Abstract
Developmentally regulated transcription often depends on physical interactions between distal enhancers and their cognate promoters. Recent genomic analyses suggest that promoter–promoter interactions might play a similarly critical role in organizing the genome and establishing cell-type-specific gene expression. The Igf2/H19 locus has been a valuable model for clarifying the role of long-range interactions between cis-regulatory elements. Imprinted expression of the linked, reciprocally imprinted genes is explained by parent-of-origin-specific chromosomal loop structures between the paternal Igf2 or maternal H19 promoters and their shared tissue-specific enhancer elements. Here, we further analyze these loop structures for their composition and their impact on expression of the linked long non-coding RNA, Nctc1. We show that Nctc1 is co-regulated with Igf2 and H19 and physically interacts with the shared muscle enhancer. In fact, all three co-regulated genes have the potential to interact not only with the shared enhancer but also with each other via their enhancer interactions. Furthermore, developmental and genetic analyses indicate functional significance for these promoter–promoter interactions. Altogether, we present a novel mechanism to explain developmental specific imprinting of Nctc1 and provide new information about enhancer mechanisms and about the role of chromatin domains in establishing gene expression patterns.
INTRODUCTION
Transcription of developmentally regulated or tissue-specific genes often depends on promoter activation by cell-type-specific enhancers. Although promoters and their cognate enhancers may be separated by great distances on the linear chromosome, studies on several model systems confirm that transcriptional activation is invariably associated with the formation of DNA loop structures that bring the promoter and enhancer into close physical proximity (1). Recent genomic analyses suggest that enhancers often regulate multiple promoters simultaneously and indicate that interactions between co-regulated promoters and shared enhancers organize the genome into functional domains (2).
Using genome-wide chromatin interaction analysis, Li et al. (3) recently established that cell-type-specific promoter–promoter interactions are also widespread in animal cells. Moreover, their data suggest that these interactions might play important regulatory roles. Specifically, they proposed that promoter–promoter interactions can act cooperatively to activate gene expression and that promoter–promoter interactions might be critical components of the chromosomal structures that underlie coordinated transcription in eukaryotic cells.
The Igf2/H19 locus has proven to be a particularly useful model system for elucidating the molecular details and functional significance of DNA loop structures in regards to coordinate gene expression via tissue-specific enhancers (4). Expression of Igf2 and H19 is each dependent on shared downstream enhancers. The best-characterized enhancers are centered at +92 and +108 kb (all base pairs are given relative to the start site for Igf2 isoform 1) and drive expression in liver and in skeletal muscle, respectively (Figure 1) (5–11). Additional tissue-specific enhancers are located even further downstream (5,8). Chromatin conformation configuration (3C) analyses of fetal liver provided early support for the importance of physical interactions of promoter and enhancer elements in gene activation (12–14).
Figure 1.
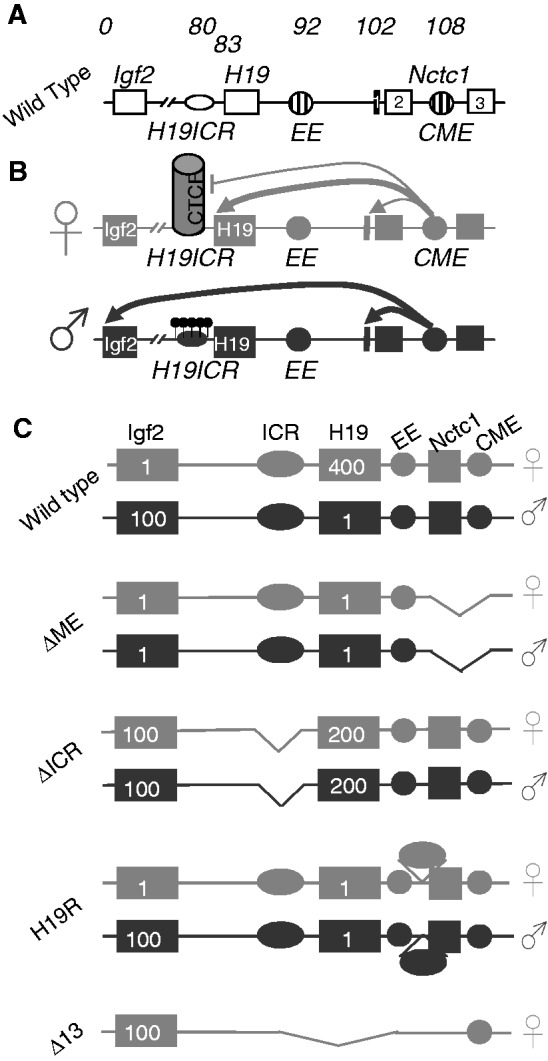
The Igf2/H19/Nctc1 locus. (A) Organization of the wild-type locus. Open rectangles denote the Igf2, H19 and Nctc1 genes. Open oval denotes the H19ICR. Striped circles denote the endoderm (EE) and CME. Numbers above the line indicate kilobase relative to the Igf2 isoform 1 transcriptional start site. (B) The insulator model for imprinted expression at the Igf2/H19 locus. On the maternal (gray) chromosome, the H19ICR is not methylated and binds the transcriptional insulator, CTCF, preventing activation of the distal Igf2 promoters. On the paternal (black) chromosome, methylation of the CpGs (black lollipops) within the H19ICR prevents CTCF binding and thus allows enhancer activation of Igf2. In addition, H19ICR epigenetic changes at the adjacent H19 promoter prevent its expression (32). (C) Structures of the ΔME, ΔICR, H19R and Δ13 mutant alleles. Numbers inside the gene boxes indicate the approximate relative expression levels in muscle cells for H19 and Igf2 on these maternal (gray) and paternal (black) chromosomes as determined in the references cited above. ΔME (8) carries a deletion that removes the shared CME (41) and exons 1 and 2 of the Nctc1 gene. ΔICR (32) carries a deletion that removes the 2.4-kb H19ICR but leaves the adjacent H19 promoter intact. H19R (29) carries an insertion of the 2.4-kb H19ICR at +10 kb, between the endodermal (EE) and mesodermal enhancers (CME). Δ13 (33) carries a deletion that removes the entire H19 RNA-coding region plus 10 kb of upstream sequences including the H19 promoter and the H19ICR.
Igf2 and H19 are reciprocally imprinted. H19 is expressed only when maternally inherited, whereas Igf2 expression is paternal in origin (15). This imprinting phenotype can be of real practical advantage in transcriptional studies: in a single cell type one can directly compare active and inactive alleles of the same gene and thus identify epigenetic marks and resultant DNA loop structures and chromatin domains that either promote or disfavor gene expression. In fact, analysis of the Igf2/H19 locus has been especially useful not only in understanding the role of alternative DNA loop structures in regulating gene expression but also in deciphering the molecular and genetic mechanisms that establish these alternative loop domains (Figure 2).
Figure 2.
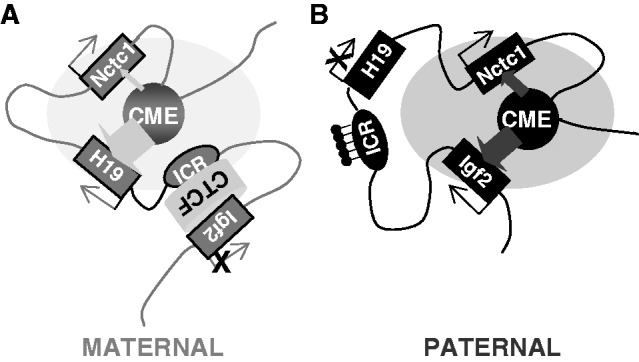
Parent-of-origin specific structures mediate gene expression at the Igf2/H19/Nctc1 imprinted locus in muscle cells. (A) On the maternal chromosome, a CTCF-dependent insulator organizes the DNA loops between distal cis-regulatory elements to favor H19 expression and to prevent interactions between the Igf2 promoters and the shared downstream CME. Recent work from Nativio et al. (17) demonstrates a critical importance for cohesin in establishing these maternal-specific chromosomal structures. Also, see Zhang et al. (25) for detailed mechanisms describing maternal ICR–CTCF–Igf2 interactions. (B) Paternal-specific methylation of CpGs within the ICR prevents CTCF binding, resulting in DNA loop structures that favor Igf2 promoter interactions with the shared enhancer. The loss of CTCF binding also results in a spread of DNA methylation and heterochromatin from the ICR into the adjacent H19 promoter region to H19 transcription. Here, we propose that Nctc1 levels are regulated by competition with H19 and Igf2 promoters for activation by the transcriptional complexes assembling at the shared enhancer. For simplicity, this model does not describe Igf2 differentially methylated regions (DMRs) located near the Igf2 promoters. DMR1 is an muscle-specific repressor of Igf2 expression that is required for complete full postnatal repression of both maternal and paternal chromosomes (62). DMR2 is a tissue non-specific positive regulatory element (63). In muscle, deletion of DMR2 results in modest decreases in Igf2. The participation of these two elements in DNA looping structures has been extensively analyzed (12,13,64) but to date, only in endodermal cells.
Imprinting at the Igf2/H19 locus is dependent on the 2.4-kb H19 imprinting control region (H19ICR) that lies between the two genes, just upstream of the H19 promoter (Figure 1A). Deletion of the H19ICR results in loss of imprinting and biallelic expression of both genes (16). On the maternal chromosome, CTCF protein binds to the ICR and through cohesin (17–19) establishes a transcriptional insulator that organizes the chromosome into loops that favor H19 expression but block interactions between the maternal Igf2 promoters and the downstream shared enhancers, thus preventing maternal Igf2 expression. Upon paternal inheritance, the CpGs within the ICR are methylated, which prevents binding of the CTCF protein so that a transcriptional insulator is not established. Thus, paternal Igf2 promoters and the shared enhancers do interact via DNA loops and expression of paternal Igf2 is facilitated. In addition, the methylated ICR drives developmentally programmed changes at the adjacent H19 promoter to silence its expression (Figures 1B and 2) (9,12,14,20–25).
Not only is the ICR necessary for imprinting at the Igf2/H19 locus but also the 2.4-kb element is sufficient to establish imprinting at any locus. That is, when inserted into ectopic sites in the genome, the ICR organizes adjacent chromosomal domains into parent-of-origin distinct conformations and thus establishes distinct maternal and paternal expression patterns for proximal genes (26–31).
While parent-of-origin loop structures explain expression of Igf2 and H19, their impact on other genes in the locus has not been investigated. Here, we analyze the expression of the muscle-specific long non-coding RNA, Nctc1, that lies downstream of H19 and is transcribed across the core muscle enhancer (CME) shared by Igf2 and H19 (Figure 1A). We show that Nctc1 expression is also dependent on this CME. Concordantly, the CME physically associates with the Nctc1 promoter just as it does with the maternal H19 and paternal Igf2 promoters. Further, we show that all the promoters in this transcriptional domain physically interact with each other depending on their association with the shared enhancer. Thus, interactions between an enhancer and one promoter do not preclude interactions between that enhancer and another promoter or between promoters. Finally, we provide evidence that these promoter–promoter interactions impact gene expression. Genetic and developmental data support a model wherein the need for the Nctc1 promoter to share the enhancer and to compete for RNA polymerase complexes assembling at the CME determines Nctc1 transcription levels and imposes a paternal bias on Nctc1 expression in neonatal animals. In adult animals, repression of H19 and Igf2 eliminates the need for sharing and Nctc1 imprinting is mitigated. Thus altogether, we provide a novel mechanism to explain developmentally specific imprinting of Nctc1 and new insights into the role and importance of chromatin domains in regulating gene expression.
MATERIALS AND METHODS
Animal studies
Generation of H19R (29), ΔME (8), ΔICR (32) and Δ13 (33) mutations and of C/C congenic strains (34) has been described. All animal work was done according to the National Institutes of Health (NIH) Policy and was approved by the National Institute of Child Health and Human Development (NICHD) Animal Care and Use Committee.
Quantitative PCR
cDNA samples prepared with and without reverse transcriptase were analyzed using SYBR Green (Roche) on the Roche Cycler 480. Primers are described in Supplementary Table S1.
Primary myoblast culture
Tissues were isolated from p3 to p5 pups and myoblasts isolated as described (35).
Chromatin immunoprecipitation analysis
Myoblasts were isolated from primary neonatal skeletal muscle tissue and differentiated into myotubes in culture for 24 h. Chromatin immunoprecipitation (ChIP) analysis was performed as previously described (14) using anti-Ser-5(P)-RNA polymerase (RNAP) antibody from Abcam (#ab5131) and anti-rabbit IgG antibody from Santa Cruz Biotechnology (#sc2027). ChIP-purified DNA was quantified and normalized as a percentage of input controls by Real-Time quantitative Reverse Transcription PCR (qRT-PCR). Primers are listed in Supplementary Table S1.
Chromatin conformation capture
3C analysis was performed as described (14) using primers and restriction fragment length polymorphisms (RFLPs) described in Supplementary Table S1 and Supplementary Figure S2. Primer efficiencies were tested using bacterial artificial chromosome DNAs as described (36), using clones 198J15 and 11 301 (34) that together cover the entire locus from upstream of Ins2 to 100 kb downstream of Nctc1 (Supplementary Figure S2).
Allele-specific assays
Allele-specific expression of Nctc1 heterogeneous nuclear RNA (hnRNA) was analyzed by melting analysis (37) using a BanI RFLP and PCR products generated with primers described in Supplementary Table S1. DNA melting was performed using a Roche LightCycler 480. The quantitative nature of the assay is demonstrated in Supplementary Figure S4.
DNA methylation analysis
DNA sequences were screened for CpG islands using programs from the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/emboss/cpgplot/) and the following parameters: window = 100, step = 1, minimum observed to expected ration of C + G to CpG = 0.6, minimum percentage of CpG in a set of 10 windows = 40%, minimum length = 100.
DNA methylation was determined by bisulfite sequencing as described (38,39). Briefly, two independent DNA samples were prepared from primary myocytes isolated from D/C neonates and subjected to cytosine conversion. Two independent nested PCRs were performed (40) on each DNA sample. See Supplementary Table S1 for primer sequences. PCR products were cloned and individual clones sequenced.
RESULTS
Co-regulation of Nctc1 with Igf2 and H19 by a shared muscle-specific enhancer
Nctc1 is a non-coding RNA whose transcript is located within a 12-kb chromosomal region defined by targeted mouse deletions and transgenic studies as the enhancer that is necessary and sufficient for expression of Igf2 and H19 in skeletal muscle (Figure 1) (7–9,41,42). Two Nctc1 isoforms have been identified: a minor isoform includes exons 1, 2 and 3, while the major splice variant includes only exons 1 and 2.
We performed an analysis of Nctc1 expression using qRT-PCR of RNAs isolated from wild-type neonatal animals using primers that would detect both Nctc1 isoforms. As already reported for adult tissues (43), Nctc1 expression in neonates is highly restricted with significant mRNA levels only in skeletal muscle (Figure 3A). These are also the cell types where we see an essential role for the mesoderm enhancer region (as defined by the ΔME chromosomal deletion) in activating H19 and Igf2 in vivo (Figure 3B). Thus, there is a strict correlation between demonstrated activity of the shared mesoderm enhancer in vivo and expression of Nctc1.
Figure 3.
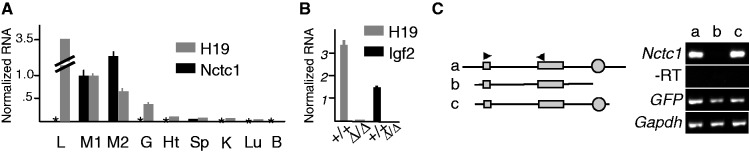
Co-regulation of Igf2, H19 and Nctc1 in muscle cells. (A) Tissue-specific expression of the Nctc1 gene. RNAs isolated from wild-type neonates were analyzed by qRT-PCR for Nctc1 and H19. Expression in hind limb muscle (M1) was set to 1. Other tissues analyzed were liver (L), tongue (M2), gut (G), heart (Ht), spleen (Sp), kidney (K), lung (Lu) and Brain (B). Asterisk denotes no detectable Nctc1. (B) The ΔME deletion affects Igf2 and H19 expression only in tissues where Nctc1 is expressed. RNAs from +/+ and from ΔME/ΔME primary myocytes were analyzed by qRT-PCR for expression of H19 and Igf2 normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Mutant cells display >1000-fold decreases. No effect of the deletion was seen in liver, gut, heart, kidney, lung and brain. (C) Expression of Nctc1 depends on the CME. ΔME/ΔME primary myoblasts were transfected with DNA constructs a, b, or c and a plasmid carrying GFP. After 24-h growth in differentiation media, RNAs were prepared and analyzed by qRT-PCR for Nctc1, GAPDH and GFP. On construct maps, Nctc1 exons 1 and 2 are depicted as filled rectangles and the CME is shown as a filled circle.
To more precisely map the Igf2/H19 CME, Alzhanov et al. (41) used transient transfection analyses and identified a 0.3-kb element (the CME in Figure 1) as necessary and sufficient for enhancer activity in cultured muscle cells. In Figure 3C, we show that this element is also essential for Nctc1 expression in primary myocytes. Thus altogether, these results indicate that Nctc1 is part of the Igf2/H19 regulatory complex in that expression of each gene in muscle is dependent on a shared enhancer element, the CME.
Note that, while Nctc1, H19 and Igf2 transcription are each dependent on the CME, their RNA levels are very different. Altogether, our expression and stability data suggest that the H19 promoter is somewhat more active (3–4×) than the Igf2 promoters and significantly more active (∼1000×) than Nctc1 (Supplementary Figure S1).
Distinct ternary DNA looping structures on maternal and paternal chromosomes mediate imprinting of Nctc1
We analyzed chromatin preparations from wild-type primary myocyte cultures using 3C (44) to determine the DNA looping structures associated with the CME. In each experiment, we used reciprocal crosses and chromosomes marked with single-nucleotide polymorphisms so that we could distinguish paternal and maternal chromosomes and therefore follow the effect of parental origin on loop formation. Thus, we can directly compare domain structures for active and inactive alleles. As expected, activation of Igf2 and H19 promoters by the CME is invariably associated with physical interactions between the promoters and this enhancer (Figure 4A). Consistent with their imprinted expression, Igf2 and H19 promoter–enhancer interactions are restricted to the paternal and maternal chromosomes, respectively. For example, the left panel in Figure 4A analyzes loop formation between the CME and the Igf2 promoter. In C/D cells (maternal C/paternal D alleles), the amplicon is only D in origin. In D/C cells (maternal D/paternal C alleles), the amplicon is only C in origin. (D and C are each wild-type alleles distinguishable by RFLP analysis as described in Supplementary Figure S2.) Together, these experiments reveal that a DNA loop is formed across the 100 kb between the Igf2 promoter and the CME but only on the paternal chromosome. Similarly, the H19 promoter–CME interactions are almost all maternal in origin (Figure 4A, center panel). In contrast, Nctc1 promoter and enhancer interactions occur on both chromosomes (Figure 4A, right panel).
Figure 4.
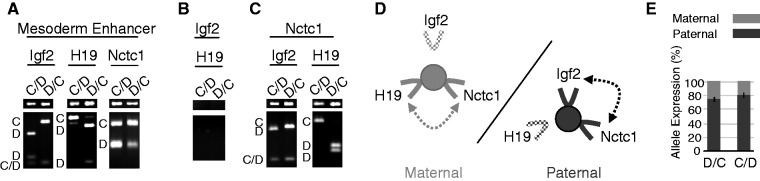
Long-range interactions between the Igf2, H19 and Nctc1 promoters and the shared muscle enhancer and Nctc1 imprinting on wild-type maternal and paternal chromosomes. (A–C) Chromatin was prepared from wild-type primary myocyte cultures and analyzed for long-range DNA interactions by 3C. (A) Interactions between the mesoderm enhancer and the Igf2, the H19 or the Nctc1 promoter regions. (B) Interactions between the Igf2 and H19 promoters. (C) Interactions between the Nctc1 promoter and the Igf2 or H19 promoter regions. In each case, the narrow top gel displays the PCR amplicon indicative of the DNA loop formation. The bottom gel displays these amplicons after restriction enzyme digestion to reveal the allelic origin(s) of the products as marked along the left margin: C, castaneus; D, domesticus and C/D, castaneus and domesticus. In describing genotypes, we use the convention: maternal allele/paternal allele. Primers and the amplicon sizes before and after digestion are described in Supplementary Table S1 and Supplementary Figure S2. For each analysis at least three independent chromatin preparations were assayed. (D) Summary of long-range interactions on maternal (left, gray) or paternal (right, black) chromosomes. On both chromosomes, transcripitionally active promoters (solid lines) interact with the CME (filled circle) and with each other while silenced promoters (stippled lines) do not interact. (E) Parent-of-origin specific expression of Nctc1. To quantitate the effects of parental origin, allele-specific expression was quantitated for RNAs isolated from D/C and from C/D neonates. Error bars show standard deviations.
While the Nctc1 promoter and CME are only 5.6 kb apart, their physical interaction is specific: 3C analyses do not detect association between the enhancer and comparably distant downstream sites (Supplementary Figure S3).
To gain a clearer understanding of the composition of these DNA loop structures, we next looked for interactions between the three promoter regions. We detected no interactions between the Igf2 and H19 promoters (Figure 4B). However, we did note interactions between Nctc1 and Igf2 promoters (but only on the paternal chromosome) and between Nctc1 and H19 promoters (but only on the maternal chromosome) (Figure 4C). These results are consistent with the presence of two distinct ternary structures containing the CME plus each of the promoters that it activates in cis: CME + H19 promoter + Nctc1 promoter on the maternal chromosome or CME + Igf2 promoter + Nctc1 promoter on the paternal chromosome (Figure 4D). Thus, Nctc1 transcription is dependent on parent-of-origin distinct chromosomal structures.
To determine if there were consequences to the Nctc1 promoter’s parent-of-origin-specific configurations, we developed assays to quantitate allele-specific expression of Nctc1 (Supplementary Figure S4). In fact, the paternal chromosome accounts for 80% of Nctc1 RNA transcripts (Figure 4E). Thus, maternal and paternal loop domains are not equivalently active in regard to Nctc1 transcription. Rather, much more Nctc1 RNA transcription is coming from CME–Igf2–Nctc1 (paternal) complexes than from CME–H19–Nctc1 (maternal) complexes.
We could not detect any parent-of-origin differences in DNA methylation near the Nctc1 gene that might account for its imprinting (Supplementary Figure S5). However, Nctc1 imprinting is dependent on the distant H19ICR (Figure 5A). Surprisingly, either maternal (n = 4, P = 0.004) or paternal ICR (n = 4, P = 0.004) deletions each result in loss of imprinting at Nctc1. In contrast, the ICR is a chromosome-specific silencer for Igf2 and for H19 (9,16).
Figure 5.
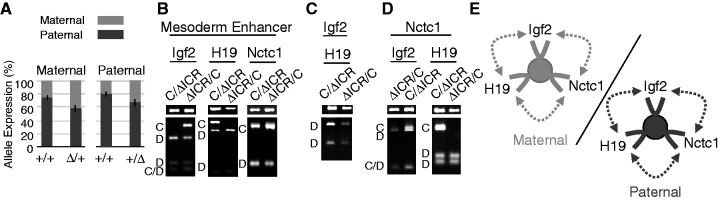
Nctc1 imprinting and chromatin loop structures are altered on ΔICR chromosomes. (A) Parent-of-origin specific expression of Nctc1. To quantitate the effects of maternal inheritance, allele-specific expression was measured for RNAs isolated from D/C and of ΔICR/C neonates. To quantitate the effects of paternal inheritance, C/D and C/ΔICR pups were compared. (B–D) Chromatin was prepared from primary myocytes isolated from mice carrying a paternal (C/ΔICR) or a maternal deletion (ΔICR/C) of the H19ICR and analyzed for long-range DNA interactions by 3C. (B) Interactions between the mesoderm enhancer and the Igf2, the H19 or the Nctc1 promoter regions. (C) Interactions between the Igf2 and H19 promoters. (D) Interactions between the Nctc1 promoter and the Igf2 or H19 promoter regions. (E) Summary of long-range interactions on maternal (left, gray) or paternal (right, black) ΔICR chromosomes. On both chromosomes, Igf2, H19 and Nctc1 promoters are all active and interact with the CME (filled circle) and with each other. See Figure 4 for additional details.
Chromosome-specific 3C analyses of ΔICR chromosomes confirm that the shared enhancer is capable of interacting with multiple promoters in a way that brings all active promoters within its domain into contact with each other (Figure 5B and D). Thus, the maternal and the paternal ΔICR chromosomes each generate a tertiary looping structure that includes the CME interacting with the Igf2, H19 and Nctc1 promoters while these promoters are also interacting with each other (Figure 5E). In sum on wild-type and on ΔICR chromosomes, co-expressed promoters interact not only with the shared enhancer but also with each other.
Nctc1 expression inversely correlates with H19/Igf2 promoter activity
It is not straightforward to understand how both maternal and paternal deletion of the H19ICR could each have the same effect on Nctc1 allelic bias. Given that the Nctc1 promoter is organized into loop structures that also contain the Igf2 and H19 promoters, we considered the possibility that ICR regulation of Nctc1 imprinting was indirect and secondary to its effects on H19 and Igf2 transcriptional activity (see Figure 1C for summary of ICR deletion phenotypes). To first test this hypothesis, we took advantage of the fact that H19 and Igf2 are each strongly down-regulated during postnatal development (>30-fold for H19, P < 0.001 and >200-fold for Igf2, P < 0.001) (Figure 6A). In contrast, Nctc1 expression increased 2.5-fold (P < 0.001) (Figure 6A) with a significant loss in imprinting (neonate = 79 ± 3% paternal, adult = 60 ± 5% paternal, P < 0.0001) or a switch in bias from 4:1 to 1.5:1 (Figure 6B). Thus, decreased Igf2 + H19 activation is associated with increased Nctc1 transcription as well as with Nctc1 loss of imprinting.
Figure 6.
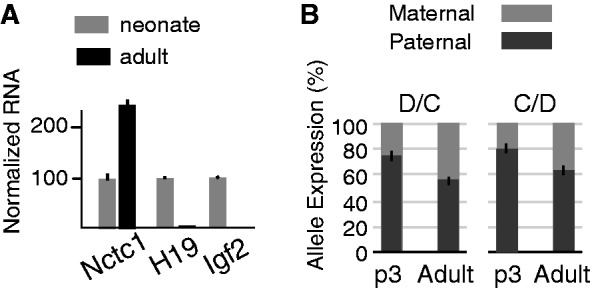
Developmental regulation of Nctc1, H19 and Igf2 gene expression and of Nctc1 imprinting. (A) RNAs were prepared from hind limbs of neonatal (p3) and of adult (12 weeks) animals and analyzed by qRT-PCR for Nctc1, H19 and Igf2. The relative expression for each gene in neonates was set to 100. (B) To ascertain the effect of development on Nctc1 imprinting, allelic frequencies were measured in RNAs isolated from D/C and C/D wild-type animals as described in Figure 4. Error bars show standard deviations.
Next, we took a genetic approach and analyzed Nctc1 expression from the mutant chromosome, H19R, that carries an insertion of the 2.4-kb H19ICR at the +10 kb position (Figure 1C). Maternal inheritance of the ectopic ICR insulates the H19 promoter from the CME and blocks maternal H19 expression in muscle (Figure 7A) (14). Thus, the H19R mutation allows us to examine Nctc1 expression specifically in the absence of H19 transcription in cis. Maternal inheritance of H19R increases total Nctc1 RNA (Figure 7A) and moreover, abrogates normal Nctc1 imprinting (Figure 7B, left panel) (+/+ = 26±3% maternal, H19R/+ = 78±4% maternal, P = < 0.001). Thus, in the absence of H19 transcription, expression of Nctc1 from the maternal chromosome is up-regulated to the point that imprinting is reversed. 3C analyses demonstrate that maternal H19R chromosomes show a binary enhancer looping structure that includes only the CME and the Nctc1 promoter and not the H19 promoter (Figure 7C, D and E). That is, the H19R insertion prevents interactions of not only the H19 promoter and the CME but also of the H19 and Nctc1 promoters, consistent with the idea that the promoters interact via the shared enhancer.
Figure 7.
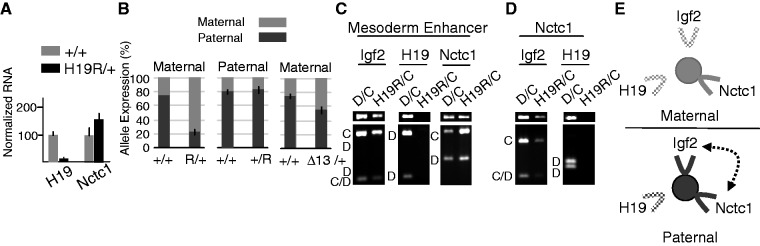
Nctc1 expression, imprinting and chromatin loop structures are altered on maternal H19R chromosomes. (A) Transcription of H19 and Nctc1 in H19R/+ muscle cells was quantitated by qRT-PCR and normalized to GAPDH RNA levels. Expression of each gene in wild-type (+/+) cells is set at 100. (B) Parent-of-origin specific expression of Nctc1 on H19R and on Δ13 chromosomes. To quantitate the effects of maternal inheritance of H19R, allele-specific expression was measured for RNAs isolated from D/C and of H19R/C neonates. To quantitate the effects of paternal inheritance of H19R, C/D and C/ΔICR pups were compared. To quantitate the effects of maternal inheritance of Δ13, allele-specific expression was measured for RNAs isolated from D/C and of Δ13/C neonates. (C–D) Chromatin was prepared from primary myocytes isolated from wild-type mice (D/C) and from mice carrying a maternally inherited copy of the H19R insertion mutation (H19R/C) and analyzed by 3C. (C) Interactions between the mesoderm enhancer and the Igf2, the H19 or the Nctc1 promoter regions. (D) Interactions between the Nctc1 promoter and the Igf2 or H19 promoter regions. (E) Summary of long-range interactions on maternal (top, gray) or paternal (bottom, black) H19R chromosomes. Maternal inheritance of the ICR insertion prevents H19 promoter interactions with the CME (filled circle) and with the Nctc1 promoter. See Figure 4 for additional details.
Upon paternal inheritance of H19R, the ectopic ICR is methylated so that CTCF does not bind and insulator activity is not established (14). Thus, H19 and Igf2 transcription is unaffected. Accordingly, paternal inheritance of H19R has no effect on Nctc1 imprinting (+/+ = 80 ± 4% paternal, +/H19R = 82 ± 5% paternal, P = 0.46) (Figure 7B, center panel) or on chromosome loop domain structures (data not shown and Figure 7E).
Analysis of the H19R chromosomes indicates that Nctc1 expression and imprinting is a downstream consequence of H19 transcriptional levels. We suggest that this is related to the fact that on a wild-type maternal chromosome, the promoters concurrently interact with and share the CME enhancer and are in some sense competing for transcriptional activation. Alleviating this competition by mutation or normal development then results in increased maternal Nctc1. However, the maternal H19R phenotype is also consistent with transcriptional interference with Nctc1 by the H19 RNA. To distinguish these two mechanisms, we analyzed RNAs isolated from Δ13/+ neonatal muscle. Δ13 is a deletion that removes the entire H19 gene and also deletes the ICR (Figure 1C) (33). Thus, Δ13 is like H19R in that maternally inherited chromosomes will not express any H19. Therefore, if repression of maternal Nctc1 is via RNA interference, then Δ13/+ muscle should present the H19R/+ phenotype, i.e. an inversion of normal Nctc1 imprinting and switch to maternal bias. However, unlike H19R, maternally inherited Δ13 chromosomes express maternal Igf2 promoters at levels similar to those seen on wild-type paternal chromosomes (Figure 1C). If repression of maternal Nctc1 is via promoter competition, then Δ13/+ cells should show no imprinting since maternal Δ13 and paternal wild-type chromosomes are equivalent in terms of activities of the upstream H19 and Igf2 promoters. What we actually see is the simple loss of imprinting in Δ13/+ animals (Δ13/+ = 46 ± 5% paternal, n = 4, P < 0.01) (Figure 7B, right panel), predicted by promoter competition.
RNAP II association with Nctc1 inversely correlates with H19 transcriptional activation
We wanted to identify molecular correlates that might explain Nctc1 imprinting and to test if promoter competition occurs at the level of assembly of transcriptional machinery at the Nctc1 promoters. Therefore, we used ChIP to compare binding of total RNAP II and activated RNAP II [Ser-5(P)-RNAP] to the Nctc1 promoter in wild-type (+/+) and in H19R/+ myocytes (Figure 8A). Consistent with increased Nctc1 RNA levels in H19R/+ myocytes, we noted a 3-fold enrichment in RNAP at the promoter in these cells. Moreover, this increase is entirely due to increased assembly on the maternal chromosome (Figure 8B).
Figure 8.
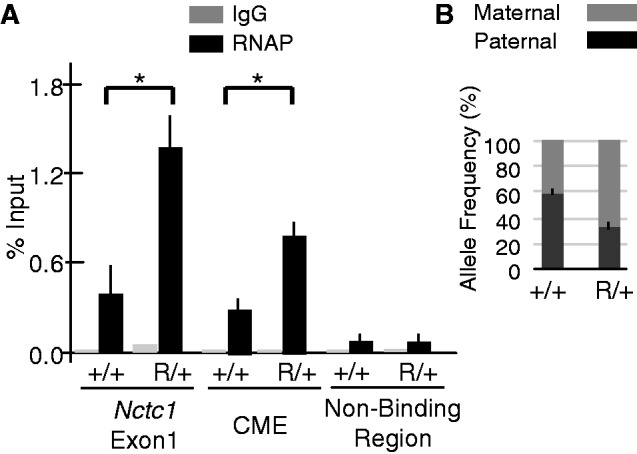
Allele-specific Ser-5(P)-RNAP II binding at the Nctc1 gene in wild-type versus H19R/+ mice. Primary myoblasts were isolated from wild-type and H19R/+ mice and differentiated in culture for 24 h. (A) Total Ser-5(P)-RNAP association at the Nctc1 gene. ChIP analysis was conducted to measure binding at both Nctc1 exon1 and the CME in +/+ and in H19R/+ mutant cells. An intergenic region between H19 and Igf2 was assayed as a control for non-specific binding. (B) Allele-specific frequency of Ser-5(P)-RNAP binding at the Nctc1 gene. ChIP samples for Nctc1 exon 1 from panel A were further analyzed for allele frequency content using DNA melting analysis. Error bars represent standard errors. P < 0.05, n = 3.
We also saw a significant binding of RNAP at the CME that further increases in H19R/+ compared with +/+ cells (Figure 8A). Thus, in H19R/+ cells, the absence of interaction with the H19 promoter and the consequent loss in H19 transcription results in an accumulation of RNAP complexes at the CME. As described in the ‘Discussion’ section, these results are consistent with the idea that enhancers serve as primary recruitment/assembly docks for RNAP that is then dispersed to promoters via complexes we characterize as DNA loops. We suggest that on H19R chromosomes, the removal of the H19 as a competitive gene makes the RNAP complexes more available for Nctc1 activation.
DISCUSSION
Strong correlative evidence supports the idea that promoter activation by distal enhancers is invariably associated with DNA loop formation between those elements (1,45). Genome-wide analyses indicate that promoter–enhancer interactions are a fundamental aspect of chromosomal organization (2). Independent genomic studies have also recently indicated that promoter–promoter interactions are common and may play a critical role in coordinating gene expression and organizing active transcriptional domains (3,46).
Here, we use a model system approach to assay and analyze the functional role of promoter–enhancer and promoter–promoter interactions in organizing the Igf2/H19/Nctc1 transcriptional domain. We provide support for the idea that interactions between an enhancer and one promoter do not preclude interactions between that enhancer and other promoters. Instead, as suggested by transcriptional factory models (47), co-regulated promoters are brought into physical proximity to each other via their shared enhancer. We also provide genetic and developmental analyses indicating that these promoter–promoter interactions can have important consequences on gene expression levels. That is, one promoter can regulate another.
Specifically, we characterized expression of the long non-coding RNA, Nctc1, which overlaps the muscle-specific enhancer shared by the Igf2 and H19 genes and show that Nctc1 is imprinted in a developmentally regulated fashion. In neonates, expression is biased toward the paternal chromosome, while adult animals show nearly equal levels of paternal and maternal allele activities. Expression of Igf2, H19 and Nctc1 in muscle is dependent on a shared muscle core enhancer. Genetic and developmental analyses suggest that Nctc1 imprinting is a side effect of the enhancer sharing. Our data indicate that physical interactions between the Nctc1, Igf2 and H19 promoters and the CME are not mutually exclusive but that co-regulated promoters can physically interact with each other via their interaction with the shared enhancer. These interactions offer a mechanism for promoters to communicate with and regulate one another. The Nctc1 allelic bias we see on wild-type and mutant chromosomes can be explained by Nctc1 promoter competition, especially with H19, for the shared enhancer. Thus, loss of imprinting in adult animals is not because of changes in the imprinting-dependent epigenome, but instead is a side effect of the developmentally programmed decrease in H19 and Igf2 gene expression.
To understand the molecular meaning of promoter–promoter interactions, we quantitated the recruitment and activation of RNAP to the Nctc1 promoter in +/+ cells and H19R/+ cells, where the Nctc1 promoter has sole access to the shared enhancer. ChIP experiments identified an increase in RNAP binding, demonstrating that competition directly affects recruitment and/or assembly of the active basal transcription complex. We were particularly interested to note that polymerase also accumulated at the CME in H19R/+ animals. As suggested in previous reports, it is plausible that the enhancer serves as a recruiting platform or loading dock for RNAP that it transfers to target promoters for transcription initiation (48,49). Precedent for this notion also exists in studies of the β-globin locus, where it is known that multiple enhancers recruit RNAP independent of the globin promoter (50). Later studies indicated that the enhancers transfer RNAP from the enhancers to the globin promoter via loop formation in a manner that can be blocked by a CTCF-bound insulator (51). By this model, the paternal bias at Nctc1 is mediated by the high efficiency with which the H19 promoter downloads RNAP at the cost of activation of the maternal Nctc1 promoter.
There are already several model systems indicating that one promoter can regulate another. The INS2 promoter (located just upstream of IGF2) physically associates with and positively regulates the SYT8 gene in pancreatic β-cells (52). Negative regulation or promoter competition has been perhaps most extensively studied in regard to vertebrate β-globin transcription, where evidence suggests that competition plays a role in developmental changes in the choice of promoter gene activation (53). Does this competition occur only at the level of loop formation? That is, is the interaction between an enhancer and a promoter exclusive so that it precludes that enhancer’s interaction with (and therefore activation of) a second gene? Our report provides indirect but strong evidence that this is not necessarily the case but that at least part of the competition occurs at some step after loop formation. Previous studies already provided evidence that the H19ICR can interact simultaneously with multiple cis-elements (54). The DNA loops that we identify and the relative levels of each of these structures are most consistent with the idea that ternary structures or quaternary structures also form around the enhancer. Altogether, analysis of this locus shows that DNA loops are a necessary precondition for gene expression but the rate of loop formation is not sufficient to explain all aspects of gene expression levels. The mechanisms by which long-range interactions activate promoter activity remain largely enigmatic (1,55). This study suggests that rate-limiting steps sometimes occur after loop formation and presents a mechanism for gene regulation likely to affect many co-regulated genes.
The biological function of the Nctc1 long non-coding RNA was not directly addressed in this study. In mammalian species, only Nctc1 promoter and not Nctc1 exonic sequences are conserved (7), suggesting that it is the transcription of the Nctc1 RNA but not the RNA product itself that is of primary evolutionary significance. In this regard, several recent genome-wide analyses have demonstrated a frequent overlap of long non-coding RNAs and tissue-specific transcriptional enhancers and several authors have hypothesized a functional importance for this association (56–60). In fact, our preliminary data suggest a crucial role for Nctc1 promoter activity in regulating enhancer function and Igf2/H19 transcription.
In sum, as described in Figure 2, the ICR regulates imprinting at the Igf2/H19 locus by at least two mechanisms. For Igf2 and H19, the ICR generates monoallelic expression directly by preventing physical contact between the shared enhancer and the Igf2 and H19 promoters on one of the two parental chromosomes (12–14,61). For Nctc1, the ICR acts indirectly by modulating H19 and Igf2 promoters, which in turn compete with Nctc1 for enhancer activation after DNA loop formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
The Eunice Kennedy Shriver National Institute of Child Health and Human Development, Division of Intramural Research, National Institutes of Health [HD001804-17 to K.P.]. Funding for open access charge: Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Vickie Carter and Theresa Hernandez for animal husbandry.
REFERENCES
- 1.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcriptional enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y, Yue F, McCleary D, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov V, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, Ruan X, Auerbach R, Sandhu K, Zheng M, Wang P, Poh H, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace J, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–417. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ainscough JF-X, Dandola L, Surani MA. Appropriate expression of the mouse H19 gene utilises three or more distinct enhancer regions spread over more than 130 kb. Mech. Dev. 2000;91:365–368. doi: 10.1016/s0925-4773(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 6.Brunkow ME, Tilghman SM. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 1991;5:1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- 7.Ishihara K, Hatano N, Furuumi H, Kato R, Iwaki T, Miura K, Jinno Y, Sasaki H. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in in Igf2/H19 imprinting. Genome Res. 2000;10:664–671. doi: 10.1101/gr.10.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaffer C, Grinberg A, Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 2001;21:8189–8196. doi: 10.1128/MCB.21.23.8189-8196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaffer CR, Srivastava M, Park K, Ives E, Hsieh S, Batlle J, Grinberg A, Huang SP, Pfeifer K. A transcriptional insulator at the imprinted H19/Igf2 Locus. Genes Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- 10.Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 11.Yoo-Warren H, Pachnis V, Ingram RS, Tilghman SM. Two regulatory domains flank the mouse H19 gene. Mol. Cell. Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurukuti S, Tiwari V, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 14.Yoon Y, Jeong S, Rong Q, Park K-Y, Chung J, Pfeifer K. Analysis of the H19ICR insulator. Mol. Cell. Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartolomei M, Ferguson-Smith A. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a002592. pii: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nativio R, Wendt K, Ito Y, Huddleston J, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters J-M, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stedman W, Kang H, Lin S, Kissil J, Bartolomei M, Lieberman P. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendt K, Yoshida K, Itah T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–803. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 20.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 21.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 22.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C, Wolffe A, Ohlsson R, Lobanenkov V. Functional association of CTCF with the insulator upstream of the H19 gene is parent-of-origin specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Ju J-F, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu T, Hoffman A. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabo P, Tang S, Rentsendorj A, Pfeifer G, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Niu B, Hu J, Ge S, Wang H, Li T, Ling J, Steelman B, Qian G, Hoffman A. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J. Cell. Biol. 2011;193:475–487. doi: 10.1083/jcb.201101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cranston M, Spinka T, Elson D, Bartolomei M. Elucidation of the minimal sequence required to imprint H19 transgenes. Genomics. 2001;73:98–107. doi: 10.1006/geno.2001.6514. [DOI] [PubMed] [Google Scholar]
- 27.Gebert C, Kunkel D, Grinberg A, Pfeifer K. H19 imprinting control region methylation requires an imprinted environment only in the male germ line. Mol. Cell. Biol. 2010;30:1108–1115. doi: 10.1128/MCB.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzaki H, Okamura E, Shimotsurm A, Fukamizu A, Tanimoto K. A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independent of its establishment in germ cells. Mol. Cell. Biol. 2009;29:4595–4603. doi: 10.1128/MCB.00275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park K, Sellars E, Grinberg A, Huang S, Pfeifer K. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol. Cell. Biol. 2004;24:3588–3595. doi: 10.1128/MCB.24.9.3588-3595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiramali S, Srivastava S, Varma G, Grinberg A, Pfeifer K, Srivastava M. An ectopic CTCF dependent transcriptional insulator influences the choice of V beta gene segments on VDJ recombination at the TCR beta locus. Nucleic Acids Res. 2012;40:7753–7765. doi: 10.1093/nar/gks556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanimoto K, Shimotsuma M, Matsuzaki H, Omori A, Bungert J, Engel J, Fukamizu A. Genomic imprinting recapitulated in the human beta-globin locus. Proc. Natl Acad. Sci. USA. 2005;102:10250–10255. doi: 10.1073/pnas.0409541102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava M, Hsieh S, Grinberg A, Williams-Simon L, Huang S-P, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting element. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 33.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 34.Gould TD, Pfeifer K. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum. Mol. Gen. 1998;7:483–487. doi: 10.1093/hmg/7.3.483. [DOI] [PubMed] [Google Scholar]
- 35.Bois P, Grosveld G. FKHR (FOX01a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 2003;22:1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palstra R-J, Tohhuis B, Spliner E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 37.Yu H, Koo I, Jeong S. Relative quantitation of restriction fragment length polymorphic DNAs via DNA melting analysis provides an effective way to determine allele frequencies. Genomics. 2009;94:355–361. doi: 10.1016/j.ygeno.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani M. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 39.Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei MS, Walter J, et al. Epigenetic modification in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 40.Tremblay KD, Duran KL, Bartolomei MS. A 5' 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alzhanov D, McInerney S, Rotwein P. Long range interactions regulate Igf2 gene transcription during skeletal muscle differentiation. J. Biol. Chem. 2010;285:38969–38977. doi: 10.1074/jbc.M110.160986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishihara K, Kato R, Furuumi H, Zubair M, Sasaki H. Sequence of a 42-kb mouse region containing the imprinted H19 locus: identification of a novel muscle-specific transcription unit showing biallelic expression. Mamm. Genome. 1998;9:775–777. doi: 10.1007/s003359900863. [DOI] [PubMed] [Google Scholar]
- 43.Ishihara K, Kato R, Furuumi H, Zubair M, Sasaki H. Sequence of a 42-kb mouse region containing the imprinted H19 locus: identification of a novel muscle-specific transcription unit showing biallelic expression. Mamm. Genome. 1998;9:775–777. doi: 10.1007/s003359900863. [DOI] [PubMed] [Google Scholar]
- 44.Dekker J. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 45.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev. 2011;22:79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanyal A, Lajoie B, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne C, Chakalova L, Brown K, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell J, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leach K, Nightingale K, Igarashi K, Engel PLJ, Becker P, Bungert J. Reconstitution of human beta-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol. Cell. Biol. 2001;21:2629–2640. doi: 10.1128/MCB.21.8.2629-2640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson K, Christensen H, Zhao B, Bresnick E. Distinct mechanisms control RNA Polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 51.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat. Struct. Mol. Biol. 2011;18:372–378. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palstra R-J, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Z, Tavoosidana G, Sjoelinder M, Goendoer A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu K, Singh W, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 55.Ong C-T, Corces V. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi B, Muller H, Ragoussis J, Wei C-L, Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLOS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Djebali S, Davis C, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim T, Hemberg M, Gray J, Costa A, Bear D, Wu J, Harmin D, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulating enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koch F, Andrau J-C. Initiating RNA Polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription. 2011;2:263–268. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowalczyk M, Hughes J, Garrick D, Lynch M, Sharpe J, Sloane-Stanley J, McGowan S, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 61.Vu T, Nguyen A, Hoffman A. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum. Mol. Genet. 2010;19:901–909. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Constancia M, Dean W, Lopes S, Moore T, Kelsey G, Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 63.Murrell A, Hesson S, Bowde L, Constancia M, Dean W, Kelsey G, Reik W. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2001;2:1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Court F, Baniol M, Hagege H, Petit J, Lelay-Taha MN, Carbonell F, Weber M, Cathala G, Forne T. Long-range chromatin interactions at the mouse Igf2/H19 locus reveal a novel paternally expressed long non-coding RNA. Nucleic Acids Res. 2011;39:5893–5906. doi: 10.1093/nar/gkr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


