Abstract
In Escherichia coli, the filament of RecA formed on single-stranded DNA (ssDNA) is essential for recombinational DNA repair. Although ssDNA-binding protein (SSB) plays a complicated role in RecA reactions in vivo, much of our understanding of the mechanism is based on RecA binding directly to ssDNA. Here we investigate the role of SSB in the regulation of RecA polymerization on ssDNA, based on the differential force responses of a single 576-nucleotide-long ssDNA associated with RecA and SSB. We find that SSB outcompetes higher concentrations of RecA, resulting in inhibition of RecA nucleation. In addition, we find that pre-formed RecA filaments de-polymerize at low force in an ATP hydrolysis- and SSB-dependent manner. At higher forces, re-polymerization takes place, which displaces SSB from ssDNA. These findings provide a physical picture of the competition between RecA and SSB under tension on the scale of the entire nucleoprotein SSB array, which have broad biological implications particularly with regard to competitive molecular binding.
INTRODUCTION
In Escherichia coli, the highly cooperative polymerization of RecA on single-stranded DNA (ssDNA) is essential for recombinational DNA repair. The RecA nucleoprotein filament catalyses recombination-like reactions in the presence of Adenosine 5′-triphosphate (ATP) or its analogues [deoxyadenosine triphosphate (dATP), Adenosine 5′-3-thiotriphosphate (ATPγS)] (1,2). The formation of the RecA nucleoprotein filament on ssDNA involves two kinetically distinct steps: a slower nucleation step, followed by a faster directional polymerization step. ATP hydrolysis makes the RecA nucleoprotein filament a dynamic structure, the stability of which is determined by polymerization and de-polymerization on ssDNA (3–5).
Much of our knowledge of the formation of active RecA nucleoprotein filament is based on bulk biochemical and biophysical studies (5–9). The recent development of a combination of single-molecule manipulation techniques has begun to contribute to a better understanding of the formation and stability of the RecA nucleoprotein filament (3,4,10). The kinetics of RecA nucleation, directional polymerization, and de-polymerization on ssDNA have been measured directly at high spatial and temporal resolutions (3,4,10). For example, magnetic and optical tweezers as well as single-molecule fluorescence resonance energy transfer (FRET) experiments have been used to investigate the mechanism of homology search and alignment by the RecA nucleoprotein filament in real time (11–14). However, despite such advances, gaps persist in our understanding of how the formation of RecA nucleoprotein filament is modulated by recombination accessory proteins. For example, the in vivo substrate for the formation of the RecA nucleoprotein filament as well as for other DNA damage signalling and processing proteins is not naked ssDNA, but rather it is ‘smooth’ or ‘beaded’ forms of ssDNA–SSB complexes (15–18).
RecA actively competes with ssDNA-binding protein (SSB) for binding sites on ssDNA (15). SSB proteins bind to ssDNA non-specifically with multiple modes and high affinity (15). Although the effects of SSB on RecA binding to DNA have been extensively studied (15–19), the consequences of competition between RecA and SSB on the formation of RecA nucleoprotein filaments have not been investigated at single-DNA molecule resolution in which tension can affect this competition. The ssDNA-binding properties of SSB tetramer (20,21) and its impact on RecA polymerization have been studied at a single-molecule resolution (22), but the results of these studies may not apply to the densely packed SSB tetramers that bind ssDNA in vivo where sliding of individual tetramers along ssDNA is unlikely during recombinational DNA repair. The properties of tightly packed SSB tetramers and their impact on the formation of RecA nucleoprotein filaments are poorly understood at single molecule resolution.
To address these unknowns, here we studied the effects of pre-formed SSB–ssDNA complex and free SSB in solution on the formation of RecA nucleoprotein filaments by stretching a ssDNA tether using high force magnetic tweezers (23,24). Additionally, we studied the effects of free SSB in solution on the stability of a pre-formed RecA nucleoprotein filament by monitoring its de-polymerization from ssDNA.
MATERIALS AND METHODS
A high force vertical magnetic tweezers (23) set up was used to perform all the experiments in this study. It can apply force in the range from 0.1 pN to >100 pN on a paramagnetic bead of a diameter of 2.8 µm (Dynal M-280 bead, Invitrogen). Analysis of bead images gives a spatial resolution of ∼2 nm in all three dimensions and a temporal resolution of 2 ms. E. coli SSB was purchased from Promega Corporation (The catalog number is M3011). E. coli RecA protein was purchased from New England BioLabs, USA (The catalog number is M0249L) or purified as previously described (25). The 576 bp DNA construct was generated by PCR using DreamTaq DNA polymerase (Fermentas) from 992 to 1552 bp on lambda DNA. Thiol and biotin were labelled on the same strand of this DNA construct. All the experiments in the article were performed at 23°C in the standard RecA assay buffer containing 20 mM Tris (pH 7.4), 50 mM KCl and 10 mM MgCl2. For experiments where RecA was present, except for Figure 3C where 1 mM ATPγS was used, all the other data were obtained in the presence of 1 µM RecA, 1 mM ATP and 1 × ATP regeneration system that maintains a stable ATP concentration of 1 mM. Additional details of thiol and biotin labelling of DNA ends, tether formation, the ATP regeneration system recipe, the force-extension curve measurement for short ssDNA tethers, force control during buffer exchange, and the ssDNA extension dynamics obtained by quick force-jumping method can be found in Supplementary Information.
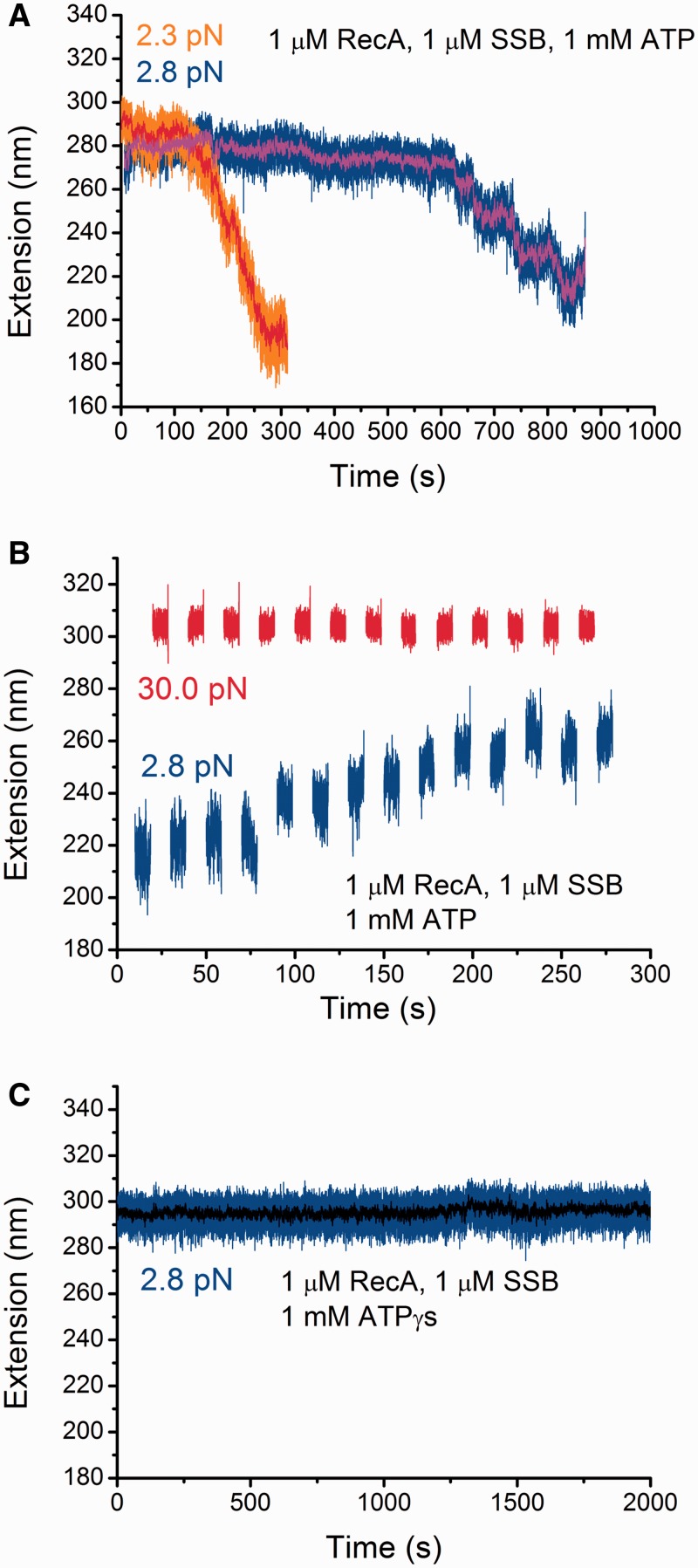
Figure 3.
Effects of SSB, ATP hydrolysis and force on the stability of pre-formed RecA nucleoprotein filaments. (A) Extension change of two different pre-formed RecA nucleoprotein filaments after introduction of protein mixture containing 1 µM RecA, 1 µM SSB, and 1 mM ATP, 1 × ATP regeneration system. Both exhibit a lag phase followed by a quick extension decreasing phase at low forces <3 pN. Raw data are in orange and blue, while the smoothed data by 40-points fast Fourier transformation are in red and magenta, respectively. (B) Extension change of a partially de-polymerized RecA nucleoprotein filament (the same ssDNA as indicated by blue in Figure 3A) during multiple cycles of jumping between a high force (∼30 pN) and a low force (∼2.8 pN). In general, these force-jump cycles lead to elongation of the ssDNA. (C) On the same ssDNA indicated by blue in Figure 3A, repeating the experiments described in Figure 3A but with a replacement of ATP with ATPγS. The result shows that the presence of SSB does not cause de-polymerization of the pre-formed RecA nucleoprotein filament with ATPγS.
RESULTS
Force responses of ssDNA in RecA-binding solution
We studied different force responses of ssDNA, ssDNA bound with RecA and ssDNA bound with SSB tetramers. To suppress thermal noise from the longitude conformational fluctuation of the flexible ssDNA, we used a short ssDNA tether, which is long enough to form a stable RecA filament and an array of SSB proteins. In this work, our ssDNA has 576 nucleotides, which is much longer than the minimal size of the RecA nucleation site (26,27), and can accommodate ∼8 SSB in the (SSB)65-binding mode and ∼16 SSB in the (SSB)35-binding mode. We first determined the force response of the 576-nucleotide ssDNA tether formed between a 2.8-µm-streptavidin-coated paramagnetic bead (Dynalbeads M-280 Streptavidin, Invitrogen) and a sulfo-SMCC-coated coverglass (Supplementary Text: ‘DNA constructs, end-labeling, tether formation, and ATP regeneration system recipe’). The ssDNA tether was produced by melting a 576 bp double-stranded DNA (dsDNA) tether at a tension of around 65 pN through a ‘strand-peeling’ transition (24,28,29) using high force magnetic tweezers (23) (Figure 1A).
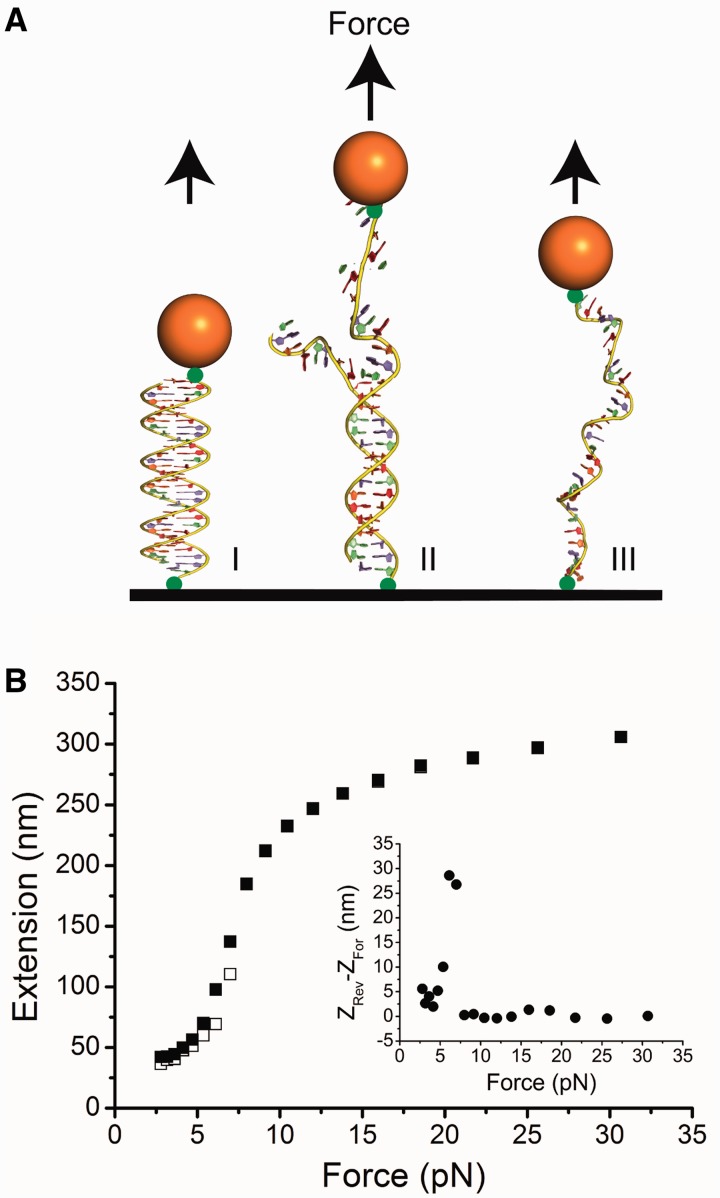
Figure 1.
Force response of ssDNA. (A) Schematic of the experimental set up in which an ssDNA tether was generated from a dsDNA tether by overstretching the DNA at ∼65 pN force applied to one of the DNA strands by magnetic tweezers (Supplementary Text: Force responses of dsDNA and ssDNA). After transition, only one strand is left under tension. (B) Force-extension curves for ssDNA in the RecA-binding buffer. At each force, the DNA was held for 5 s. The black open squares represent data obtained in the force-increase scan and the black solid squares represent data obtained in force-decrease scan through the same set of forces. Inset shows the hysteresis which is extension obtained in the force-decrease scan minus that obtained in the force-increase scan.
To characterize the effects of force on a 576-nucleotide ssDNA tether, we measured its force-extension curve. In the vertical magnetic tweezers, what is actually measured is the height of the bead from the cover glass. At each force, the equilibrium bead position is determined by force balance and torque balance (23). At different forces, the changes in torque balance will cause rotation of the bead, thus leading to change in the bead height. For a short DNA tether, such bead rotation will cause large errors in extension measurement.
Using a property that the bead adopts a unique orientation corresponding to a unique height at each force with an error of <10 nm (Supplementary Figure S1), we developed a method to measure the force-extension curves of the short ssDNA tether: we first measured the extension difference between the dsDNA before the force-induced strand-peeling transition (Figure 1A) and the ssDNA after the transition at the same set of forces, in which the height change owing to bead rotation was eliminated with error <10 nm (Supplementary Figure S1). Then, the ssDNA force-extension curve was obtained by adding back the theoretical dsDNA force-extension curve based on the worm-like-chain polymer model of dsDNA (details in Supplementary Text: Force responses of dsDNA and ssDNA).
Figure 1B shows the force-extension curve of an ssDNA tether recorded during a force-decrease scan (solid squares) followed by a force-increase scan (open squares) at 23°C in the standard RecA assay buffer containing 20 mM Tris (pH 7.4), 50 mM KCl and 10 mM MgCl2. At each force, the extension of DNA was averaged for data recorded over 5s (all the force-extension curves in the article were measured using 5-s time window unless specifically stated). Potentially, secondary structures can form between complementary DNA sequences at low force, causing hysteresis in extension between the force-decrease and force-increase scans. To quantify the level of hysteresis in 576-nucleotide ssDNA, we plotted the value of the extension in the force-decrease scan minus that recorded in the force-increase scan (Figure 1B inset). At forces <8 pN, the non-zero positive value suggests that secondary structures formed with a peak hysteresis value of ∼30 nm at ∼7 pN. Overall, the force response of the 576-nucleotide ssDNA and its dependence on the monovalent and magnesium concentrations are consistent with previous studies of ssDNA force responses (30–33) (Supplementary Figure S2A and B). To minimize the additional extension reduction from the secondary structure formation, in the following force-extension curves, we only plot the data obtained in the force-decrease scan unless specifically stated, as DNA has less time to form secondary structure during the force-decrease scan compared with the force-increase scan.
Distinct effects of RecA and SSB on the force response of ssDNA
To investigate the effects of RecA and SSB on the force response of ssDNA, we compared the force-extension curves of ssDNA in the presence and absence of RecA or SSB. Figure 2A shows the force-extension curves of an ssDNA before RecA was introduced (squares) and after RecA nucleoprotein filament was fully polymerized (circles). The RecA nucleoprotein filament was formed by holding the DNA at >15 pN for >100 s in 1 µM RecA and 1 mM ATP, 1 × ATP regeneration system, in the standard RecA assay buffer (see Methods). In 1 µM RecA concentration and over the time scale, RecA nucleoprotein filament fully polymerized along the DNA and reached a steady state (Supplementary Figure S3). Note that no free ssDNA was present in solution; therefore, the RecA (and SSB in the results described subsequently) was always in excess to ssDNA-binding sites.
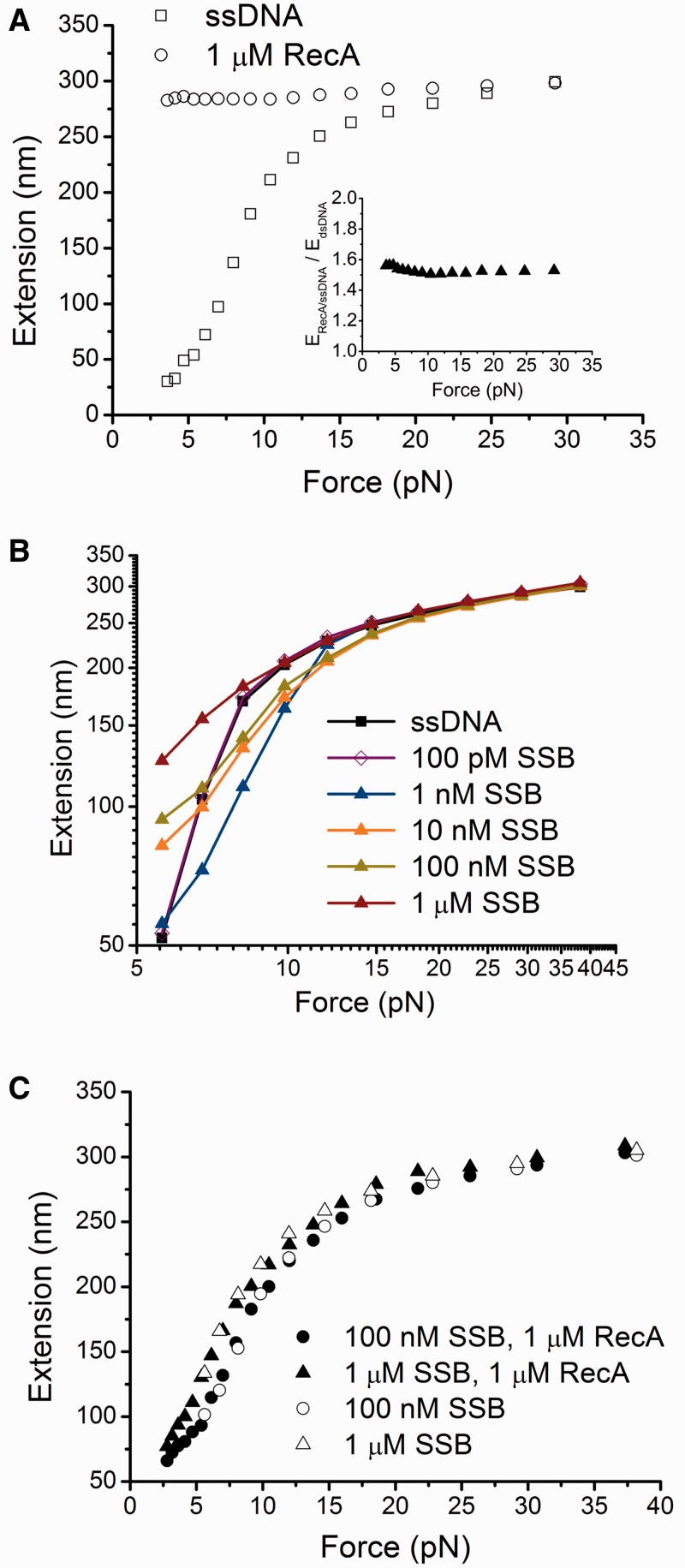
Figure 2.
Effects of RecA and SSB on force responses of ssDNA. (A) Black open circles indicate the force-extension curve of ssDNA bound by a fully coated RecA nucleoprotein filament formed with ATP. Black open squares indicate the force-extension curve of the same ssDNA before RecA binding. Inset shows the extension of a fully polymerized RecA nucleoprotein filament measured in experiments divided by the theoretical extension of a dsDNA of equal number of base pairs (576 bp) at the corresponding force values based on the worm-like-chain model of DNA (Eq. S1) (34). (B) Force responses of ssDNA incubated with different concentrations of SSB. (C) Force-extension curves of ssDNA obtained with incubation with either 1 µM RecA and 100 nM SSB, or 1 µM RecA and 1 µM SSB in the presence of ATP. Data in 100 nM SSB or 1 µM SSB from (B) are plotted together for comparison. In each panel, the force-extension curves were recorded during the force-decrease scans. Data in (A–B) and data indicated by the open symbols in (C) were recorded after holding the ssDNA at large force (>15 pN) for >100 s in the protein solutions. Data indicated by the solid symbols in (C) were recorded after the protein solution was slowly introduced at low force (<5 pN).
Compared with the force-extension curve of the same ssDNA before RecA was introduced, the ssDNA bound by the RecA was always longer at forces up to ∼30 pN (Figure 2A). This finding is consistent with the stiff structure of the RecA nucleoprotein filament, which has a bending persistence length of ≥1 µm (10,35). Furthermore, our results are in accord with a previous study that showed that ssDNA bound by the RecA is ∼50% longer than dsDNA of an equal number of base pairs (Figure 2A, inset) (10).
The force-extension curves of ssDNA in the presence of SSB differ from those in the presence of RecA (Figure 2B). In the presence of 1–100 pM SSB, the force-extension curves of ssDNA were not distinguishable from curves obtained in the absence of SSB (therefore only data obtained in 100 pM SSB are shown) (Figure 2B), suggesting no or very few SSB proteins were bound to DNA at <100 pM SSB. However, SSB at concentrations ranging from 1–1000 nM significantly affected the force-extension curve of ssDNA (Figure 2B). In this concentration range, the SSB binding to DNA nearly reached a steady state over the time scale of our force scans (Supplementary Figure S4). At forces below ∼7 pN, and SSB concentrations >10 nM, the extension of ssDNA bound by SSB was longer than naked ssDNA, which is likely to be caused by the removal of secondary structures in ssDNA and repulsive interactions between adjacent units of SSB bound to ssDNA. In contrast, at forces of 8–18 pN where the secondary structures do not exist (Figure 1B inset) and in the presence of 1–100 nM SSB, the extension of ssDNA bound by SSB is shorter than naked ssDNA. This effect could be attributed to wrapping of ssDNA around the SSB tetramers (20,21). At forces >18 pN, the extension of ssDNA bound by SSB equals the extension of naked ssDNA for all the SSB concentrations studied. As at the high force, the ssDNA is still associated with SSB proteins (Supplementary Figure S4B), these results suggest that at high force SSB binds to ssDNA in a manner that does not cause significant wrapping of ssDNA. At the highest SSB concentration (1µM tested, shortening of ssDNA is not observed at any force. This finding suggests that ssDNA becomes saturated with bound SSB in a less-wrapped mode, thereby may provide potential binding sites for additional SSB tetramers.
These findings are consistent with previously reported multiple binding modes of SSB that wrap different numbers of nucleotide residues in ssDNA (16,20,21): (i) wrapped modes (SSB)56 and (SSB)65, in which ∼56 or ∼65 nt of ssDNA is wrapped around SSB subunits, and (ii) the least wrapped mode (SSB)35, in which ∼35 nt of ssDNA are bound by an average of two SSB subunits (16,21). The selection of the SSB-binding mode depends on many factors including monovalent salt, pH, magnesium, and protein concentration (20,21,36). According to previous reports, the least wrapped (SSB)35 mode is favoured at low salt concentrations at high SSB-binding density on ssDNA (16,21,37,38). In our experiments, the more wrapped modes (SSB)56 or (SSB)65 might occur at low SSB-binding density in low SSB concentration. This can explain the ssDNA shortening at low SSB concentrations (e.g. 1 nM) observed in our experiments. On the other hand, the increase in extension that we observed at 1 µM SSB, regardless of the amount of force, suggests that at this SSB concentration, ssDNA is in the least wrapped state due to high binding density at higher SSB concentrations. This is consistent with the recent single-molecule FRET studies in similar buffer solution conditions that reported much reduced FRET values and fluctuations at high SSB concentrations likely due to binding of more SSB (22). At this concentration, the ssDNA extension is longer than the naked ssDNA, which is possibly due to the repulsive interactions, such as electrostatic and steric interactions, between positively changed large SSB tetramers densely packed on ssDNA.
Our findings show that binding of RecA and SSB to ssDNA affects extension of ssDNA differently, depending on the concentrations of the accessory proteins and applied force. Here we emphasize that as no free ssDNA was present in solution, in all SSB concentrations, the SSB protein is always in excess of ssDNA. Therefore, the SSB concentration dependence of the ssDNA force response likely indicates the concentration-dependent binding density of SSB on ssDNA and the switch of binding modes.
SSB outcompetes RecA binding to ssDNA and inhibits RecA nucleation
In E. coli, RecA binds to ssDNA under conditions in which SSB concentrations are in the range of a few hundred nM (15,39). To study the competition between RecA and SSB for binding sites on ssDNA, we introduced 1 µM RecA together with either 100 nM or 1 µM SSB in Standard RecA assay buffer with 1 mM ATP, 1× ATP regeneration system. The introduction of RecA–SSB mixture was slow, over a period of about 30 min, to avoid exerting large drag force to the bead (Supplementary Text: Force control during buffer change). Figure 2C shows that, in the presence of 100 nM and 1 µM SSB, the RecA filament did not form at any increment of force, and the force-extension curves are almost identical to those generated in presence of SSB without addition of RecA (Figure 2B), indicating that SSB outcompetes RecA for binding sites on ssDNA. To determine whether RecA polymerization can occur at large force over a longer time scale, we held the tether at ∼37 pN for >80 s or many cycles between a lower force of ∼11 pN and a higher force ∼49 pN, but RecA polymerization did not occur at the high force in our time scale (Supplementary Figure S5A–D). These results show that RecA nucleation on ssDNA is completely inhibited at these SSB concentrations over the force range and time scale in our solution conditions.
SSB- and ATP hydrolysis-dependent de-polymerization of pre-formed RecA nucleoprotein filament at low force and re-polymerization at high force
Having established that SSB competitively inhibits the binding of RecA to ssDNA under our experimental conditions, we sought to measure its effect on the maintenance of a pre-formed RecA nucleoprotein filament. Firstly, a fully coated RecA nucleoprotein filament was formed at ∼30 pN in the presence of 1 µM RecA and 1 mM ATP, 1× ATP regeneration system. Subsequently, a mixture of 1 µM RecA, 1 µM SSB, and 1 mM ATP, 1 × ATP regeneration system was introduced, after which the force was decreased to ∼2.8 pN (blue in Figure 3A). An initial lag phase was observed for ∼600 s, during which the extension fluctuated around a constant average of ∼290 nm, suggesting a nearly steady RecA nucleoprotein filament during this time. The lag phase was followed by a rapid extension decreasing phase, during which the extension of ssDNA decreased by ∼70 nm in around 200 s, indicating dynamic de-polymerization of RecA from the ssDNA. As RecA nucleoprotein filament is stable at such force in the absence of SSB (Supplementary Figure S6), this de-polymerization is dependent on the presence of SSB. To examine the reproducibility of such SSB-dependent RecA de-polymerization, we performed similar experiment on a different ssDNA (orange in Figure 3A). Similarly, a lag phase followed by a rapid extension decreasing phase was observed. However, both the duration of the lag phase and the speed of de-polymerization in the extension decreasing phase were significantly different from the first DNA, suggesting high stochasticity of the two steps. SSB-dependent de-polymerization of pre-formed RecA nucleoprotein filament was also observed at the lower SSB concentration of 100 nM (Supplementary Figure S7)
To investigate whether the partially de-polymerized RecA filament can re-polymerize at higher force, for the partially de-polymerized RecA filament (the same ssDNA as indicated in blue in Figure 3A), multiple cycles of jumping between a high force (∼30 pN) and a low force (∼2.8 pN) were performed (Figure 3B). At each force, the ssDNA was held for 5 s. After each cycle, the ssDNA became longer in most of the cycles. These observations demonstrate that a partially de-polymerized RecA filament at low force can re-polymerize at higher forces.
We also performed similar experiments using 1 mM ATPγS to ascertain the role of ATP hydrolysis in the formation of RecA nucleoprotein filament. We first formed a fully coated RecA nucleoprotein filament at high force in 1 µM RecA and 1 mM ATPγS. Subsequently, a mixture of in 1 µM RecA, 1 mM ATPγS and 1 µM SSB was introduced, and the force was decreased to a lower force of ∼2.8 pN, and the extension was monitored for >2000 s (Figure 3C). No extension decrease was observed over the time, in contrast to the rapid extension decreasing phase followed by a lag phase of several hundreds of seconds. This result indicates that, in addition to the dependence on SSB, the de-polymerization at low force also depends on ATP hydrolysis.
SSB- and ATP hydrolysis-dependent de-polymerization of pre-formed RecA nucleoprotein filament at low force and re-polymerization at high force were also independently investigated by examination of the hysteresis in the force-extension curves recorded in force-increase and force-decrease scans. Consistent results were obtained, as shown in Supplementary Figure S8A and B.
DISCUSSION
In this study, we examined the formation of RecA nucleoprotein filament and its stability with both free SSB and ssDNA-bound SSB as well as on tension. Our data suggest a model for force-dependent RecA and SSB competitive inhibition on ssDNA (Figure 4). At >100 nM SSB concentrations and 1 µM RecA, SSB outcompetes RecA binding to ssDNA, resulting in formation of an array of SSB tetramers (Figure 4A). This tightly packed beaded complex of SSB–ssDNA imposes a barrier for RecA nucleation, or against other proteins, such as RecO (1,40). For a pre-formed RecA nucleoprotein filament, at >100 nM SSB and 1 µM RecA, our results revealed de-polymerization of RecA at low force in an ATP hydrolysis- and SSB-dependent manner, and re-polymerization of a partially de-polymerized RecA filament at high force. The high force applied in our experiments is close to the force range of 20–40 pN that can be generated by a single RNA polymerase (41) or DNA polymerase (42). Thus, our results provide important mechanistic understanding of how nucleation and polymerization of RecA might occur in vivo and how the stability of RecA nucleoprotein filament is regulated by SSB and tension.
Figure 4.
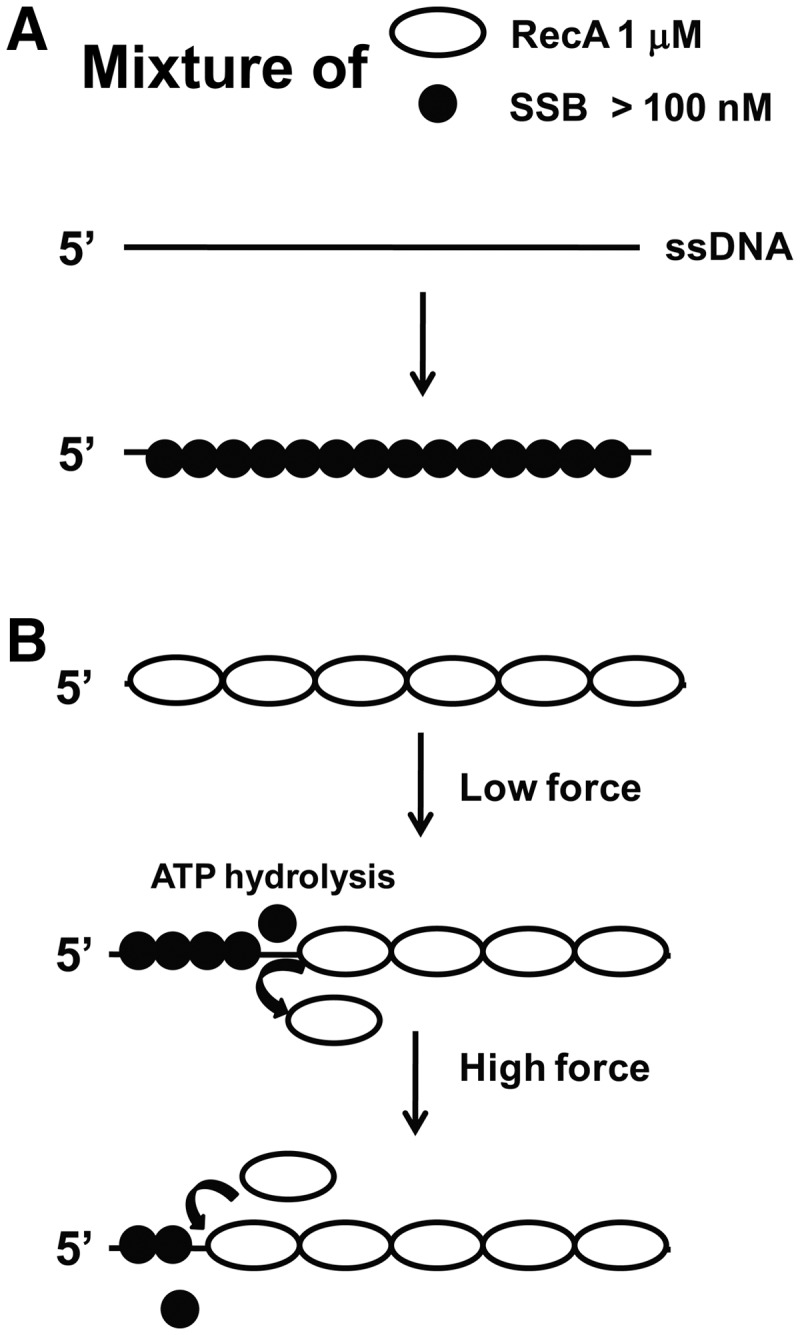
Model for force-dependent regulation of the formation of RecA nucleoprotein filament by SSB. (A) More than 100 nM SSB outcompetes 1 µM RecA, resulting in the formation of a pure SSB–ssDNA nucleoprotein array. (B) In >100 nM SSB and 1 µM RecA, net de-polymerization of a pre-formed RecA nucleoprotein filament occurs at low force in an ATP hydrolysis- and SSB-dependent manner. Increasing force results in re-polymerization of RecA nucleoprotein filament likely with the direction from 3′ to 5′ of the ssDNA. Note that the geometric objects indicating ssDNA, RecA and SSB do not represent their real conformations.
We show that SSB in the range of 100 nM–1µM outcompetes the binding of RecA to ssDNA, leading to the formation of fully coated SSB–ssDNA complex, which suggests that the nucleation of RecA is inhibited at these SSB concentrations over the experimental time scale (Figure 2). Formation of a RecA nucleoprotein filament involves two kinetic steps: (a) formation of a stable RecA nucleoprotein filament composed of four or five RecA monomers each bound to three nucleotides of ssDNA, and (b) 5′ to 3′ extension of the RecA nucleoprotein filament (1,2). We speculate that high concentrations of SSB bind ssDNA faster than RecA, thereby depleting the amount of ssDNA available for RecA nucleation.
When 1 µM RecA is mixed with >100 nM SSB with ATP and magnesium, the RecA nucleoprotein filaments are slowly de-polymerized at low levels of force and this de-polymerization depends on ATP hydrolysis (Figure 3). It has been suggested that ATP hydrolysis in a RecA nucleoprotein filament primarily occurs in the 5′ to 3′ direction on ssDNA (43). Because the RecA nucleoprotein filament is unstable in the presence of bound ADP (1), ATP hydrolysis may cause de-polymerization of a patch of RecA filament from the 5′ end, freeing areas of ssDNA to be occupied by SSB. At low tension, SSB binds to the RecA-free ssDNA region, hence blocks re-polymerization of RecA, resulting in net slow de-polymerization of RecA (Figure 3). Therefore, our results suggest that the SSB-dependent net de-polymerization of RecA is not due to active displacement of RecA by SSB; instead, it is a consequence of freed ssDNA sites for SSB to bind once RecA dissociates from the filament induced by ATP hydrolysis. These results are consistent with a previous study that demonstrated competitive binding between SSB and RecA and the lack of a direct SSB–RecA protein interaction (19).
It has been widely accepted that RecA nucleoprotein filaments form on ssDNA during a process of 5′ to 3′ polymerization, whereas RecA de-polymerization can also occur, mainly from the 5′ end, in a process requiring ATP hydrolysis (1,43). Thus, during SSB-dependent de-polymerization of RecA, the ssDNA vacated by RecA is expected to be at the 5′ end and SSB is expected to bind to those areas of ssDNA, whereas the rest ssDNA at the 3′ end is still occupied by RecA (Figure 4B, middle). This picture predicts an SSB nucleoprotein array at the 5′ side of the ssDNA and a RecA nucleoprotein filament at the 3′ side of the ssDNA after the partial de-polymerization of the original RecA filament at low force in 1 µM RecA, 1 mM ATP and >100 nM SSB.
Interpretation of the re-polymerization of RecA in 1 µM RecA, 1 mM ATP and >100 nM SSB at high force is somewhat complicated. Here we discuss three possibilities. (i) At high force, RecA can invade and nucleate inside the SSB-coated region (the 5′ side of the ssDNA), resulting in polymerization in the 5′ to 3′ direction. However, this possibility is not supported by our results that for an SSB-coated ssDNA in >100 nM SSB without pre-existing RecA filament clusters inside, RecA polymerization could not happen even at high force (Figure 2C, Supplementary Figure S5). Thus, the re-polymerization of RecA observed in Figure 3B and Supplementary Figure S8A and B should depend on the pre-existing, partially de-polymerized RecA. This is also consistent with an earlier study reporting that RecA displaces a single SSB bound to a poly-dT ssDNA supported by a pre-formed RecA nucleation cluster (4). (ii) A second possibility is that de-polymerization may occur at the 3′-end, such that re-polymerization would occur in the 5′ to 3′ direction. However, this mechanism is not consistent with extensive previous studies showing that RecA de-polymerization primarily occurs at the 5′-end (1,43). (iii) A more likely possibility is that de-polymerization of RecA primarily occurs at the 5′-end, whereas re-polymerization may occur from the end of pre-existing RecA filament that occupies the 3′ side of ssDNA in the 3′ to 5′ direction (Figure 4B). In most previous experiments, only 5′ to 3′ polymerization has been observed likely because it occurs much faster than the 3′ to 5′ polymerization. Although this interpretation is in contrast to the widely accepted unidirectional polymerization of RecA in a 5′ to 3′ direction (1,2), it is consistent with two previous experiments based on single-molecule FRET assay of the RecA dynamics at the 5′-end and the measurements of the lifetime of synaptic intermediates during homologous searching (4,44).
We found that a partially de-polymerized RecA nucleoprotein filament at low force could re-polymerize at higher tension (Figure 3B). Although the mechanisms of RecA polymerization at larger force are unclear, it can be qualitatively understood based on mechanical differences between the SSB nucleoprotein array and the RecA nucleoprotein filaments. The RecA nucleoprotein filament is helical, rigid and longer than both the naked ssDNA (45) and the SSB nucleoprotein complex formed under our experimental conditions (Figure 2). Therefore, the work done by force favours RecA polymerization. At sufficiently large forces, the energy supplied by force and polymerization energy of RecA may exceed the energy needed to displace SSB tetramers from ssDNA, resulting in polymerization.
In summary, our results have important biological implications. Our work elucidates how the nucleation, polymerization and stability of RecA nucleoprotein filaments are regulated by SSB or ssDNA in a force-dependent manner. In view of tight binding of SSB to ssDNA, these findings provide important insights into our understanding of the formation of RecA nucleoprotein filaments in vivo. Our work underscores the importance of tensile force and its resultant effects on ssDNA in evaluating the relative binding affinities of RecA versus SSB. Importantly, force-dependent competitive binding was also observed in protein–dsDNA interactions and in protein–protein interactions, which play important roles in regulating gene transcription and the mechanosensing of cells (46,47).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Text, Supplementary Figures 1–8 and Supplementary References [48–51].
FUNDING
National Research Foundation of Singapore through the Mechanobiology Institute at National University of Singapore (to J.Y.); Ministry of Education of Singapore [MOE2008-T2-1-096]; J. C. Bose National Fellowship from Government of India, New Delhi (to K.M.). Funding for open access charge: National Research Foundation of Singapore through the Mechanobiology Institute at National University of Singapore (to J.Y.).
Conflict of interest statement. I declare that the authors have no competing interests or other interests that might be perceived to influence the results and/or discussion reported in this article.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Michael Cox (University of Wisconsin, Madison, USA), Timothy M. Lohman (Washington University School of Medicine, USA) and Stephen Kowalczykowski (UC Davis, USA) for stimulating discussions.
REFERENCES
- 1.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 3.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 4.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 5.Cox JM, Abbott SN, Chitteni-Pattu S, Inman RB, Cox MM. Complementation of one RecA protein point mutation by another Evidence for trans catalysis of ATP hydrolysis. J. Biol. Chem. 2006;281:12968–12975. doi: 10.1074/jbc.M513736200. [DOI] [PubMed] [Google Scholar]
- 6.Stasiak A, Stasiak AZ, Koller T. Visualization of RecA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harb. Symp. Quant. Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- 7.Flory J, Tsang SS, Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc. Natl Acad. Sci. USA. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 9.Norden B, Elvingson C, Kubista M, Sjoberg B, Ryberg H, Ryberg M, Mortensen K, Takahashi M. Structure of RecA-DNA complexes studied by combination of linear dichroism and small-angle neutron scattering measurements on flow-oriented samples. J. Mol. Biol. 1992;226:1175–1191. doi: 10.1016/0022-2836(92)91060-3. [DOI] [PubMed] [Google Scholar]
- 10.van Loenhout MT, van der Heijden T, Kanaar R, Wyman C, Dekker C. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37:4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol. Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Ragunathan K, Joo C, Ha T. Real-time observation of strand exchange reaction with high spatiotemporal resolution. Structure. 2011;19:1064–1073. doi: 10.1016/j.str.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forget AL, Kowalczykowski SC. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature. 2012;482:423–427. doi: 10.1038/nature10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagi D, Tlusty T, Stavans J. High fidelity of RecA-catalyzed recombination: a watchdog of genetic diversity. Nucleic Acids Res. 2006;34:5021–5031. doi: 10.1093/nar/gkl586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith JD, Harris LD, Register JIII. Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb. Symp. Quant. Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 17.Muniyappa K, Williams K, Chase JW, Radding CM. Active nucleoprotein filaments of single-stranded binding protein and recA protein on single-stranded DNA have a regular repeating structure. Nucleic Acids Res. 1990;18:3967–3973. doi: 10.1093/nar/18.13.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muniyappa K, Shaner SL, Tsang SS, Radding CM. Mechanism of the concerted action of recA protein and helix-destabilizing proteins in homologous recombination. Proc. Natl Acad. Sci. USA. 1984;81:2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalczykowski SC, Clow J, Somani R, Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA. Demonstration of competitive binding and the lack of a specific protein-protein interaction. J. Mol. Biol. 1987;193:81–95. doi: 10.1016/0022-2836(87)90629-2. [DOI] [PubMed] [Google Scholar]
- 20.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 21.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 22.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys. J. 2011;100:517–523. doi: 10.1016/j.bpj.2010.12.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu H, Chen H, Zhang X, Qu Y, Marko JF, Yan J. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Res. 2011;39:3473–3481. doi: 10.1093/nar/gkq1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaze MB, Muniyappa K. RecA protein of Mycobacterium tuberculosis possesses pH-dependent homologous DNA pairing and strand exchange activities: implications for allele exchange in mycobacteria. Biochemistry. 1999;38:3175–3186. doi: 10.1021/bi9819125. [DOI] [PubMed] [Google Scholar]
- 26.Bianco PR, Weinstock GM. Interaction of the RecA protein of Escherichia coli with single-stranded oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:4933–4939. doi: 10.1093/nar/24.24.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenner SL, Mitchell RS, Morrical SW, Neuendorf SK, Schutte BC, Cox MM. Reca protein-promoted Atp hydrolysis occurs throughout Reca nucleoprotein filaments. J. Biol. Chem. 1987;262:4011–4016. [PubMed] [Google Scholar]
- 28.Fu H, Chen H, Marko JF, Yan J. Two distinct overstretched DNA states. Nucleic Acids Res. 2010;38: 5594–5600. doi: 10.1093/nar/gkq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Chen H, Fu H, Doyle PS, Yan J. Two distinct overstretched DNA structures revealed by single-molecule thermodynamics measurements. Proc. Natl Acad. Sci. USA. 2012;109:8103–8108. doi: 10.1073/pnas.1109824109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 31.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 32.Saleh OA, McIntosh DB, Pincus P, Ribeck N. Nonlinear low-force elasticity of single-stranded DNA molecules. Phys. Rev. Lett. 2009;102:068301. doi: 10.1103/PhysRevLett.102.068301. [DOI] [PubMed] [Google Scholar]
- 33.McIntosh DB, Saleh OA. Salt Species-Dependent Electrostatic Effects on ssDNA Elasticity. Macromolecules. 2011;44:2328–2333. [Google Scholar]
- 34.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:12. [Google Scholar]
- 35.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146: 222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bujalowski W, Lohman TM. Escherichia-Coli single-strand binding-protein forms multiple, distinct complexes with single-stranded-DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 38.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia-Coli single-strand binding protein-single-stranded DNA complexes-cation, anion, Ph, and binding density effects. J. Biol. Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 39.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 42.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 43.Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mani A, Braslavsky I, Arbel-Goren R, Stavans J. Caught in the act: the lifetime of synaptic intermediates during the search for homology on DNA. Nucleic Acids Res. 2010;38:2036–2043. doi: 10.1093/nar/gkp1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 46.Walthers D, Li Y, Liu Y, Anand G, Yan J, Kenney LJ. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 49.Cocco S, Yan J, Leger JF, Chatenay D, Marko JF. Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2004;70:011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Zhu X, Cong P, Sheetz MP, Nakamura F, Yan J. Differential mechanical stability of filamin a rod segments. Biophys. J. 2011;101:1231–1237. doi: 10.1016/j.bpj.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bustamante C, Smith SB, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.