of statistical methods for expression quantitative trait loci mapping. Mamm. Genome: Off. J. Int. Mamm. Genome Soc. 2006;17:509–517. doi: 10.1007/s00335-005-0189-6. [DOI] [PubMed] [Google Scholar]
- 34.Botta M, Haider S, Leung IXY, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol. Syst. Biol. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajapakse I, Scalzo D, Tapscott SJ, Kosak ST, Groudine M. Networking the nucleus. Mol. Syst. Biol. 2010;6:395. doi: 10.1038/msb.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erdös P, Renyi A. On random graphs. 1959:290–297. [Google Scholar]
- 37.Artzy-Randrup Y, Stone L. Generating uniformly distributed random networks. Phys. Rev. E. 2005;72:1–7. doi: 10.1103/PhysRevE.72.056708. [DOI] [PubMed] [Google Scholar]
- 38.Basler G, Ebenhöh O, Selbig J, Nikoloski Z. Mass-balanced randomization of metabolic networks. Bioinformatics (Oxford, England) 2011;27:1397–1403. doi: 10.1093/bioinformatics/btr145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Z, Dai X. Nuclear colocalization of transcription factor target genes strengthens coregulation in yeast. Nucleic Acids Res. 2012;40:27–36. doi: 10.1093/nar/gkr689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 41.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 42.De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol. 2011;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fudenberg G, Getz G, Meyerson M, Mirny LA. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat. Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorsett D, Ström L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr. Biol. 2012;22:R240–R250. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ocampo-Hafalla MT, Uhlmann F. Cohesin loading and sliding. J. Cell Sci. 2011;124:685–691. doi: 10.1242/jcs.073866. [DOI] [PubMed] [Google Scholar]
- 48.Merkenschlager M. Cohesin: a global player in chromosome biology with local ties to gene regulation. Curr. Opin. Genet. Dev. 2010:555–561. doi: 10.1016/j.gde.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang SC-H, Rhind N, Bechhoefer J. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol. Syst. Biol. 2010;6:404. doi: 10.1038/msb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.