Abstract
Ribosome biogenesis is a multi-step process that couples cell growth with cell proliferation. Although several large-scale analysis of pre-ribosomal particles have identified numerous trans-acting factors involved in this process, many proteins involved in pre-rRNA processing and ribosomal subunit maturation have yet to be identified. Las1 was originally identified in Saccharomyces cerevisiae as a protein involved in cell morphogenesis. We previously demonstrated that the human homolog, Las1L, is required for efficient ITS2 rRNA processing and synthesis of the 60S ribosomal subunit. Here, we report that the functions of Las1 in ribosome biogenesis are also conserved in S. cerevisiae. Depletion of Las1 led to the accumulation of both the 27S and 7S rRNA intermediates and impaired the synthesis of the 60S subunit. We show that Las1 co-precipitates mainly with the 27S rRNA and associates with an Nsa1 and Rix1-containing pre-60S particle. We further identify Grc3 as a major Las1-interacting protein. We demonstrate that the kinase activity of Grc3 is required for efficient pre-rRNA processing and that depletion of Grc3 leads to rRNA processing defects similar to the ones observed in Las1-depleted cells. We propose that Las1 and Grc3 function together in a conserved mechanism to modulate rRNA processing and eukaryotic ribosome biogenesis.
INTRODUCTION
The synthesis of ribosomes is intimately linked to cell growth and cell cycle progression in eukaryotes (1–5). Limiting ribosome biogenesis has been shown to result in cell size decrease in several organisms. In budding yeast, impaired ribosome synthesis delays progression through START by extending the length of the early G1-phase of the cell cycle (1–4). Thus, the rate of ribosome biogenesis couples the regulation of both cell size and passage through the cell cycle. The process of eukaryotic ribosome biogenesis has been studied comprehensively in the budding yeast Saccharomyces cerevisiae. Synthesis originates in the nucleolus on tandem repeats of ribosomal DNA (rDNA) where RNA Polymerase I (Pol I) transcribes a 35S pre-rRNA transcript. As this 35S pre-rRNA is transcribed it undergoes co-transcriptional and post-transcriptional modifications to direct the processing of the 35S pre-rRNA into the mature 18S, 5.8S and 25S rRNAs [reviewed in (6–11)]. These processing events occur co-transcriptionally or as the pre-rRNA is incorporated into the 90S pre-ribosomal particle (12). Cleavage at site A2 produces the 40S (containing the 18S rRNA) and 60S (containing the 5.8S and the 25S rRNAs) pre-ribosomal subunits from the 90S particle. The subunits undergo further independent modifications and are being processed as they move toward the cytoplasm. The pre-ribosomes are then exported through the nuclear pore complex and into the cytoplasm where the final steps of processing occur to form the mature 40S and 60S ribosomal subunits. Co-sedimentation experiments on density gradients and tandem affinity purification (TAP) followed by mass spectrometry in S. cerevisiae have led to the identification of several pre-ribosomal particles and a large number of non-ribosomal factors that participate in the synthesis of eukaryotic ribosomes (13–19). However, potentially due to the dynamic nature of the association of several proteins with these pre-ribosomal particles, the identities of many factors involved in eukaryotic ribosome biogenesis remain to be determined.
Las1 (Lethal in the Absence of SSD1-v1) was first isolated in a genetic screen for mutations that required the SSD1-v allele for viability in S. cerevisiae (20). Deletion of Las1 resulted in a G1 arrest with 80% of the cells unbudded, whereas overexpression of Las1 produced large cells with multiple bud projections, indicating that Las1 could be involved in regulating cell growth and cell cycle progression (20). We recently characterized the putative human homolog of Las1, Las1-Like (LAS1L), as a protein required for cell proliferation and ribosome biogenesis (21). Depletion of LAS1L results in a p53-dependent G1-phase cell cycle arrest, defects in pre-rRNA processing, and failure to synthesize mature 60S ribosomal subunits (21). LAS1L co-sediments with the pre-60S ribosomal particles and interacts with the mammalian homologs of the Rix1 complex (PELP1, WDR18, TEX10), the SUMO protease SENP3, and the polynucleotide kinase NOL9 (22,23). Although Las1 shares regions of sequence homology with LAS1L (21), a function for Las1 in pre-rRNA processing or ribosome synthesis had not been described in S. cerevisiae.
Grc3 was first described as an essential gene whose expression is cell cycle regulated (24). A large-scale genomic study later demonstrated that Grc3 was required for pre-rRNA processing and efficient synthesis of the mature 25S rRNA (18). Recent studies have further described Grc3 as a polynucleotide kinase whose activity is required for proper RNA Pol I transcription termination by the exonuclease Rat1 (25). In addition, the human homolog of Grc3, NOL9, was also recently described as a polynucleotide kinase involved pre-rRNA processing and synthesis of the 60S ribosomal subunit (26).
In this study, we demonstrate that Las1 is required for efficient ITS2 processing and production of the mature 25S and 5.8S rRNAs. Purification of different maturing particles revealed that Las1 associates with the 27S rRNA and both the Nsa1 and Rix1-containing pre-60S particles. We further demonstrate that Las1 interacts with the polynucleotide kinase Grc3, whose depletion results in rRNA processing defects similar to those observed in strains with reduced Las1 levels. We propose that Las1 and Grc3 act together to regulate the synthesis of the 25S rRNAs. Our findings demonstrate an evolutionarily and functionally conserved role of Las1 and Grc3 in pre-rRNA processing and eukaryotic ribosome biogenesis.
MATERIALS AND METHODS
Strains and microbiological techniques
Standard procedures were used for the growth and maintenance of S. cerevisiae (27). A complete list of strains used in this study can be found in Table 1. tetO7 promoter strains were cultured in YPD (1% yeast extract, 2% peptone, 2% dextrose) or synthetic dextrose (SD) minimal media containing 2% glucose and grown to an OD600 0.4–0.8. The Las1-Myc strain was constructed using one-step PCR as previously described (28). Transformants were selected on SD-His minimal media with 2% glucose and confirmed by PCR. To construct genomic LAS1 plasmids, LAS1 including 212 bp 5′ and 150 bp 3′ flanking sequences were amplified by PCR as EcoRI-SalI fragments and cloned into pRS413. To construct genomic GRC3 plasmids, GRC3 including 494 bp 5′ and 499 bp 3′ flanking sequences were amplified by PCR as BamHI-SalI fragments and cloned into pRS415. The FLAG-GRC3 plasmid was constructed by introducing a FLAG-Tag between the promoter and coding sequence of GRC3 via a two-step PCR procedure with the following primer sets: PCR 1: 5′-CCACTGCGGCCGC TTGTTGCGCACTAGGTACG3′ and 5′- CAAGTGGATCCCTTGTCATCGTCATCTTTATAATCCATAGCGGTAGAATATAATAGAA-3′. The first PCR fragment was cloned in pRS413 in the NotI-BamHI sites. PCR 2: 5′-CCACTGGATCC GTGATAGATTCCAAACAGG-3′ and 5′- CTCAAGTGTCGACCGATGTTGATTTTGAAGAAATTATC-3′. The second PCR was ligated to the PCR1 using the BamHI and SalI restriction sites. p426GPD-Flag-GRC3 was constructed using a two-step PCR process using the FLAG-GRC3 pRS415 plasmid as template. The GRC3 K252A/S253 mutant was constructed from pRS413-GRC3 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The pRS411-Sik1-RFP is described in (16). The p426GPD plasmid was obtained from the American Type Culture Collection. The tetO7 parental strain R1158, tetO7-LAS1 and tetO7-GRC3 strains used in this study along with the BY4741 strain were obtained from Open Biosystems. The Las1-GFP strain was obtained from Invitrogen.
Table 1.
Yeast strains used and constructed in this study
| Strain | Genotype | Reference |
|---|---|---|
| Wild type (R1158) | URA::CMV-tTA MATa his3-1 leu2-0 met15-0 | OpenBiosystems |
| Wild Type + pRS413 | URA::CMV-tTA MATa his3-1 leu2-0 met15-0; pRS413 | This Study |
| tetO7-LAS1 | URA::CMV-tTA LAS1::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0 | OpenBiosystems |
| tetO7-LAS1 + pRS413 | URA::CMV-tTA LAS1::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0; pRS413-HIS3 | This Study |
| tetO7-LAS1 + LAS1 | URA::CMV-tTA LAS1::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0; pRS413-LAS1-HIS3 | This Study |
| tetO7-LAS1 + Flag-Grc3 | URA::CMV-tTA LAS1::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0; pRS413-Flag-GRC3-HIS3 | This Study |
| tetO7-GRC3 | URA::CMV-tTA GRC3::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0 | OpenBiosystems |
| tetO7-GRC3 + pRS413 | URA::CMV-tTA GRC3::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0; pRS413-HIS3 | This Study |
| tetO7-GRC3 + GRC3 | URA::CMV-tTA GRC3::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0; pRS413-GRC3-HIS3 | This Study |
| tetO7-GRC3 + GRC3 K252A/S253A | URA::CMV-tTA GRC3::kanR-tetO7-TATA MATa his3-1 leu2-0 met15-0; pRS413-GRC3-K252A/S253A-HIS3 | This Study |
| tetO7-GRC3 Las1-Myc | URA::CMV-tTA GRC3::kanR-tetO7-TATA LAS1-13 MYC::HIS3MX6 MATa leu2-0 met15-0 | This Study |
| tetO7-IPI1 Las1-Myc + Flag-Grc3 | URA::CMV-tTA IPI1::kanR-tetO7-TATA LAS1-13 MYC::HIS3MX6 MATa leu2-0 met15-0; pRS415-Flag-GRC3-LEU2 | This Study |
| BY4741 | MATa his3-1 leu2-0 met15-0 ura3-0 | OpenBiosystems |
| Las1-Myc | LAS1-13MYC::HIS3MX6 MATa leu2-0 met15-0 ura3-0 | This Study |
| Las1-Myc + p426GPD | LAS1-13MYC::HIS3MX6 MATa leu2-0 met15-0 ura3-0; p426GPD-URA3 | This Study |
| Las1-Myc + Flag-Grc3 | LAS1-13MYC::HIS3MX6 MATa leu2-0 met15-0 ura3-0; p426GPD-FLAG-GRC3-URA3 | This Study |
| Las1-GFP | LAS1-GFP(S65T)::HIS3MX MATa leu2-0 met15-0 ura3-0 | Invitrogen |
| Las1-GFP + Sik1-RFP | LAS1-GFP(S65T)::HIS3MX MATa leu2-0 met15-0 ura3-0; pRS411-SIK1-mRFP-MET15 | This Study |
| Rix1-TAP Las1-Myc + Flag-Grc3 | RIX1-TAP::HIS3MX6 LAS1-13MYC::KANMX MATa leu2-0 ura3-0; pRS416-Flag-GRC3-URA3 | This Study |
| Las1-Myc Ssf1-TAP | LAS1-13MYC::KANMX SSF1-TAP::HIS3MX6 MATa leu2-0 ura3-0 | This Study |
| Las1-Myc Nsa1-TAP | LAS1-13MYC::KANMX NSA1-TAP::HIS3MX6 MATa leu2-0 ura3-0 | This Study |
| Las1-Myc Rix1-TAP | LAS1-13MYC::KANMX RIX1-TAP::HIS3MX6 MATa leu2-0 ura3-0 | This Study |
| Las1-Myc Arx1-TAP | LAS1-13MYC::KANMX ARX1-TAP::HIS3MX6 MATa leu2-0 ura3-0 | This Study |
| Las1-Myc Enp1-TAP | LAS1-13MYC::KANMX6 ENP1-TAP::HIS3MX6 MATa leu2-0 ura3-0 | This Study |
Cell proliferation assays and cell cycle analysis
For the growth curve assays, cells were grown in YPD or YPD with 20 µg/ml doxycycline for 24 h. 250 000 cells/ml were then added to either YPD or YPD with 20 µg/ml doxyclycline. Cells were harvested every 90 min and the OD600 was measured. For the dilution plating assays, cells were grown in YPD or SD-His minimal media and diluted to an OD600 of 0.05. 1:10 serial dilutions were plated on the respective media with or without 10 µg/ml doxycycline and incubated at 30°C for 48 h. For cell cycle analysis, cells were grown in YPD with 10 µg/ml doxycycline, washed with cold water and fixed in ethanol at a 70% final concentration for 16 h at 4°C. Cells were then washed in 50 mM sodium citrate pH 7.4, and resuspended in 50 mM sodium citrate pH 7.4 containing 250 μg/ml RNase A and incubated at 50°C for 1 h. Proteinase K (ThermoFisher) was added to a final concentration of 10 μg/ml and incubated at 50°C for an additional hour. Cells were then sonicated for 20S and propidium iodide was added to a final concentration of 16 μg/ml. Cells were incubated in the dark for 30 min and subjected to FACS analysis.
α-Factor synchronization assay
Cells were grown in YPD with 10 µg/ml doxycycline for 24 h and washed twice with water. Cells were then resuspended in YPD containing 10 µg/ml doxycycline and 2 µg/ml α-factor (ThermoFisher) at a density of 106 cells/ml. Cells were incubated with agitation at 30°C for 1 h and an additional 1 µg/ml α-factor was added for 1 h. Bud percentage was monitored periodically, and once >90% of cells were budded, cells were released from α-factor-induced arrest by washing twice with water and resuspended in YPD with 10 µg/ml doxycycline. Cells were harvested every 30 min and subjected to FACS analysis as described above.
Co-immunoprecipitation experiments, TAP and WB analysis
For mass spectrometry analysis of Las1-interacting proteins, the BY4741 and Las1-Myc strains were grown to log phase and lysed by vortexing in ELB buffer (50 mM HEPES pH 7.2, 300 mM NaCl, 2 mM EDTA, 0.5% NP-40) plus protease inhibitors (aprotinin, leupeptin, AEBSF) and glass beads. Lysates were cleared by centrifugation at 22 000g for 10 min at 4°C and the supernatant was incubated with 1 μg of antibody for 1 h with rotation at 4°C. Protein G-sepharose beads (Invitrogen) were added for another 1 h with rotation 4°C. Precipitated complexes were eluted in SDS-PAGE sample buffer and separated on a 10% SDS-PAGE. The gel was silver stained (Pierce silver stain kit) and proteins present only in the Las1-Myc lane were cut out, digested with trypsin and analysed by mass spectrometry. The TAP of the pre-60S particle was performed as described in (14). Purified proteins were separated by SDS-poly-acrylamide gel electrophoresis (SDS-PAGE) and 25% was transferred to a nitrocellulose membrane (Bio-Rad) for western blotting (WB) analysis. The rest of the purified complexes was separated on another SDS-PAGE and silver stained to evaluate the quality of the purifications (Pierce silver stain kit). The following antibodies were used: anti-TAP (Open Biosystems), anti-Myc 9E10 (Developmental Studies Hybridoma Bank), anti-FLAG M2 (Sigma) and anti-PSTAIRE (Santa Cruz Biotechnology).
RNA extraction, northern hybridization and primer extension analysis
For RNA extraction, cells were harvested by centrifugation at 3000g for 3 min at 4°C, washed twice in AE buffer (50 mM sodium acetate pH 5.2, 10 mM EDTA), and resuspended in AE buffer with 1% SDS. RNA was extracted by the addition of acidic phenol pH 5.2 equilibrated in AE buffer. Cells were frozen in a dry ice ethanol bath for 5 min, incubated at 65°C for 5 min and vortexed for 30 sec. Freeze/thaw/vortex cycles were repeated twice and then centrifuged at 16 000g for 15 min at 4°C. The aqueous layer was isolated and RNA was extracted with a 25:24:1 phenol/chloroform/isoamyl alcohol mixture. Samples were centrifuged at 16 000g for 15 min, and RNA was precipitated from the aqueous layer using sodium acetate and 100% ethanol at − 80°C for 20 min. RNA was pelleted at 16 000g for 20 min at 4°C, and the pellet was washed with 75% ethanol before resuspension in water.
For northern hybridization, RNA was separated on a 1.2% agarose-formaldehyde gel, rinsed with water and incubated in 0.01 N NaOH/3 M NaCl for 20 min. RNA was then transferred to a Hybond N + membrane (GE Healthcare) overnight. Oligonucleotide probes were 5′-end labeled using T4 polynucleotide kinase (New England Biolabs) in the presence of 32P-γ-ATP (MP Biomedical). The following probes were used for analysis of high molecular weight rRNAs: Probe A: 5′-GGAAATGCTCTCTGTTCAAAAAGCTTTTACACTCTTGACCAGCGCACTCC-3′; Probe B: 5′-TATCTTAAAAGAAGAAGCAACAAGCAGTAAAAAAGAAAGAAACCGAAATC-3; Probe D: 5′-GAAAGAAACTTCACAGCCTAGCAAGACCGCGCACTTAAGCGCAGGCCCGG-3′; Probe E: 5′-TACCTCTGGGCCCCGATTGCTCGAATGCCCAAAGAAAAAGTTGCAAAGAT-3′; Probe I: TCCAATGAAAAGGCCAGCAATTTCAAGTTAACTCCAAAGAGTATCACTCA-3′; Probe J: TTCCCAAACAACTCGACTCTTCGAAGGCACTTTACAAAGAACCGCACTCC-3′; probe C: 5′-CAGAAGGAAAGGCCCCGTTGGAAATCCAGTACACGAAAAAATCGGACCGG-3′; Probe F: 5′-CCAGTTACGAAAATTCTTG-3′. For analysis of low molecular weight rRNAs, equal amounts of total RNA were separated on an 8% denaturing polyacrylamide gel and transferred to a Hybond N + membrane (GE Healthcare). The following probes were used for low molecular weight analysis of rRNAs: Probe G: 5′-GCGTTGTTCATCGATGC-3′; Probe H: 5′-TGAGAAGGAAATGACGCT-3′; 5S: 5′-CTACTCGGTCAGGCTC-3′. The membranes were pre-hybridized in Church buffer (1% [wt/vol] bovine serum albumin, 1 mM EDTA, 0.25 M NaPO4 [pH 7.2], 7% [wt/vol] SDS) containing 100 µg/ml salmon sperm DNA at 65°C for 1 h. Labeled probes were then incubated with the membrane in Church buffer containing 100 µg/ml salmon sperm DNA at 65°C overnight. Membranes were washed in 0.1× SSC/ 0.1% SDS and exposed to autoradiography film.
Primer extension analysis was performed using the primer extension system—AMV reverse transcriptase from Promega. Products were separated on an 8% denaturing polyacrylamide gel and expose to an autoradiography film.
Sucrose gradient analysis
Cells were grown to an OD600 of 0.3. 100 ug/ml of cyclohexhimide was added to the culture for 10 min. Cells were washed in cold lysis buffer (50 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide, 7 mM BME) and lysed by vortexing with glass beads. Nine OD260 units were layered on top of a 7–47% sucrose gradient in lysis buffer and centrifuged at 40 K rpm for 2.5 h at 4°C in a Beckman SW-40 rotor. 600 µl fractions were then collected and the A260 measured for each fraction. Proteins were precipitated from 250 µl of each fraction using TCA, washed twice in 100% ethanol and resuspended in urea sample buffer (62.5 mM Tris–HCl pH 6.8, 3 M Urea, 2% SDS, 0.01% bromophenol blue, 0.1 M DTT). Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane for WB analysis. The anti-Myc 9E10 antibody was from the Developmental Studies Hybridoma Bank.
Pulse-chase analysis
Cells were grown in YPD media and doxycycline was added to a final concentration of 10 μg/ml for 12 h. Cells were grown to an OD600 of 0.8 in and washed twice in SD-Met media containing doxyclycline (10 μg/ml). Cells were resuspended in SD-Met media containing 10 µg/ml doxycycline and incubated for 30 min at 30°C. Cells were centrifuged at 3000g for 3 min and resuspended in 1 ml SD-Met media with 10 μg/ml doxycycline. 250 μCi of 3H-methyl-Methionine was added for 1 min. Chase was initiated by diluting 250 μl aliquots of labeled cells in 1.75 ml of prewarmed SD media containing 1 mg/ml methionine and 10 µg/ml doxycycline. Cells were harvested at 0, 2, 5 and 15 min, washed in ice cold water, and frozen on dry ice. RNA was harvested as described above and separated on a 1.2% agarose gel. RNA was transferred to a Hybond N + membrane (GE Healthcare) overnight. The membrane was then treated with En3Hance (Perkin-Elmer) and CCl4 and exposed to autoradiography film. Pulse-chase analysis of smaller RNAs was performed as described in (12) using [5,6-3H]-uracil.
Fluorescence microscopy
Cells were grown in SD-Met minimum media to log phase, washed with water and mounted on slides coated in poly-l-lysine. Fluorescence microscopy was performed on a Nikon TiE Wide field fluorescence microscope with a 100X Plan-Apo/1.4 NA Oil objective. Images were acquired using the NIS Elements AR 64 bit 3.10, SP7, Hotfix9 (Build 550) software. All microscopy was performed at room temperature and all images were prepared in Adobe Photoshop and Adobe Illustrator.
RESULTS
Las1 is required for cell cycle progression
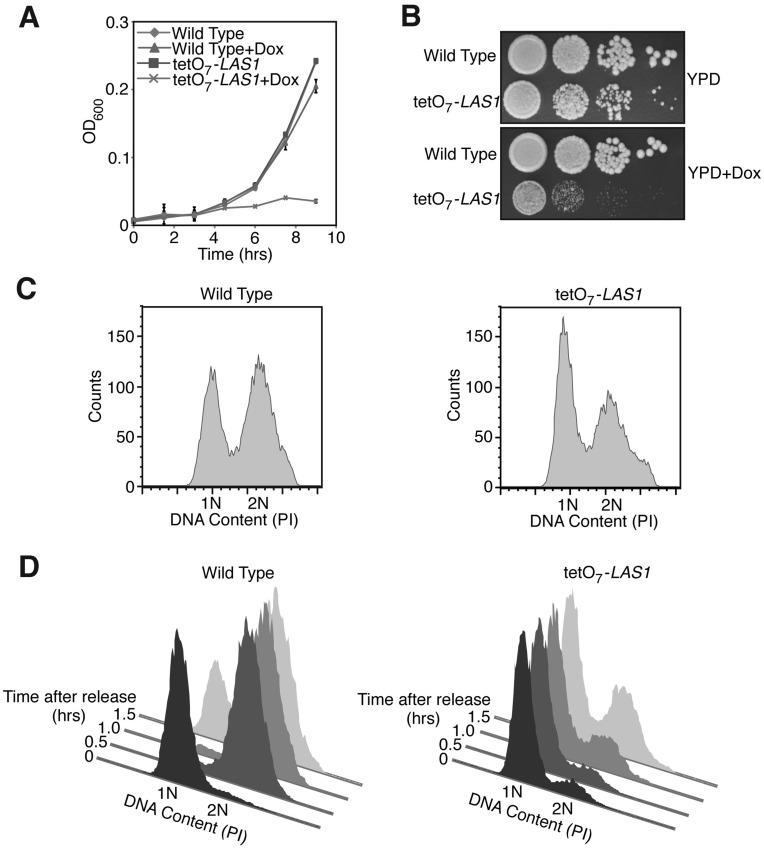
In order to further investigate the role of Las1 in S. cerevisiae, a tetO7-LAS1 strain, wherein the LAS1 allele is under the control of a tetracycline-regulated promoter, was utilized (29). To determine if Las1 is required for cell proliferation, wild type and tetO7-LAS1 strains were grown with or without doxycycline and proliferation rates were compared. Wild type and tetO7-LAS1 strains grown without doxycycline exhibited similar proliferation rates (Figure 1A). However, when grown in the presence of doxycycline, the tetO7-LAS1 cells displayed severely impaired proliferation rates compared to wild type (Figure 1A). This slow growth phenotype was also observed by serial dilution plating of wild type and tetO7-LAS1 strains, further strengthening the observation that Las1 is required for cell proliferation (Figure 1B). To investigate whether or not this lower proliferation rate is a result of a cell cycle arrest, wild type and tetO7-LAS1 strains were grown in the presence of doxycycline, and DNA profiles were examined by FACS analysis. In the presence of doxycycline the parental strain exhibited typical G1-phase and G2-phase profiles (Figure 1C). In contrast, tetO7-LAS1 cells grown in doxycycline exhibited a much larger G1-phase population compared to the G2-phase population, indicating that cells lacking Las1 have a prolonged G1-phase of the cell cycle (Figure 1C). To determine whether or not this protracted G1-phase is a result of a failure to progress from G1-phase into S-phase, wild type and tetO7-LAS1 strains were synchronized in G1 by the presence of alpha factor and released into the cell cycle. Various time points were collected and cell cycle profiles were determined by FACS analysis. Wild-type cells passed from G1-phase, through S-phase, and into G2-phase within 30 min of release into the cell cycle (Figure 1D). In addition, a G1 population was beginning to accumulate by 1.5 h after release (Figure 1D). However, tetO7-LAS1 cells failed to advance from G1 into the cell cycle within 30 min (Figure 1D). After 1.5 h only a small fraction of the cells had passed through S-phase into G2-phase (Figure 1D). These data suggest that Las1 is required for cells to progress from G1 into S-phase, and it is, this failure, to progress through the cell cycle that impairs cell proliferation in the absence of Las1.
Figure 1.
Las1 is required for cell cycle progression. (A) Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 24 h. Cells were then diluted into YPD with 10 µg/ml doxycycline and OD600 was measured at the indicated time points. (B) Wild type and tetO7-LAS1 strains were grown in YPD or YPD with 10 µg/ml doxycycline and then diluted to an OD600 of 0.05. Ten-fold serial dilutions were performed and 5 µl was plated on YPD plates with or without 10 µg/ml doxycycline and incubated at 30°C for 48 h. (C) Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline to log phase, stained with propidium iodide, and subjected to FACS analysis. (D) Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 24 h and cells were then arrested by the addition of α-factor. Cells were released into the cell cycle by washing with water and diluted into YPD with 10 µg/ml doxycycline. Indicated time points were collected and cells were stained with propidium iodide and analysed by FACS analysis.
Las1 is essential for pre-rRNA processing
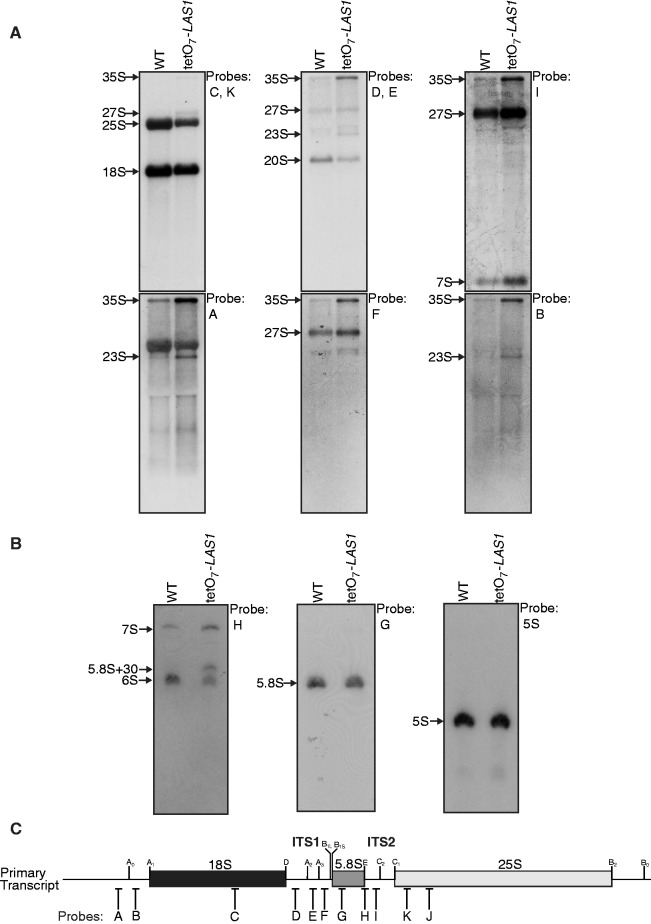
RNA Pol I transcribes a 35S pre-rRNA transcript that will generate the mature 18S, 5.8S and 25S rRNAs after several modification and processing steps that are summarized in Supplementary Figure S1 [(12), reviewed in (7)]. Depletion of factors involved in any of the steps in pre-rRNA processing results in accumulation of aberrant pre-rRNA species that can be identified by northern blot analysis. To determine if Las1 is involved in any of these pre-rRNA processing steps, wild type and tetO7-LAS1 strains were grown in the presence of doxycycline and total RNA was harvested and subjected to northern blot analysis with probes complementary to specific regions of pre-rRNA (probe locations are depicted in Supplementary Figure S1). Investigations using probes specific for the 25S (probe J), E-C2 (probe I), and A3-B1L (probe F) regions indicated an increase in the 27S and 7S rRNA intermediates and a decrease in the 25S mature rRNA upon Las1 depletion suggesting a defect in the endonucleolytic processing of the ITS2 region (Figure 2A). In addition, analysis of low molecular weight rRNAs using a probe specific for the E-C2 (probe H) region revealed accumulation of the 7S and 5.8S + 30 rRNAs and a loss of the 6S rRNA, suggesting that Las1 is required for the exonucleolytic processing of these rRNA intermediates (Figure 2B). Depletion of Las1 also resulted in the accumulation of the 35S pre-rRNA and the appearance of a 23S pre-rRNA, a species resulting from impaired A0, A1 and A2 cleavage (Figure 2A) (11,30–34). Both the accumulated levels of the 35S pre-rRNA and the presence of the 23S aberrant pre-rRNA upon Las1 depletion are likely due to a regulatory feedback mechanism often observed upon depletion of factors involved in pre-60S biogenesis. We also detected a slight loss of the 20S pre-rRNA and 18S mature rRNA upon Las1 depletion (Figure 2A). However the decrease in the 18S level was not as prominent as the loss of the mature 25S rRNA suggesting that Las1 is mainly involved in the maturation of the 60S particle rRNAs.
Figure 2.
Las1 is required for pre-rRNA processing. (A) Northern blot analysis of steady-state pre-rRNAs. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 12 h. Equal amounts of RNA were hybridized with probes specific for the indicated region of pre-rRNA. The position of each pre-rRNA is indicated on the left of each panel. Note that the signal detected with probe A above the 23S band corresponds to unspecific binding of the probe to the 25S rRNA. (B) Northern blot analysis of low molecular weight RNAs. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 12 h. Equal amounts of total RNA were separated on an 8% denaturing acrylamide gel and hybridized with probes specific for the indicated region of pre-rRNA. The positions of each pre-rRNA are indicated on the left of each panel. (C) Schematic representation of the rRNA primary transcript in budding yeast. The position of the probes utilized for the northern blotting analyses in A and B are indicated with letters.
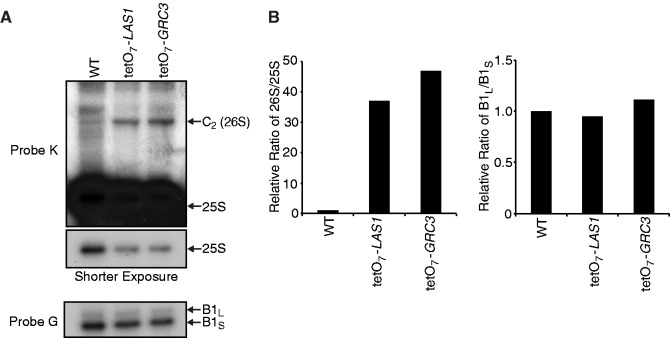
Primer extension analysis using an oligo-nucleotide located at the 5′-end of the 25S rRNA (probe K) revealed the accumulation of a 26S rRNA intermediate upon Las1 depletion, indicating that Las1 is also required for the last maturation step of the 25S rRNA after the C2 cleavage (Figure 3A and B). Extension with a primer located at the 5′-end of the 5.8S rRNA (probe G) did not detect any differences in the levels of the 5.8S B1L and B1S species upon Las1 depletion indicating that Las1 does not play a major role in ITS1 region processing (Figure 3A and B).
Figure 3.
Depletion of Las1 and Grc3 leads to the accumulation of a 26S rRNA intermediate. (A) Primer extension analysis. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 12 h. Equal amounts of RNA were subjected to primer extension analysis using probes as indicated on the left side of each panel. The position of each pre-rRNA is indicated on the right of each panel. (B) Densitometry analysis of the relative 26S/25S and B1L/B1S ratios for the primer extension analysis in (A).
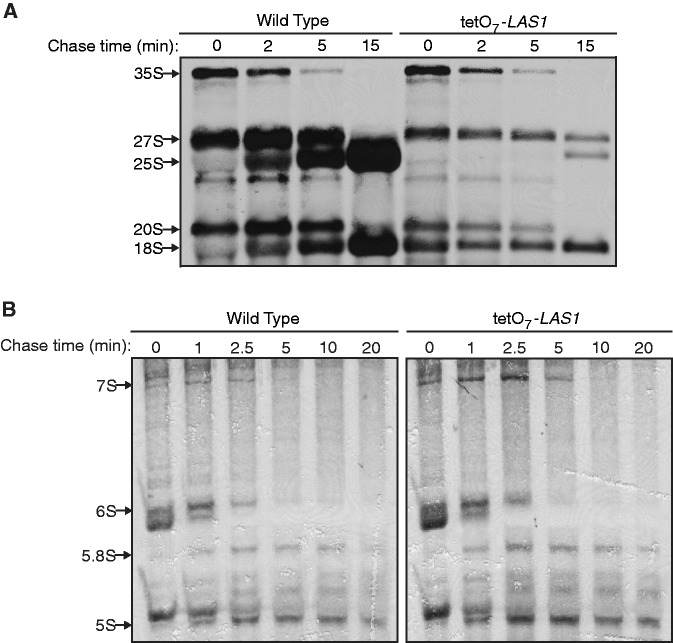
To confirm a Las1 requirement for synthesis of the mature 25S rRNA, wild type and tetO7-LAS1 strains were pulse labeled with 3H-methyl-methionine and chased for different time points to measure the level of newly synthesized rRNA. Depletion of Las1 resulted in delayed processing of the 27S rRNA accompanied by a severe defect in 25S rRNA production, confirming that Las1 is required for 27S processing and 25S rRNA synthesis (Figure 4A). Pulse-chase analysis of the smaller rRNAs revealed mainly the accumulation of the 7S rRNA also seen by northern blot analysis with Las1 depletion (Figures 2B and 4B). Collectively, these observations further suggest that Las1 is required specifically for rRNA processing at both the 5′- and 3′-ends of ITS2.
Figure 4.
Depletion of Las1 impairs 25S rRNA synthesis. (A) Pulse-chase analysis of newly synthesized pre-rRNAs. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 6 h. Cells were starved of methionine for 30 min and then pulse labeled with 3H-methyl-methionine for 1 min. Cells were then chased for the indicated time points with an excess of cold methionine and total RNA was harvested. 20 000 cpm was separated on a 1.2% agarose-formaldehyde denaturing gel, transferred to a nylon membrane, and visualized by autoradiography. (B) Pulse-chase analysis of newly synthesized small pre-rRNAs. Wild type and tetO7-LAS1 strains were grown in uracil-depleted media with 10 µg/ml doxycycline for 6 h. Cells were pulse labeled with [5,6-3H]-uracil for 1 min and then chased for the indicated time points with an excess of cold uracil and total RNA was harvested. Equal counts were loaded and separated on an 8% acrylamide-formaldehyde denaturing gel, transferred to a nylon membrane, and visualized by autoradiography.
Las1 interacts with the Grc3 polynucleotide kinase
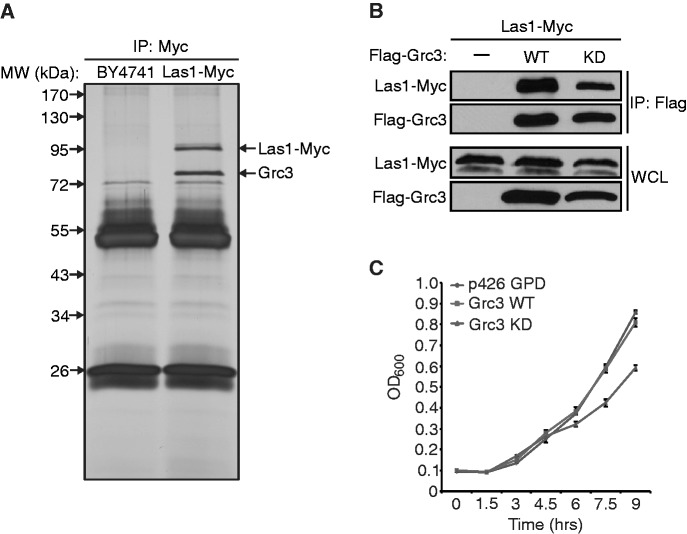
We previously demonstrated that the human homolog of Las1, LAS1L, interacts with members of the Rix1 complex (PELP1, TEX10, WDR18), the SUMO deconjugating enzyme SENP3, and the polynucleotide kinase NOL9 (23). To determine if these interactions were conserved and to further elucidate the role of Las1 in S. cerevisiae we employed a proteomic approach to identify its interacting proteins. Lysates from a strain expressing Las1-Myc and its parental strain BY4741 were immuno-precipitated using an anti-Myc antibody and Las1-interacting proteins were identified by mass spectrometry analysis. The two prominent bands observed were identified as Las1 and the polynucleotide kinase Grc3 by LC-MS/MS (Figure 5A). In order to verify the mass spectrometry data, Flag-Grc3 was overexpressed in the Las1-Myc strain and lysates were immunoprecipitated with an anti-Flag antibody. WB analysis using an anti-Myc antibody confirmed that Las1 forms a complex with Grc3 (Figure 5B). The immunoprecipitation analyses were performed in relatively high salt concentration (300 mM) with detergents, which suggests that the interaction between Las1 and Grc3 is stable and does not depend on the integrity of the pre-ribosomal particles. These observations indicate that Las1 and Grc3 could act together in a complex involved in regulating rRNA processing.
Figure 5.
Las1 interacts with Grc3 polynucleotide kinase. (A) Mass spectrometry analysis of Las1-associated proteins. BY4741 and Las1-Myc strains were grown to OD600 of 0.6 and proteins were immunoprecipitated from lysates with an α-Myc antibody. Bands were excised and proteins were identified by LC-MS/MS. Identified proteins are marked with an arrow. (B) The Las1-Myc + p426GPD and Las1-Myc + Flag-Grc3 strains were grown in SD-Ura to an OD600 of 0.6. Lysates were then immunoprecipitated with an α-Flag antibody. Immunoprecipitated proteins were separated by SDS-PAGE and analysed by WB using an anti-Flag (Grc3) or an anti-Myc (Las1) antibody. (C) BY4741 + empty vector (p426 GPD), overexpressing Grc3 WT or Grc3 KD were grown and diluted into SD-Ura and OD600 was measured at the indicated time points.
Because Grc3 seems to be a major binding partner of Las1, we sought to determine the requirement of each protein for the other’s stability. A tetO7-GRC3 strain expressing Las1-Myc under its endogenous promoter was grown with doxycycline to turn off Grc3 expression and Las1 protein levels were determined at various time points by WB analysis. Las1 protein levels began to drop at around 2 h after the addition of doxycycline (Figure 6A). Likewise, similar experiments using a tetO7-LAS1 strain exogenously expressing Flag-Grc3 under its endogenous promoter demonstrated that Las1 is required to promote Grc3 protein stability (Figure 6B). However, we did not observe any drop in Las1 or Grc3 protein levels upon depletion of Ipi1, a member of the Rix1 complex know to be required for 60S particle synthesis (Figure 6C). This suggests that the loss of Las1 and Grc3 proteins observed in Figure 3A and B are not due to a downstream effect of global inhibition of ribosome biogenesis. RT-qPCR analysis using total RNA from the 4 h time point in doxycycline confirmed that the decrease in protein levels was not due to a decrease in Las1 or Grc3 mRNA transcription after depletion of Grc3 and Las1, respectively (Figure 6D). This data further suggest that Las1 and Grc3 form a stable complex and that these proteins require each other to perform their role in rRNA processing.
Figure 6.
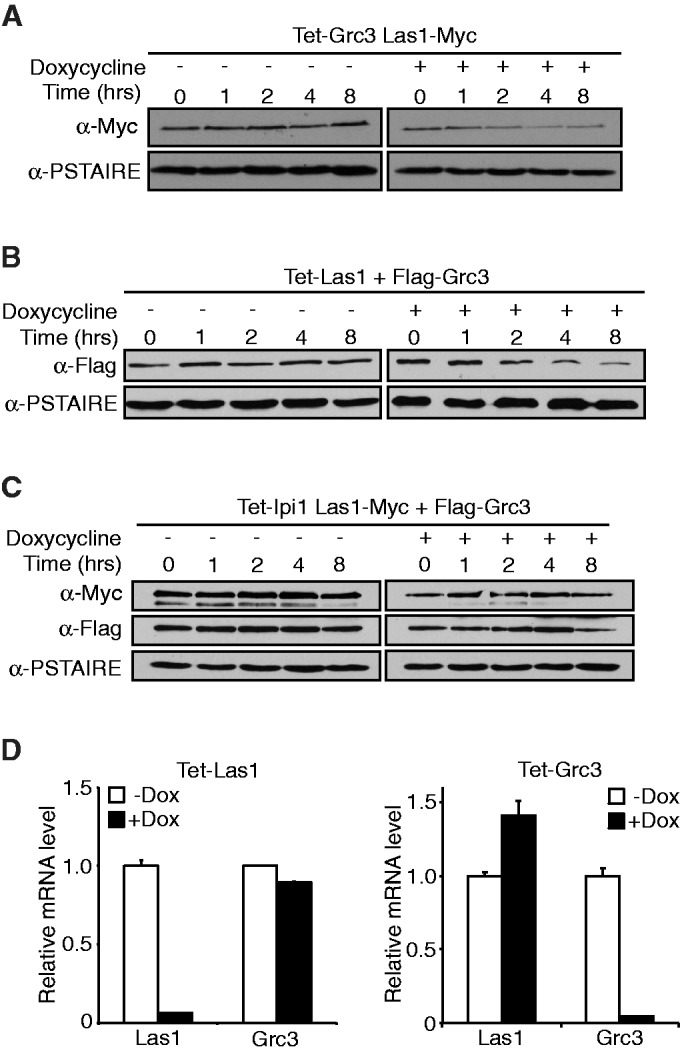
Las1 is required for Grc3 protein stability. (A) The tetO7-GRC3 Las1-Myc strain was grown to an OD600 of 0.1 and treated with (+) or without (−) 10 µg/ml doxycycline for the indicated period of time. Cells were harvested and equal amounts of protein were separated by SDS-PAGE and analysed by WB with an anti-Myc antibody (Las1) and PSTAIRE (loading control). (B) The tetO7-Las1 strain expressing Flag-Grc3 was grown to an OD600 of 0.1 and treated with (+) or without (−) 10 µg/ml doxycycline for the indicated period of time. Cells were harvested and equal amounts of protein were separated by SDS-PAGE and analysed by WB with an anti-Flag antibody (Grc3) and PSTAIRE (loading control). (C) The tetO7-Ipi1 Las1-Myc strain expressing Flag-Grc3 was grown to an OD600 of 0.1 and treated with (+) or without (−) 10 µg/ml doxycycline for the indicated period of time. Cells were harvested and equal amounts of protein were separated by SDS-PAGE and analysed by WB with an anti-Myc antibody (Las1), anti-Flag antibody (Grc3) and PSTAIRE (loading control). (D) Total RNA was harvested from the 2 h time points from the experiments in (A) or (B) (left and right panel, respectively). RT-qPCR analysis was performed to evaluate Las1 or Grc3 mRNA levels as indicated.
Grc3 kinase is required for ITS2 pre-rRNA processing
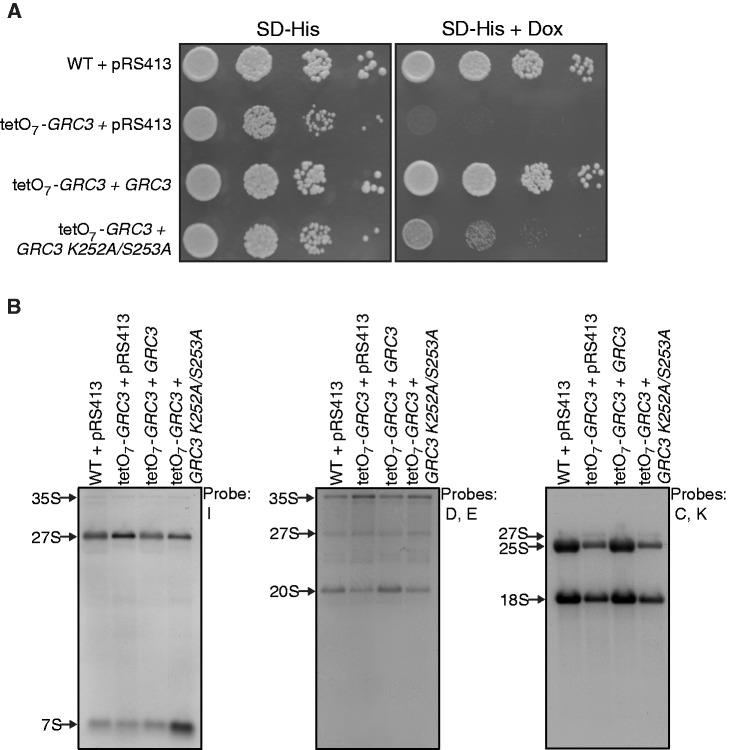
A large-scale screen for yeast non-coding rRNA processing mutants suggested a role for Grc3 in ITS2 rRNA processing (18). In a more recent study, Grc3 has been characterized as a polynucleotide kinase whose activity is required for efficient termination of Pol I transcription by the ‘torpedo’ action of Rat1 (25). We next investigated more precisely the role of Grc3 in rRNA processing to evaluate if depletion of Grc3 would lead to defects similar to the ones observed in the absence of Las1 (Figure 7A). Northern blot analysis revealed that depletion of Grc3 or expression of a Grc3 K252A/S253A kinase dead (KD) mutant (25) both caused accumulation of the 35S pre-rRNA and the 27S pre-rRNA accompanied by a loss of the 25S mature rRNA as we observed for Las1 depletion (Figure 7B). In addition, depletion of Grc3 or expression of the KD mutant also resulted in a minor loss of both the 20S pre-rRNA and 18S mature rRNA (Figure 7B). Primer extension analysis confirmed the decrease in the mature 25S rRNA synthesis and revealed the accumulation of a 26S intermediate upon depletion of Grc3 (Figure 3). Interestingly, only the expression of the Grc3 KD mutant resulted in an increase of the 7S pre-rRNA similar to what we observed upon Las1 depletion. This suggests that Grc3 may not be essential for efficient exonucleolytic processing of the 7S rRNA. It is possible that the Grc3 KD mutant sequesters Las1 and other processing factors required for that processing step into an inactive complex. To test that hypothesis, we sought to determine if the KD Grc3 could still interact with Las1. Immuno-precipitation experiments in the Las1-Myc strain overexpressing a KD Grc3 revealed that Las1 interacts with Grc3 regardless of kinase activity (Figure 2B). In addition, growth curve analysis in the Las1-Myc strain overexpressing Flag-Grc3 or Flag-Grc3 KD demonstrated that overexpressing the KD Grc3 decreases cell proliferation (Figure 2C), suggesting that the KD Grc3 could act as a dominant negative that sequesters Las1 into inactive complexes. Taken together, our data indicate that the kinase activity of Grc3 is required for ITS2 pre-rRNA processing and synthesis of the mature 25S rRNA. Furthermore, the pre-rRNA processing defects observed upon depletion of Grc3 are nearly identical to the processing defects exhibited by Las1 depletion, strengthening our hypothesis that Las1 and the Grc3 polynucleotide kinase function together in regulating pre-rRNA processing and eukaryotic ribosome biogenesis.
Figure 7.
Grc3 kinase activity is required for cell proliferation and pre-rRNA processing. (A) Wild type + pRS413, tetO7-GRC3 + pRS413, tetO7-GRC3 + GRC3, and tetO7-GRC3 + GRC3 K252A/S253A strains were grown in SD-His or SD-His with 10 µg/ml doxycycline and then diluted to and OD600 of 0.05. Ten-fold serial dilutions were performed and 5 µl was plated on SD-His plates with or without 10 µg/ml doxycycline and incubated at 30°C for 48 h. (B) Northern blot analysis of steady-state pre-rRNAs. Wild type + pRS413, tetO7-GRC3 + pRS413, tetO7-GRC3 + GRC3, and tetO7-GRC3 + GRC3 K252A/S253A strains were grown in SD-His with 10 µg/ml doxycycline for 12 h. Equal amounts of RNA were hybridized with probes specific for the indicated region of pre-rRNA. The position of each pre-rRNA is indicated on the left of each panel.
Las1 and Grc3 are required for 60S ribosomal particles synthesis
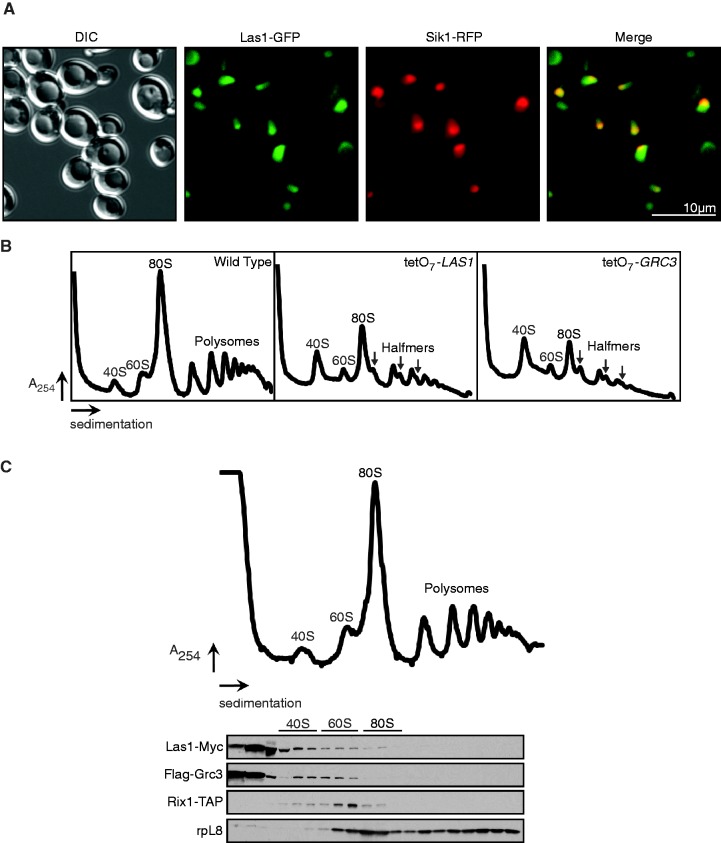
Our previous findings have indicated that LAS1L, the human putative homolog of Las1, is a predominantly nucleolar protein required for ribosome biogenesis (21). To determine the sub-cellular localization of Las1 in S. cerevisiae, we employed fluorescent microscopy using a Las1-GFP tagged strain. Sik1-RFP expressed on an exogenous plasmid was used as a marker for the nucleolus (16,35). Cells were grown to log phase and the sub-cellular localization of Las1-GFP was examined. Las1 exhibited a strong co-localization with Sik1, indicating that Las1 is a nucleolar protein (Figure 8A).
Figure 8.
Las1 localizes to the nucleolus and co-fractionates with pre-60S ribosomal particles. (A) Las1-GFP + Sik1-RFP strain was grown in SD-Met to log phase and GFP and RFP signals were analysed by fluorescence microscopy. DAPI was used to costain the nucleus. (B) The tetO7-Parental (WT), tetO7-LAS1 or tetO7-GRC3 strains were grown in YPD to an OD600 of 0.3. Cellular extracts were separated on a 7–47% sucrose gradient and A254 were recorded. The positions of the 40S and 60S pre-ribosomal subunits, 80S ribosomes and polysomes or halfmers are indicated on the profile. (C) The Las1-Myc Rix1-TAP strain expressing Flag-Grc3 was grown in YPD to an OD600 of 0.3. Cellular extracts were separated on a 7–47% sucrose gradient, OD240 were recorded and 600 µl fractions were collected. Proteins were precipitated from each fraction, separated by SDS-PAGE, and analysed by WB with anti-Myc, anti-TAP, anti-Flag or anti-rpL8 antibodies as indicated. The positions of the 40S and 60S pre-ribosomal subunits, 80S ribosomes and polysomes are indicated on the profile.
Our previous studies have determined that LAS1L is required for the formation of the 60S pre-ribosomal particle (21). Moreover, the ITS2 rRNA processing defects and dramatic loss of the mature 25S rRNA observed in Las1 and Grc3 depleted strains suggest that these two proteins are required specifically for synthesis of the 60S subunit. To confirm a role for Las1 and Grc3 in the synthesis of the 60S pre-ribosomal particles, polysome profile analyses were performed on whole cell extracts of the wild type, tetO7-LAS1 and tetO7-GRC3 strains. As expected, depletion of Las1 and Grc3 resulted in a reduction of the 60S particles but did not affect formation of the 40S subunit (Figure 8B). Moreover, sucrose gradient centrifugation performed on extracts from a Rix1-TAP Las1-Myc strain expressing Flag-Grc3 revealed that both Las1 and Grc3 co-sedimented with pre-60S subunits (Figure 8C). The functionality of the Flag-Grc3 construct was confirmed by its ability to rescue the growth defect phenotype observed upon Grc3 depletion (Supplementary Figure S2D). Although their sedimentation patterns were not identical, Las1-Myc and Flag-Grc3 also co-sedimented with fractions overlapping with Rix1 and with lighter fractions probably corresponding to free proteins of smaller sub-complexes (Figure 8C). Taken together, these analyses indicate that Las1 and Grc3 are factors involved in processing of pre-rRNA specific to the 60S ribosomal particle.
Las1 associates with Nsa1 and Rix1-containing pre-60S particles
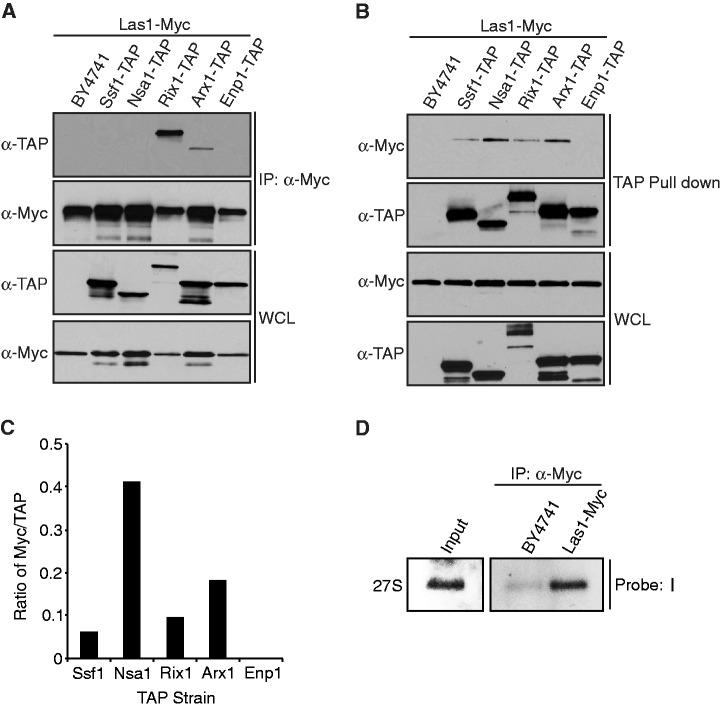
The use of TAP methods has lead to the isolation of different pre-60S particles ranging from early nucleolar to cytoplasmic intermediates (14,17,36,37). Each of these particles is characterized by the presence of specific proteins (Ssf1, Nsa1, Rix1 and Arx1) and different rRNA intermediates along the pathway of maturation [reviewed in (8)]. To determine which pre-60S particle(s) Las1 associates with, Las1-Myc was immunoprecipitated from lysates of the Ssf1-TAP, Nsa1-TAP, Rix1-TAP, Arx1-TAP and Enp1-TAP (control for 40S subunit) strains separately in conditions that preserve the integrity of the pre-ribosomal particles. WB analysis using an anti-TAP antibody revealed that Las1 associates mainly with a particle containing Rix1 (Figure 9A). This data are consistent with previous observations that human Las1 associates with the homologs of the Rix1 complex (PELP1, TEX10 and WDR18) (22,23). However, the reverse experiment using TAP pulldown on IgG sepharose beads demonstrated that Las1 associates also with the Nsa1 particle and to a lesser extent with Rix1 and Arx1 (Figure 9B and C, and Supplementary Figure S3). This discrepancy could be due to the inaccessibility of the tags used for pulldown on certain particles. For example, Las1-Myc could be present on the Nsa1 particle but hidden due to the binding of other factors that are not there on the Rix1 particle (Figure 9A).
Figure 9.
Las1 associates with the Nsa1 and Rix1 pre-60S ribosomal particles. (A) The Las1-Myc, Las1-Myc Ssf1-TAP, Las1-Myc Nsa1-TAP, Las1-Myc Rix1-TAP, Las1-Myc Arx1-TAP or Las1-Myc Enp1-TAP strains were grown to log phase and protein lysates subjected to immunoprecipitation in conditions that preserve the integrity of the pre-ribosomal particles using an anti-Myc antibody. Immunoprecipitated complexes were separated by SDS-PAGE and analysed by WB using an anti-TAP or an anti-Myc (Las1) antibody. WCL, whole cell lysate. (B) The Las1-Myc, Las1-Myc Ssf1-TAP, Las1-Myc Nsa1-TAP, Las1-Myc Rix1-TAP, Las1-Myc Arx1-TAP or Las1-Myc Enp1-TAP strains were grown to log phase and protein lysates subjected to TAP in conditions that preserve the integrity of the pre-ribosomal particles. Purified complexes were separated by SDS-PAGE and analysed by WB using an anti-TAP or an anti-Myc (Las1) antibody. WCL, whole cell lysate. (C) Densitometry quantification of the ratio of Myc-Las1 over each respective TAP-proteins from panel B. (D) The BY4741 and Las1-Myc strains were grown to log phase and protein lysates subjected to immunoprecipitation with an ant-Myc antibody in conditions that preserve the integrity of the pre-ribosomal particles. RNA was extracted from the immuno-purified particles and subjected to northern blot analysis using an E-C2 probe (I). Total RNA was loaded as input.
Each of the pre-60S particles tested is characterized by the presence of different maturing rRNA species [for a review see (8)]. The Nsa1 particle is characterized by the presence of the 27SB rRNA intermediate whereas, in the Rix1-associated particle the 27S pre-rRNA has been almost completely processed into mature 25S and 7S rRNAs (17). To clarify the association of Las1 to the Nsa1 and Rix1 particles we sought to determine the nature of the rRNA intermediate co-precipitating with Las1. RNA-immunoprecipiation of lysates from a Las1-Myc strain using an anti-Myc antibody revealed that Las1 associates mainly with the 27S rRNA intermediate (probe I) (Figure 9D) and to a much lesser extent with the 7S rRNA (not shown). The 27S rRNA intermediate was also detected using probes specific to the 25S and 5.8S rRNA species (not shown). No other rRNA intermediates were detected using the probes listed in Supplementary Figure S1. Taken together, this suggests that Las1 associates with an early pre-60S particle containing the 27S rRNA, Nsa1 and Rix1. As Las1 is required for efficient ITS2 cleavage it could transiently interact with a later particle containing the 7S rRNA and Rix1.
DISCUSSION
In this study, we demonstrate that the function of Las1 in regulating ribosome biogenesis is conserved in budding yeast. Indeed, our data indicate that Las1 is required specifically for the processing of the ITS2 region of the pre-rRNA and for the formation of the mature 25S rRNA that is contained in the 60S ribosomal subunit. Moreover, our analysis revealed that Las1 co-sediments with early pre-60S ribosomal particles containing Nsa1, Rix1 and the 27S rRNA intermediate. Additionally, we show that Las1 associates with the 5′ polynucleotide kinase Grc3 and that depletion of Grc3 or expression of a KD mutant leads to similar pre-rRNA processing defects. We propose that Las1 and Grc3 function together in a conserved mechanism to modulate pre-rRNA processing and the synthesis of eukaryotic ribosomes.
We previously demonstrated that the human homolog LAS1L co-sediments with the 60S pre-ribosomal particles and that its depletion leads to the accumulation of a 32S rRNA intermediate (equivalent of the 27S rRNA) and a dramatic loss of the mature 28S rRNA (21,23). Similarly, depletion of Las1 results in an accumulation of the 27S pre-rRNA species (Figures 4A and 6B), suggesting that like its human homolog, Las1 is also required for cleavage at site C2 (21). Several possibilities exist for the function of Las1 in C2 cleavage. Las1 could be responsible for recruiting the as of yet unidentified endonuclease performing the C2 cleavage to the pre-60S particle. It is also feasible that Las1 is responsible for directing rRNA modifications required for the C2 endonuclease to function or provide a docking site for processing factors involved in this cleavage. We observed that depletion of Las1 also results in accumulation of low molecular weight pre-rRNAs, specifically the 7S and 5.8S + 30 pre-rRNAs, which are 3′ extended precursors of the mature 5.8S rRNA. These intermediates are usually processed by the 3′–5′ exonucleolytic RNA exosome (30,31,38–40). It is possible that depletion of Las1 impairs the 3′ processing of these pre-rRNAs by the exosome and further experiments will be required to confirm this. Primer extension analysis also revealed the accumulation of a 26S rRNA intermediate upon Las1 depletion suggesting that Las1 is required for processing at both the 5′ and 3′ extremities of ITS2 post C2 cleavage. The 5′-3′ exoribonuclease Rat1 and its associates cofactor Rai1 have been shown to be required for the 5′-end processing of the 26S rRNA (41,42). Deletion of RAI1 was also shown to inhibit both Rat1p-dependent 5′-end processing and Rrp6p-dependent 3′-end processing of 5.8S rRNA (39). While this manuscript was in revision, Schillewaert et al. (43) also reported similar processing defects associated with Las1 depletion using thermosensitive and galactose-regulated strains. Interestingly, they demonstrated that Las1 copurifies with pre-ribosomal particles that contain Rat1 and its cofactor Rai1 (43). Taken together, this suggests that Las1 is required for both Rat1–Rai1-dependent synthesis of the 5′-end of the 25S rRNA as well as for the exosome Rrp6-dependent formation of the 5.8S rRNA 3′-end.
Using a proteomic approach, we identified Grc3 as a Las1-interacting protein. Grc3 has been previously identified as an essential protein in S. cerevisiae and a factor required for pre-rRNA processing (18,24). Moreover, Grc3 was recently characterized as a 5′-polynucleotide kinase required for transcription termination by RNA Pol I (25). In this study, we demonstrate that Grc3 and its kinase activity are required for ITS2 pre-rRNA processing. Depletion of Grc3 or expression of a Grc3 KD mutant results in accumulation of rRNA precursors also seen upon depletion of Las1, suggesting that Las1 and Grc3 kinase cooperate in the regulation of pre-rRNA processing. The exact function of Las1 in the regulation of Grc3 kinase activity remains unclear. Our data suggest that Las1 is required for stabilization of Grc3 and vice versa. Las1 could also be an RNA-binding protein that allows Grc3 to phosphorylate the pre-rRNA substrate for subsequent processing. Recent publications have also linked Grc3 with the 5′ exonuclease Rat1 and its cofactor Rai1. Braglia et al. (25) determined that the 5′ polynucleotide kinase activity of Grc3 was required for the ‘torpedo’ action of Rat1 to terminate Pol I transcription efficiently. Furthermore, Las1 and Grc3 were found by TAP and mass spectrometry analysis to associate with Rai1, which acts as a cofactor for Rat1 (43,44). Rat1 preferentially processes 5′-monophosphorylated RNAs, and it has been hypothesized that Grc3/NOL9 kinase activity would help maintain a 5′-monophosphate at the end of the post-cleaved rRNA and allow efficient processing by Rat1 (26,45). Future work will be directed toward investigating if Las1 interacts in a complex with Grc3 and Rat1 and how Las1 regulates the activity of these proteins. Of interesting note is the accumulation of the 7S precursor that does not occur with Grc3 depletion, but rather only occurs with expression of the Grc3 KD mutant (Figure 8B). Our data suggest that the Grc3 KD mutant could be sequestering Las1 and possibly Rat1 into non-functional complexes, leading to an inefficient processing of the 7S rRNA to the 5.8S rRNA. It remains to be determined if Grc3 interacts with other processing factors as well and if it also functions with the RNA exosome in processing of the 7S rRNA.
Interestingly, we and others have previously determined that LAS1L interacts with NOL9, the human homolog of Grc3, as well as with the human homologs of the Rix1 complex (PELP1, TEX10, WDR18) (22,23). Our mass spectrometry analysis of Las1-binding proteins revealed that Las1 and Grc3 were present in a complex at almost a stoichiometric ratio. The interaction detected was unlikely on the pre-60S particle as high salt concentration and detergents were used in the lysis and immuno-precipitation buffers. Indeed, we previously demonstrated that human LAS1L can interact with NOL9 both on and off the pre-60S particle (23). Surprisingly, we did not detect any interaction with the Rix1 complex as we did with LAS1L using the same immuno-precipitation conditions (23). This suggest that in S. cerevisiae, the interaction between Las1 and the Rix1 complex may not be as strong or stable as observed in human cells (23). Nevertheless, in this study we demonstrate that Las1 interacts with Rix1 and Nsa1 in conditions that preserve the integrity of the pre-60S ribosomal particles. RNA-immunoprecipitation experiments also revealed that Las1 associates mainly with a particle containing the 27S rRNA intermediate confirming a role for Las1 in C2 cleavage. We only detected a weak 7S rRNA signal coming down with Las1 suggesting that once the C2 cleavage is completed, Las1 may only transiently associate with the Rix1 particle that contains, in majority, the 25S and 7S/5.8S rRNAs.
Further insight into the functions of Grc3 and Las1 has recently been demonstrated in Schizosaccharomyces pombe (46). In S. pombe Grc3 was found to interact with Las1 and members of the Rix1 complex (46). Interestingly, Grc3, Las1 and the Rix1 complex proteins all were shown to be localized to heterochromatic regions and to be required for heterochromatin silencing in S. pombe, although no such function for Las1 or Grc3 has been described in S. cerevisiae. As post-transcriptional heterochromatic silencing in S. cerevisiae is thought to be mediated by some components of the RNA exosome (47–49), future experiments will be directed toward investigating a possible cooperation of Las1 and Grc3 with the exosome in the silencing of heterochromatic transcripts.
Using budding yeast as a model organism, our data shed further light on pre-rRNA processing pathways and the complex process of eukaryotic ribosome biogenesis. Importantly, we identify an evolutionarily conserved interaction for Las1 and Grc3 and further demonstrate a conserved role for this interaction in the modulation of pre-rRNA processing and the synthesis of eukaryotic ribosomes from S. cerevisiae to mammals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
University of Texas Health Science Center Medical School at Houston start-up funds (to C.D.); National Institutes of Health [GM53655 to A.W.J.]. Funding for open access charge: The University of Texas HSC Houston Medical School Startup Funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors like to thank Kevin Morano and Ambro van Hoof for reagents and experimental advice. They thank Ghislain Breton and Erica Cassimere for their support and critical review of the manuscript. They also thank Claire Mauvais for excellent technical assistance.
REFERENCES
- 1.Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr. Biol. CB. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 7.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochimica et biophysica acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 11.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Ann. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 12.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 14.Kressler D, Roser D, Pertschy B, Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 19.Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doseff AI, Arndt KT. LAS1 is an essential nuclear protein involved in cell morphogenesis and cell surface growth. Genetics. 1995;141:857–871. doi: 10.1093/genetics/141.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castle CD, Cassimere EK, Lee J, Denicourt C. Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol. Cell. Biol. 2010;30:4404–4414. doi: 10.1128/MCB.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castle CD, Cassimere EK, Denicourt C. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol. Biol. Cell. 2012;23:716–728. doi: 10.1091/mbc.E11-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Moghazy AN, Zhang N, Ismail T, Wu J, Butt A, Ahmed Khan S, Merlotti C, Cara Woodwark K, Gardner DC, Gaskell SJ, et al. Functional analysis of six novel ORFs on the left arm of chromosome XII in Saccharomyces cerevisiae reveals two essential genes, one of which is under cell-cycle control. Yeast. 2000;16:277–288. doi: 10.1002/(SICI)1097-0061(200002)16:3<277::AID-YEA524>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Braglia P, Heindl K, Schleiffer A, Martinez J, Proudfoot NJ. Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I. EMBO Rep. 2010;11:758–764. doi: 10.1038/embor.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heindl K, Martinez J. Nol9 is a novel polynucleotide 5′-kinase involved in ribosomal RNA processing. EMBO J. 2010;29:4161–4171. doi: 10.1038/emboj.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 28.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 30.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bax R, Raue HA, Vos JC. Slx9p facilitates efficient ITS1 processing of pre-rRNA in Saccharomyces cerevisiae. RNA. 2006;12:2005–2013. doi: 10.1261/rna.159406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl L, Bommankanti A, Li X, Hayden L, Jones A, Khan M, Oni T, Zengel JM. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA. 2009;15:1407–1416. doi: 10.1261/rna.1302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 36.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 37.Fatica A, Cronshaw AD, Dlakic M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 38.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 39.Fang F, Phillips S, Butler JS. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanchin NI, Goldfarb DS. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 1999;27:1283–1288. doi: 10.1093/nar/27.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geerlings TH, Vos JC, Raue HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′–>3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schillewaert S, Wacheul L, Lhomme F, Lafontaine DL. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol. Cell. Biol. 2012;32:430–444. doi: 10.1128/MCB.06019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 2003;23:2042–2054. doi: 10.1128/MCB.23.6.2042-2054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J. Biol. Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 46.Kitano E, Hayashi A, Kanai D, Shinmyozu K, Nakayama J. Roles of fission yeast Grc3 protein in ribosomal RNA processing and heterochromatic gene silencing. J. Biol. Chem. 2011;286:15391–15402. doi: 10.1074/jbc.M110.201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol. Cell. Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.