Abstract
The genome of influenza A viruses (IAV) is split into eight viral RNAs (vRNAs) that are encapsidated as viral ribonucleoproteins. The existence of a segment-specific packaging mechanism is well established, but the molecular basis of this mechanism remains to be deciphered. Selective packaging could be mediated by direct interaction between the vRNA packaging regions, but such interactions have never been demonstrated in virions. Recently, we showed that the eight vRNAs of a human H3N2 IAV form a single interaction network in vitro that involves regions of the vRNAs known to contain packaging signals in the case of H1N1 IAV strains. Here, we show that the eight vRNAs of an avian H5N2 IAV also form a single network of interactions in vitro, but, interestingly, the interactions and the regions of the vRNAs they involve differ from those described for the human H3N2 virus. We identified the vRNA sequences involved in five of these interactions at the nucleotide level, and in two cases, we validated the existence of the interaction using compensatory mutations in the interacting sequences. Electron tomography also revealed significant differences in the interactions taking place between viral ribonucleoproteins in H5N2 and H3N2 virions, despite their canonical ‘7 + 1’ arrangement.
INTRODUCTION
Influenza A viruses (IAVs) are a serious threat to human health (1). They are responsible for the majority of seasonal flu cases and for occasional flu pandemics that can have high case fatality and mortality rates (1). The IAV genome consists of eight single-stranded viral RNA (vRNA) segments of negative polarity ranging form 890 to 2341 nt, each encoding at least one essential protein in the antisense orientation (2). In the viral particles, each vRNA is associated with a trimeric viral polymerase complex and numerous copies of the nucleoprotein (NP), forming a viral ribonucleoprotein (vRNP) (2,3). The polymerase subunits, basic protein (PB2), PB1 and acidic protein (PA), are encoded by the three largest vRNAs, named vRNAs 1, 2 and 3, respectively. The polymerase complex recognizes the 5′ and 3′ termini of the vRNAs that are highly conserved amongst IAV strains and among the eight vRNA species (4–6). The 5′ and 3′ termini, which are 13 and 12 nt long, respectively, are partially complementary and impose a hairpin structure to the vRNPs (7,8).
While influenza B viruses almost exclusively infect humans, the main reservoir of IAVs is constituted by wild aquatic birds, such as ducks, geese and swans, and the IAVs are much more diverse in the waterfowl than in the human population (9). A human IAV pandemic can arise when a flu strain jumps from an animal species to the human species and is transmitted efficiently in the human population (1). An avian influenza virus can be transmitted directly or indirectly to humans, without undergoing major genetic changes: it is thought that such a process gave rise to the ‘Spanish flu’ in 1918. Alternatively, when an animal and a human flu strain co-infect an intermediate host, such as a pig, the vRNAs from the two flu strains can mix to generate new strains (a process named genetic reassortment) that can spread from the intermediate host to humans. If the reassortant viruses contain ‘animal’ hemagglutinin (HA) and neuraminidase (NA) segments that encode virus surface proteins for which humans have limited or no preexisting immunity, it can cause a pandemic in the human population: this happened in 1957 (the ‘Asian flu’), in 1968 (the ‘Hong Kong flu’) and in 2009 (the ‘swine flu’) (1,10).
The segmented nature of the IAV genome confers an obvious evolutionary advantage, but it generates a formidable problem for the viral RNA packaging machinery (3). Natural IAV infections are initiated at low multiplicity of infection; therefore, viral particles that do not possess a complete set of vRNAs will fail to replicate. IAVs rely on a segment-specific packaging mechanism to ensure that each viral particle contains one copy of each vRNA (3). Indeed, IAVs normally incorporate one and only one copy of each vRNA (11–14).
Defective interfering RNAs are generated in cell culture and in vivo under high multiplicity of infection conditions (15–24). They are generated during vRNA replication and contain large deletions in the central coding region(s) (15–17,19,20,22–24). Importantly, they specifically compete for packaging into the viral particles with their parental vRNA (18,19,21,23), indicating that vRNAs contain segment-specific packaging signals. Sequencing of the defective interfering RNAs indicated that these sequences usually reside within ∼100–300 nt from each end of the vRNAs. More recently, reverse genetics experiments, mainly performed on the A/WSN/33 (H1N1) strain (WSN), and to a lesser extent on the A/PR/8/34 (H1N1) strain (PR8), revealed the existence of segment-specific packaging signals in each vRNA, usually located within ∼100 nt from the vRNA ends (4,25–32). The packaging sequences appear to be multipartite and discontinuous (3). No precise boundary could be mapped in these experiments: successive deletions from the middle to the border of the coding regions resulted in a gradual decrease of the packaging efficiency. Importantly, some, but not all, conserved codons, which tend to accumulate in the vRNA terminal regions, have been shown to be important for optimal packaging of several vRNAs (33–35).
How the packaging signals ensure selective packaging of a set of eight different vRNAs is still unknown, but the existence of intermolecular interactions between vRNAs is an attractive hypothesis, especially as no viral or human protein specifically recognizing an IAV packaging signal has been identified (3,26). However, no such interactions have been identified in IAV viral particles. We therefore recently developed an in vitro approach, which showed that the eight vRNAs of the A/Moscow/10/99 (H3N2) strain (MO) are involved in a single interaction network (36). The vRNA regions involved in several interactions were identified and were found to overlap with the regions containing the WSN/PR8 packaging signals, even when they could be narrowed down to a few nucleotides, suggesting that the vRNA network detected in vitro is relevant to IAV vRNA packaging (36). Combined with electron tomography, which revealed the internal organization of MO virions, our in vitro data suggested that the eight vRNAs are selected and packaged as an organized supramolecular complex held together by direct base pairing of the packaging signals (36).
Here, we applied the same strategy to an avian IAV: the A/Finch/England/2051/91 (H5N2) strain (EN). We showed that the eight vRNAs of the EN strain are also involved in a single network of RNA interactions in vitro. We grossly defined the regions of the vRNAs involved in each interaction, and in several cases, we identified the interacting regions at the nucleotide level. Surprisingly, neither the interaction network nor the regions of the vRNAs involved in the intermolecular interactions is conserved between the EN and MO strains. Electron tomography also revealed significant differences in the organization of vRNPs in EN and MO virions. Thus, our data suggest that the packaging signals might not be conserved between these viral strains. The precise identification of the nucleotides involved in the vRNA/vRNA interactions in vitro will allow testing their role in IAV replication.
MATERIALS AND METHODS
Plasmids, cloning and site-directed mutagenesis
The EN complementary DNAs were cloned between the BsmbI sites of pUC2000 vectors starting from the corresponding reverse genetic plasmids. These vectors were obtained as previously described and contain a T7 promoter, the cloning cassette of pHW2000 containing two BsmbI sites, and a unique restriction site (Eco47III, Bsh1236I or Ecl136II) between the PstI and EcoRI sites (36). Sixty-seven mutants with deletions ranging from 11 to ∼400 nt have been obtained by polymerase chain reaction using strictly complementary primers bridging the region to be deleted. After 35 polymerase chain reaction cycles, the parental plasmids were digested with DpnI, and the amplification products were used to transform One Shot MAX efficiency DH5α-T1 (Invitrogen) Escherichia coli cells. Fifteen mutants bearing point substitutions were obtained using the same strategy.
In vitro transcription, RNA purification and electrophoretic mobility shift assay
DNA templates digested with Eco47III, Bsh1236I or Ecl136II were either co-transcribed in vitro by pairs, and RNA complexes were analysed directly, or vRNAs were synthesized individually, purified, and incubated with another vRNA, followed by analysis of the complexes by native agarose gel electrophoresis. Co-transcription experiments were performed essentially as described previously (36). Briefly, linearized plasmids (∼200 ng of each plasmid) were incubated for 3 h at 37°C and treated with ribonuclease-free DNase I before analysis of the RNA complexes. Alternatively, in vitro transcription of individual vRNAs was performed with ∼30 µg of linearized plasmids essentially as described previously (37), except that the concentration of each nucleotide triphosphate (NTP) was 8 mM, and that 0.5 µg of yeast inorganic pyrophosphatase (Roche) was included in the reaction mixture. Following DNase I treatment, vRNAs were extracted with phenol/chloroform, ethanol precipitated and purified on a TSK G2000SW column (Tosoh Bioscience) as described previously (37). The integrity of the vRNAs was routinely checked by denaturing polyacrylamide gel electrophoresis. Pairs of purified vRNAs (2 pmol each) were denaturated for 2 min at 90°C in 8 µl of water and cooled on ice. Two microlitres of 5-fold concentrated buffer (final concentration: 50 mM sodium cacodylate, pH 7.5; 40 mM KCl and 0.1 mM MgCl2) were added, and the samples were incubated for 30 min at 37°C or 55°C, then analysed on 0.8% agarose gels containing 0.01% (w/v) ethidium bromide after addition of 2 µl of loading buffer [40% (v/v) glycerol, 0.05% (w/v) xylene-cyanol and 0.05% (w/v) bromophenol blue]. Native gel electrophoresis of the RNA complexes was performed at 4°C in a buffer containing 50 mM Tris, 44.5 mM borate and 0.1 mM MgCl2. The RNA weight fraction (%) of each band in a lane was determined, and the percentage of intermolecular complex was determined by dividing the weight fraction of the corresponding band by the sum of the weight fractions of all bands in the lane.
In oligo-mapping experiments, vRNAs were incubated in the presence of 10 pmol of an oligodeoxyribonucleotide (ODN) complementary to one of the vRNAs. A trace amount (10 000–20 000 cpm) of 32P-5′-labelled ODN was included to be able to check its binding to the targeted vRNA. To that aim, after imaging under ultraviolet light, gels were fixed for 10 min in 10% (v/v) trichloroacetic acid, dried at room temperature and submitted to autoradiography.
Electron microscopy (EM) and tomography
Electron microscopy (EM) and electron tomography of EN virus budding from Madin-Darby canine kidney (MDCK) cells were performed as described previously (36,38).
RESULTS
Optimization of the conditions for studying interactions between EN vRNAs
We recently studied the interactions taking place between the vRNAs of the IAV MO strain during in vitro transcription (36). Mutational analysis of three of these interactions showed that they involved the terminal regions of the vRNAs open reading frames (36), which are known to contain packaging signals [in the case of the human PR8 and WSN IAVs (3)], suggesting that the interactions detected in vitro are relevant to vRNA packaging. When we applied the same protocol to EN vRNAs, we observed 15 intermolecular interactions, but in only three of them did the complex represent ≥20% of the RNA mass. Besides, we observed that several EN vRNAs, but no MO vRNAs, interacted with a green fluorescent protein (GFP) messenger RNA control under these conditions (data not shown). Finally, we introduced a series of ∼200-nt deletions covering the complete vRNA 8 non-structural (NS), but none of these deletions had a significant effect on its interaction with other EN vRNAs (data not shown). Thus, we concluded that the experimental conditions that allowed for specific interactions between MO vRNAs are not well suited for EN vRNAs.
We thus individually transcribed EN vRNAs in larger amounts and purified them, to be able to co-incubate them by pairs under varying conditions. Based on the work on retroviruses, which showed that different conditions have to be used to analyse in vitro dimerization of the genomic RNA of different viruses (39,40), we incubated all possible pairs of EN vRNAs at varying ionic strengths and temperatures. Only a few weak interactions were detected at 37°C (data not shown), but these interactions and additional ones were more clearly detected at 55°C. Surprisingly, they formed also more efficiently at low ionic strength. We thus analysed the interactions between EN vRNAs in a buffer containing 50 mM sodium cacodylate, pH 7.5; 40 mM KCl and 0.1 MgCl2.
EN vRNAs form a single interaction network
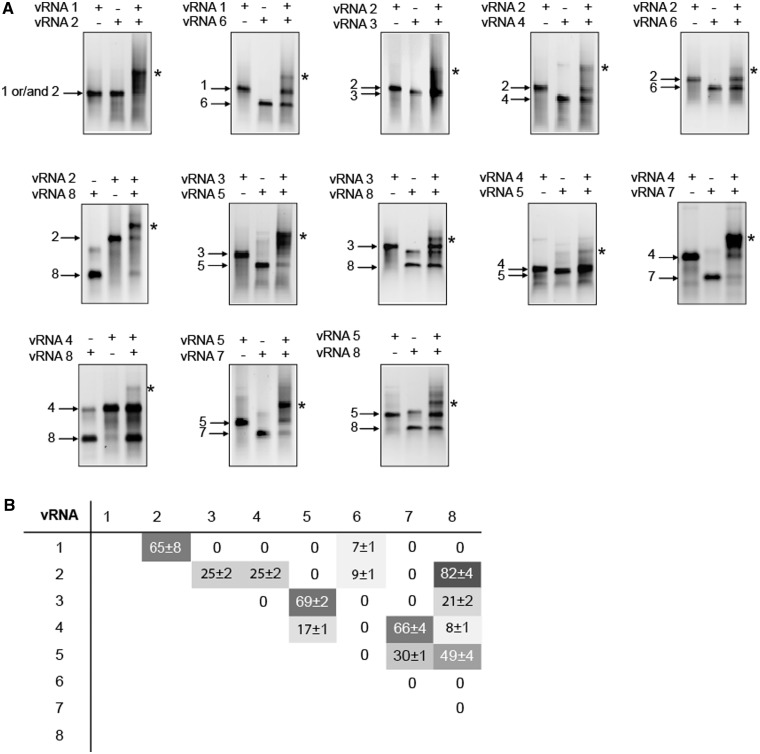
When incubated under these conditions, most individual EN vRNAs migrated as a single band corresponding to monomers (Figure 1 and Supplementary Figure S1). A significant band attributed to homodimeric species was observed with vRNA 8 (coding for NS1 and NS2/nuclear export protein (NEP)), and fainter dimeric bands were observed with vRNAs 4 (coding for HA), 5 (coding for NP) and 7 (coding for M1 and M2). Importantly, when pairs of EN vRNAs were co-incubated, 13 additional bands corresponding to intermolecular complexes were detected (Figure 1A and Supplementary Figure S1). Not all vRNA/vRNA interactions were equally strong: four complexes accounted for 7–17% of the RNA mass in the lane, five accounted for 21–49% and four accounted for 65–82% (Figure 1B). Each vRNA was involved in several interactions: three vRNAs interacted with two partners, one interacted with three partners, three interacted with four partners and one (vRNA 2) interacted with five other vRNAs (Figure 1). Remarkably, the network of interactions linking the EN vRNAs is different from the one linking the MO vRNAs (36). These two networks only share four interactions in common: those between vRNAs 2 and 6 (coding for NA), 4 and 7, 4 and 8 and 5 and 8.
Figure 1.
The eight EN vRNAs form a single interaction network in vitro. (A) Analysis of vRNA/vRNA interactions by agarose gel gelectrophoresis. Individual vRNAs are indicated by arrows and intermolecular complexes are marked by asterisks. (B) Quantification of the complexes. The weight fraction (%) of the RNA migrating as an intermolecular complex is expressed as mean ± standard error of the mean (n = 3–9).
Identification of the regions of the EN vRNAs involved in intermolecular interactions in vitro
The regions of the vRNAs containing segment-specific packaging signals have been mainly studied for WSN and PR8, two human H1N1 strains of IAV (25,26,28–35,41). These regions tend to be located towards the ends of the vRNAs: most packaging signals are located within ∼100 nt from both ends of the vRNA coding regions. In the case of MO, we recently showed that regions of the vRNAs involved in vRNA/vRNA interactions in vitro correspond to regions identified as packaging signals in WSN or/and PR8 (36). We therefore introduced systematic deletions in all EN vRNAs, except vRNA 6, to identify the regions involved in most vRNA/vRNA interactions detected in vitro. Analysis of the weakest interactions appeared technically more difficult, and we therefore did not attempt to identify the regions of vRNA 6 interacting with vRNAs 1 and 2 (Figure 1B).
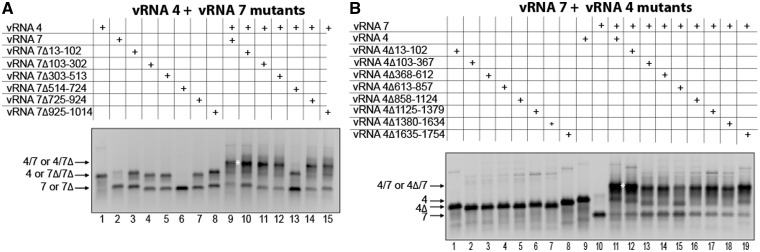
The vRNA 4/vRNA 7 interaction
We first introduced deletions close to the ends of vRNA 7, without affecting the promoter sequences, and tested their effect on the interaction with vRNA 4. Surprisingly, none of these deletions had a significant impact on the interaction with vRNA 4 (Figure 2A, lanes 10 and 15). We thus also constructed a series of deletion mutants covering the central part of vRNA 7. Remarkably, one of them, extending from nucleotides 514–724 completely abolished the interaction with vRNA 4 (Figure 2A, lane 13). In addition, we observed that all but the 514–724 deletions favoured homodimerization of vRNA 7 (Figure 2A, lanes 1 to 8). Similarly, we constructed a set of eight deletion mutants covering the entire vRNA 4 sequence, except the terminal promoters. Again, deletions close to the ends of the vRNA had no effect on the interaction between vRNAs 4 and 7 (Figure 2B, lanes 12 and 19). The most pronounced effect on this interaction was observed for the deletion covering nucleotides 613–857 of vRNA 4 (Figure 2B, lane 15). However, unlike deletion 514–724 in vRNA 7, deletion 613–857 did not totally abolish the vRNA 4/vRNA 7 interaction. This might be an indication that more than one region of vRNA 4 are involved in this interaction (see later). Accordingly, several other deletions also weakened the interaction between vRNAs 4 and 7, albeit less dramatically (Figure 2B, lanes 13–14 and 16–18).
Figure 2.
Region 514–724 of vRNA 7 and region 613–857 of vRNA 4 are required for optimal interaction between these vRNAs. Deletions were introduced in vRNA 7 (A) or vRNA 4 (B), and formation of the intermolecular complex was monitored by agarose gel electrophoresis.
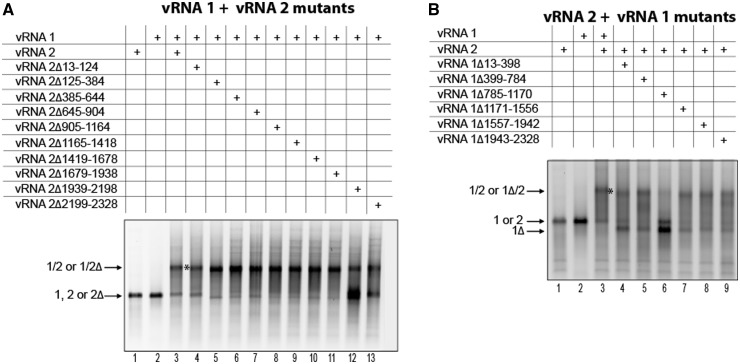
The vRNA 1/vRNA 2 interaction
We next introduced sequential deletions in vRNAs 1 and 2 to delineate the regions involved in the intermolecular interaction between these two vRNAs. None of the vRNA 1 and vRNA 2 deletion mutants formed homodimers, and they all migrated as expected based on their size (Figure 3 and data not shown). One deletion in vRNA 2, encompassing nucleotides 1939–2198 had a dramatic effect on the vRNA 1/vRNA 2 interaction (Figure 3A, lane 12). Deletion of nucleotides 785–1170 in vRNA 1 had a similar effect (Figure 3B, lane 6). These two deletions had major effects but did not completely abolish the interaction, suggesting minor contributions from other regions of both vRNAs.
Figure 3.
Region 1939–2198 of vRNA 2 and region 785–1170 of vRNA 1 are required for optimal interaction between these vRNAs. Deletions were introduced in vRNA 2 (A) or vRNA 1 (B), and formation of the intermolecular complex was monitored by agarose gel electrophoresis.
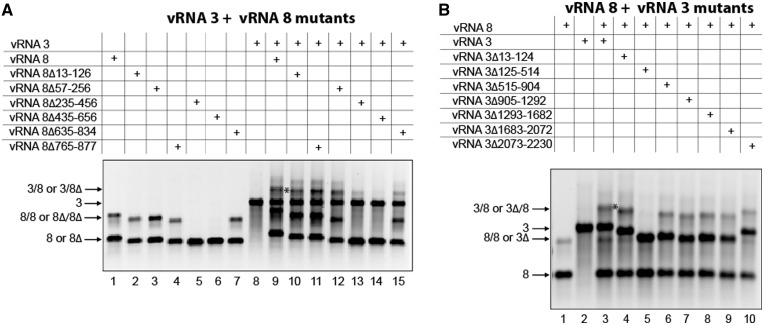
The vRNA 3/vRNA 8 interaction
We next studied the interaction between vRNAs 3 and 8 (Figure 4). As already mentioned, vRNA 8 significantly dimerized, and dimerization was also observed with all vRNA 8 mutants, except those in which nucleotides 235–456 or 435–656 were deleted (Figure 4A, lanes 1–7). Several deletions in this RNA had an impact on the interaction with vRNA 3: deletion of nucleotides 435–656 totally abolished the intermolecular interaction, whereas deletion of nucleotides 235–456 had a drastic effect, although some complex was still detected (Figure 4A, lanes 13 and 14). One possibility is that region 435–456 of vRNA 8, which was deleted in both mutants, is involved in the interaction with vRNA 3, together with downstream sequences. In addition, deletion of nucleotides 635–834 also had a noticeable, although more limited, effect on the vRNA 3/vRNA 8 interaction (Figure 4A, lane 15). Similarly, the analysis of vRNA 3 showed that deletion of nucleotides 125–514 totally abolished the intermolecular interaction (Figure 4B, lane 5), whereas deletions in the 515–2230 region had a limited but significant negative impact on this interaction (Figure 4B, lanes 6–10).
Figure 4.
Optimal interaction between vRNAs 3 and 8 requires region 435–656 of vRNA 8 and region 125–514 of vRNA 3. Deletions were introduced in vRNA 8 (A) or vRNA 3 (B), and formation of the intermolecular complex was monitored by agarose gel electrophoresis.
General features of the EN vRNA/vRNA interactions
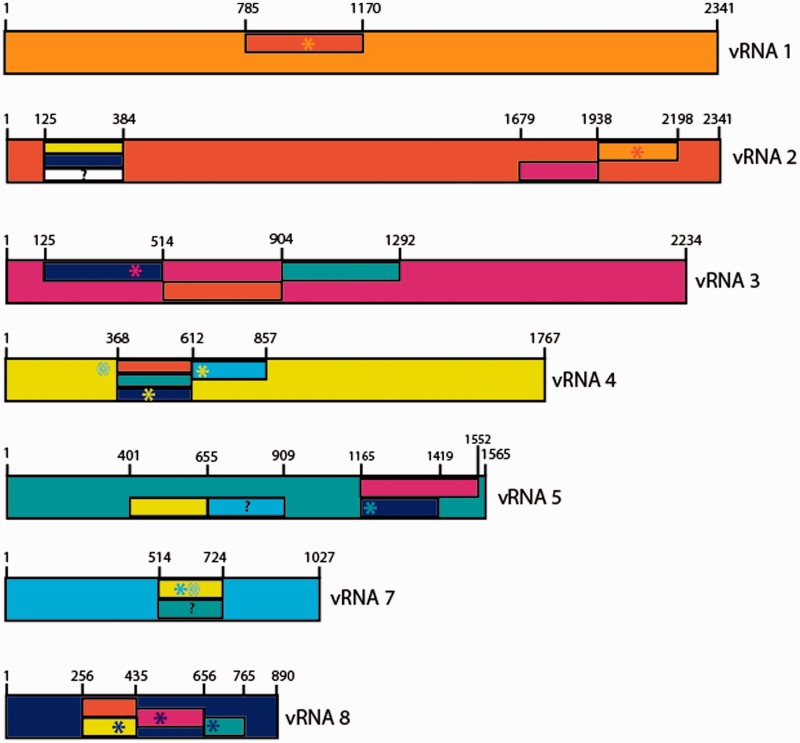
The strategy detailed previously for interactions between vRNAs 4 and 7, 1 and 2 and 3 and 8 was applied to all interactions detected in vitro, and allowed to gain information on the regions of the vRNAs involved in most of them (Figure 5, Supplementary Figures S2 and S3 and data not shown). In many cases, several (usually contiguous) deletions affect a given interaction, but one deletion has usually a more pronounced effect than the other ones, suggesting that it constitutes the main interacting domain, and only this region has been reported in Figure 5. The deletions that have less pronounced effects may either correspond to secondary interaction sites or affect the interaction indirectly by affecting the vRNA structure. However, in the case of the vRNA 3/vRNA 5 interaction, two non-overlapping adjacent deletions in vRNA 5 completely abolished the interaction with vRNA 3 (Figure 5), suggesting that the sequence of vRNA 5 interacting with vRNA 3 is located close to the junction between these two deletions (i.e. nt 1419). In contrast with our recent report on MO vRNAs (36) and with the location of the PR8 and WSN packaging regions (3), the regions involved EN vRNA/vRNA interactions are usually located >100 nt from the ends of the coding regions. The only exception could be the region of vRNA 5 interacting with vRNA 3, which remains to be delineated more precisely (see earlier in text and Figure 5). Deletion of nucleotides 635–834 of vRNA 8 abolished interaction with vRNA 5 (Figure 5 and Supplementary Figure S3), but deletion of nucleotides 765–877 did not (Supplementary Figure S3, lane 5), indicating that the region of vRNA 8 interacting with vRNA 5 is located >100 nt away from the 5′ end of the coding region of this vRNA. Remarkably, several regions of interaction are located close to the middle of the vRNAs (Figure 5). Deletion analysis of vRNA 1 did not allow to identify a region of this vRNA required for the interaction with vRNA 6 because none of the deletion significantly impaired this interaction. Deleting nucleotides 125–384 in vRNA 2 decreased the interaction with vRNA 6, but the effect was modest. As these two interactions appeared difficult to analyse, we did no attempt to identify the regions of vRNA 6 they involve.
Figure 5.
Summary of the vRNA regions involved in intermolecular vRNA interactions. vRNAs have different colours and are drawn to scale. Regions defined by deletion analysis are presented by rectangles that have the same colour as the interacting vRNA. Note that when only one of several overlapping deletions affects the interaction between two vRNAs, the region required for the interaction is shorter than this deletion, as the overlapping sequences are not required. Deletions that have limited effects are indicated by question marks. Sequences identified by ODN mapping or/and site-directed mutagenesis are indicated by asterisks.
Precise identification of the sequences of EN vRNAs involved in vRNA/vRNA interactions
We next attempted to precisely identify the nucleotides involved in several vRNA/vRNA interactions. When deletion experiments allowed us to identify the region of one vRNA involved in an intermolecular interaction, we used a series of antisense ODNs spanning this region to refine the interaction zone. Sequence analysis then usually allowed to propose a limited number of possible interactions between the zone identified by ODN scanning on one vRNA and the region identified by deletion on the other vRNA. These possible interactions were then tested with ODNs complementary to the second vRNA. When many interactions were possible, a systematic ODN scan of the longer vRNA was performed.
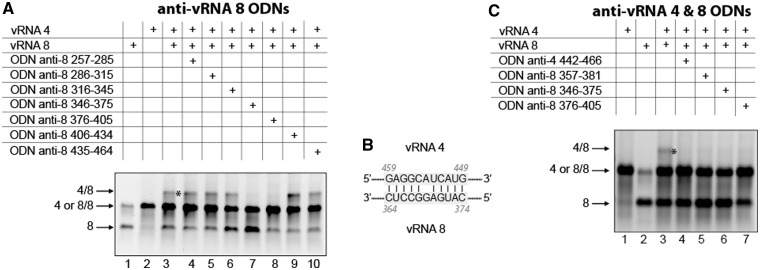
The vRNA 4/vRNA 8 interaction
A first example of this strategy is shown in Figure 6 for the interaction between vRNA 4 and 8. Deletion analysis indicated that region 257–434 of vRNA 8 interacts with vRNA 4 (Supplementary Figure S2B, lanes 6–8). Two adjacent ODNs complementary to nucleotides 346–375 and 376–405 almost totally abolished the vRNA 4/vRNA 8 interaction (Figure 6A, lanes 7–8). Knowing, that in vRNA 4, the region of interaction is located between nucleotides 368 and 612 (Supplementary Figure S2A), the likeliest interaction is between nucleotides 364–374 of vRNA8 and nucleotides 449–459 of vRNA 4 (Figure 6B): it involves six G-C and four A-U base-pairs, as well as a non-canonical G-A base pair. The existence of this interaction is inferred from the inhibitory effect of ODNs complementary to nucleotides 442–466 of vRNA 4 and to nucleotides 357–381 of vRNA 8 (Figure 6C). Note that the ODN complementary to nucleotides 346–375 of vRNA 8 also directly targets this interaction, whereas the ODN complementary to nucleotides 376–405 hybridizes immediately adjacent to that region and probably has an indirect effect (Figure 6A and C).
Figure 6.
Nucleotides 357–381 of vRNA 8 and nucleotides 442–466 of vRNA 4 are involved in the interaction between these vRNAs. (A) ODNs were used to map region 257–464 of vRNA 8. (B) Proposed interaction between vRNAs 4 and 8. (C) ODNs complementary to vRNAs 4 and 8 were used to test the proposed interaction.
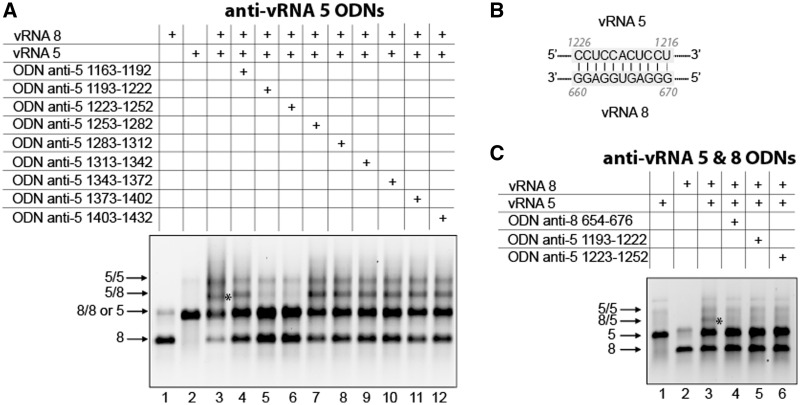
The vRNA 5/vRNA 8 interaction
Among a series of nine ODNs targeting region 1165–1419 of vRNA 5 identified by deletion analysis (Supplementary Figure S3B), two adjacent ODNs complementary to nucleotides 1193–1222 and 1223–1252, respectively, dramatically inhibited the interaction between vRNAs 5 and 8 (Figure 7A, lanes 5 and 6). Interestingly, nucleotides 1216–1226 of vRNA 5 are strictly complementary to nucleotides 660–670 of vRNA 8 (Figure 7B), which are located in region 657–764 identified by deletion analysis of this vRNA (Supplementary Figure S3A). As predicted, an anti-vRNA 8 ODN targeting nucleotides 654–676 also abolished the interaction between vRNAs 5 and 8, supporting the existence of the proposed base pairing between these vRNAs (Figure 7C).
Figure 7.
Interaction between vRNAs 5 and 8 involves nucleotides 1193–1252 of vRNA 5 and nucleotides 654–676 of vRNA 8. (A) ODNs were used to map region 1163–1432 of vRNA 5. (B) Proposed interaction between vRNAs 5 and 8. (C) ODNs complementary to vRNAs 5 and 8 were used to test the proposed interaction.
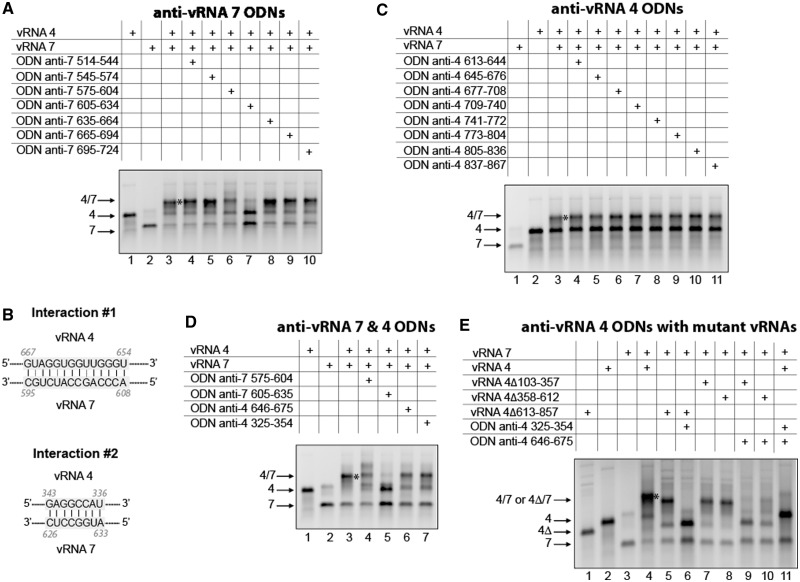
The vRNA 4/vRNA 7 interaction
Region 514–724 of vRNA 7 is involved in the interaction with vRNA 4 (Figure 2A), and ODN mapping of this region revealed that an anti-vRNA 7 ODN complementary to nucleotides 605–634 strongly inhibited this interaction (Figure 8A). Sequence analysis revealed that nucleotides 595–608 of vRNA 7 can form 14 consecutive base pairs, including 3 G-U base pairs, with nucleotides 654–667 of vRNA 4 (Figure 8B, interaction 1). However, none of the ODNs complementary to region 613–857 identified by deletion analysis (Figure 2B) had any significant effect on the interaction between vRNAs 4 and 7 (Figure 8C). Further sequence analysis revealed that nucleotides 626–633 of vRNA 7 are complementary to nucleotides 336–343 of vRNA 4 (Figure 8B, interaction 2). This interaction would also explain the inhibitory effect of the anti-vRNA 7 605–634 ODN (Figure 8A). However, an anti-vRNA 4 ODN targeting nucleotides 325–354 did not affect the interaction between vRNAs 4 and 7 (Figure 8D, lane 7). We thus suspected that interactions 1 and 2 might coexist, and this indeed appeared to be the case: the interaction between vRNAs 4 and 7 was drastically inhibited when two anti-vRNA 4 ODNs targeting, respectively, nucleotides 325–354 and 646–675 were co-incubated with the vRNAs (Figure 8E, lane 11). When substituting mutant vRNAs 4 with a deletion preventing one of the interactions for the WT vRNA 4, the anti-vRNA 4 ODN targeting the other interaction was sufficient to prevent formation of the vRNA 4/vRNA 7 complex (Figure 8E, lanes 6, 9 and 10). This represents the only case in which we identified two base pairings that are each sufficient to maintain the interaction between vRNAs at almost WT level.
Figure 8.
Alternative interactions between vRNAs 4 and 7. (A) ODNs were used to map region 514–724 of vRNA 7. (B) Proposed interactions between vRNAs 4 and 7. (C) ODNs were used to map region 613–867 of vRNA 4. (D) Anti-vRNA 4 and anti-vRNA 7 ODNs were used to test the two proposed interactions. (E) Combinations of anti-vRNA 4 ODNs or of an anti-vRNA 4 ODN and a vRNA 4 deletion mutant were used to test the coexistence of the two proposed interactions.
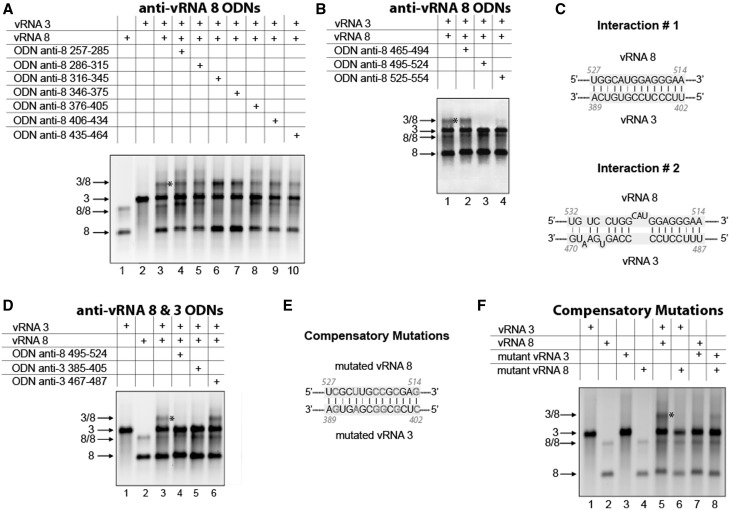
The vRNA 3/vRNA 8 interaction
Deletion analysis of vRNA 8 revealed that region 235–456, and especially nucleotides 435–656, are involved in the interaction with vRNA 3 (Figure 4A). None of the ODNs targeting the first of these regions had a potent inhibitory effect (Figure 9A), but two ODNs targeting nucleotides 495–524 and 525–554, respectively, abolished the interaction between vRNAs 3 and 8. Sequence analysis revealed two possible interactions between this region of vRNA 8 and region 125–514 of vRNA 3 (Figure 9C), which was identified by deletion mutations (Figure 4B). Anti-vRNA 3 ODNs selectively targeting interaction 1 or 2 supported the existence of interaction 1 only (Figure 9D). To further validate this result, we introduced compensatory mutations in the sequences of vRNAs 3 and 8 involved in interaction 1 (Figure 9E). As expected, introducing mutations in only one of the vRNAs abolished the interaction (Figure 9F, lanes 6 and 7), whereas combining the two mutated vRNA restored it (Figure 9F, lane 8), albeit not at the WT level.
Figure 9.
Nucleotides 389–402 in vRNA 3 interact with nucleotides 514–527 in vRNA 8. ODNs were used to map region 257–464 (A) and 465–554 (B) of vRNA 8. (C) Proposed interactions between vRNAs 3 and 8. (D) ODNs complementary to vRNAs 3 and 8 were used to test the two proposed interactions. (E) Compensatory mutations introduced in vRNAs 3 and 8 to test interaction 1. (F) Effects of mutations in vRNAs 3 or/and 8 on the intermolecular interaction.
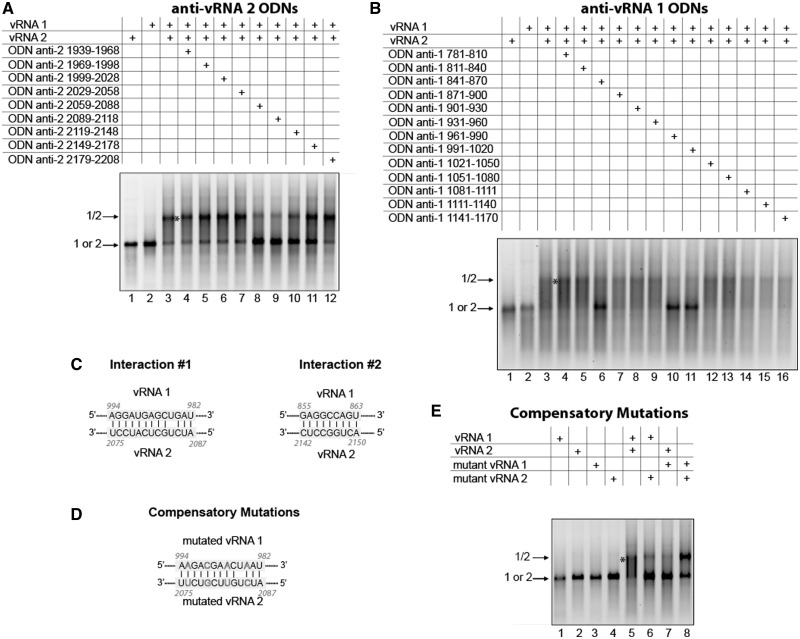
The vRNA 1/vRNA 2 interaction
Mapping of region 1939–2198 of vRNA 2, previously identified by deletion mutagenesis (Figure 3A), using anti-vRNA 2 ODNs identified region 2059–2118 as playing a role in the interaction with vRNA 1 (Figure 10A). The reciprocal experiment performed on region 781–1170 of vRNA 1 allowed us to delineate a region interacting with vRNA 2 within nucleotides 961–1020 (Figure 10B), but region 841–870 also seemed to play some role in the interaction between vRNAs 1 and 2. Bioinformatic analysis of the vRNA 1 and vRNA 2 sequences identified two interactions that could explain the effect of these regions (Figure 10C). However, the sequence of vRNA 2 involved in interaction 2 falls outside the region identified by ODN mapping (Figure 10A). To further establish the role of interaction 1 in the interaction between vRNAs 1 and 2, we introduced complementary mutations in these vRNAs (Figure 10D). When using only one mutated vRNA, the interaction was strongly reduced, but combining the two mutated vRNAs restored this interaction to WT levels (Figure 10E).
Figure 10.
Nucleotides 982–994 in vRNA 1 interact with nucleotides 2075–2087 in vRNA 2. ODNs were used to map region 1939–2208 of vRNA 2 (A) and region 781–1170 of vRNA 1 (B). (C) Proposed interactions between vRNAs 1 and 2. (D) Compensatory mutations introduced in vRNAs 1 and 2 to test interaction 1. (E) Effects of mutations in vRNAs 1 or/and 2 on the intermolecular interaction.
EM and tomography
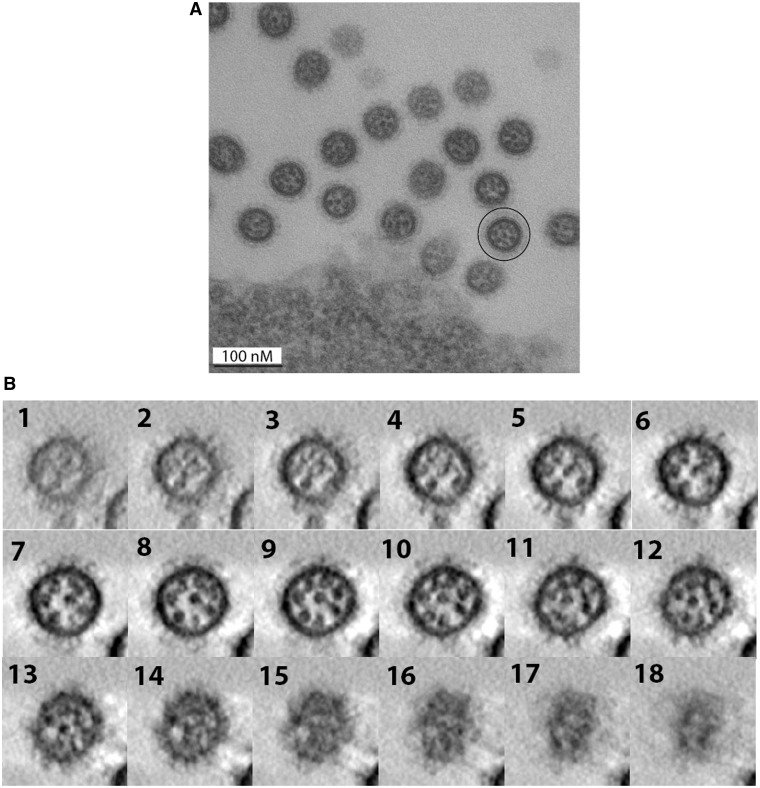
Our in vitro data showed that the network of interactions linking the EN vRNAs differs from the recently defined MO interaction network (36): different interactions that tend to involve different regions of the vRNAs were observed. If these (or at least some of these) interactions exist in virions, one would expect differences in the internal organization of EN and MO viruses. Using EM, zero to eight dots corresponding to vRNPs can be observed in transversal sections of EN viruses budding from MDCK cells (Figure 11A). When eight vRNPs are visible, seven vRNPs surround a central one (Figure 11A). These EM pictures indicate that the overall disposition of the vRNPs is similar in EN virions (Figure 11A) and in MO (36), WSN (42,43) and PR8 (42) virions.
Figure 11.
Electron microscopy (A) and electron tomography (B) of EN viral particles budding from MDCK cells. (A) Cross-sections of budding EN virions. A viral particle in which the eight individual vRNPs form a typical ‘7 + 1’ arrangement is circled. (B) Successive virtual cross-sections of an EN virion, from the bottom to the budding tip of the viral particle.
In electron tomography, cross-sections of individual EN virions revealed three different zones (Figure 11B and Supplementary Figure S4). In the central part of the virions, vRNPs appeared as individual round dots, but contacts were frequently observed between the dots (panels 5–10 of Figure 11B and panels 6–10 of Supplementary Figure S4). The apical part of the budding virions constitutes a transition zone, in which the dots corresponding to vRNPs progressively become elongated and smaller; density appears between the vRNPs and finally, just below the matrix layer, the individual vRNPs are not visible anymore (Figure 11B, panels 11–15 and Supplementary Figure S4, panels 11–15). Unexpectedly, the vRNPs also appeared as smaller elongated dots in the region of the virion that is closer from the producer cell. Numerous contacts were observed in this region (Supplementary Figure S4, panels 2–5), and it was frequently impossible to visualize the individual vRNPs (Figure 11B, panels 2–5).
DISCUSSION
Increasing evidence [reviewed in (3)] support the idea that the eight vRNPs constituting the IAV genome are selectively packaged into virions. Since the first demonstration of the existence of segment-specific packaging regions in an IAV vRNA using reverse genetics, direct interactions between vRNAs appeared as an attractive hypothesis to explain this process (26). This hypothesis is reinforced by the fact that no viral or cellular protein specifically recognizing an IAV segment-specific packaging signal has been identified so far. However, demonstrating the existence of specific vRNA/vRNA interactions in viral particles without any knowledge of the sequences that might be involved is challenging, and no such interaction has been identified, in vivo, to date.
We recently analysed the interactions taking place between the MO vRNAs in vitro (36). We found that the eight MO vRNAs are involved in a single network of interactions, and we showed that the regions of the vRNAs that are involved in three of the main interactions in vitro correspond, in the case of PR8 or WSN IAV, to regions containing packaging signals (36). We also detected many contacts between vRNPs, likely involving the packaging regions, using electron tomography, suggesting that the eight vRNPs are co-packaged as a supramolecular complex (36). Because little is known about the selective packaging of avian IAVs, we applied the same strategy to EN, an avian H5N2 IAV. As for MO, we found that the eight vRNAs are involved in a single interaction network (Figure 1).
However, marked differences between the MO and EN viruses were observed in our biochemical experiments. First, the experimental conditions allowing detection of specific interactions between vRNAs are different. The retrovirology field taught us that it is necessary to use different conditions, including temperature, to study dimerization of different viral RNAs: for instance, the optimal temperature for studying in vitro RNA dimerization is 37°C for human immunodeficiency virus-1 (39), 50°C for avian leukosis virus (44) and 55°C for murine leukemia virus (40). In our previous study on the human MO IAV, we detected specific interactions during in vitro transcription, which was performed at 37°C. In the case of the avian EN IAV, 55°C allowed specific detection of vRNA/vRNA interactions, whereas 37°C did not, but we have not systematically tested all intermediate temperatures. Incidentally, it has to be kept in mind that human IAVs replicate in the upper respiratory tract, where the temperature is 33°C, whereas avian IAVs replicate at 40°C, in the intestine of birds. Second, most interactions were observed in only one of the two viruses, and third, the regions of the vRNAs involved in these interactions are globally different. In MO, the vRNA/vRNA interactions were usually affected by deletions at the ends of the coding sequences (36), whereas here we observed that interactions between vRNAs are mediated by more central sequences (Figures 2–10 and Supplementary Figure S2 and S3). Even though the interaction networks are remarkably different between MO and EN viruses, it should be noted that some central regions might also play a role in the interaction between MO vRNAs (e.g. the central region of vRNA 4 in the vRNA 4/vRNA 7 interaction) (36).
We detected 13 intermolecular interactions between EN vRNAs (Figure 1), compared with only eight between MO vRNAs (36), and it is unclear whether all these interactions could take place within a single virion. It is possible that within virions, only a subset of these interactions, which could differ from one virion to another, do take place: this would explain the observation that the relative disposition of the eight vRNPs is not strictly conserved among MO or WSN virions (36,43). Besides, some of the interactions between EN vRNAs were fairly weak (e.g. between vRNAs 1 and 6, 2 and 6 and 4 and 8; Figure 1), and may not exist in virions, although they were reproducibly observed in vitro. However, vRNAs are packaged as vRNPs, and crystal structures of the NP protein suggested that vRNAs bind at the surface of the protein oligomers (45,46). In addition, chemical probing indicated that NP increases the accessibility of the vRNA bases (47). Thus, NP might facilitate intermolecular interactions between vRNAs, rather than prevent them.
EM and tomography revealed that the eight vRNPs are attached at the matrix layer at the tip of budding virions (36,42,43,48,49). Therefore, the interactions involving sequences located at similar distances from either ends of the two vRNAs are more likely to take place in virions: this is for instance the case of the interactions between vRNAs 2 and 3, 2 and 4, 2 and 8, 3 and 8, 4 and 7 and 4 and 8 (Figure 5). In the case of vRNAs 4 and 7, we detected two alternative interactions in vitro (Figure 8), but based on the argument developed previously, interaction 2 depicted in Figure 8B is more likely to take place in virions than interaction 1.
Electron tomography of budding EN virions (Figure 11 and Supplementary Figure S4) also revealed significant differences with MO (36) and WSN (43) virions. Towards the budding tip of the EN virions, the vRNPs appeared as small elongated dots; numerous contacts were observed between these dots, to a point that it progressively became impossible to distinguish the individual vRNPs (Figure11 and Supplementary Figure S4). This observation is reminiscent of the tomograms obtained with MO and WSN virions (36,43). In the middle of the viral particles, EN vRNPs appeared as larger round dots. Numerous contacts were also observed between vRNPs in this region of the tomograms, and the vRNPs sometimes appeared connected by a thin linker (Figure 11 and Supplementary Figure S4). Such contacts were also frequently observed in WSN virions (43), but were rare in MO virions (36). In the lower part of most tomograms of EN virions, vRNPs again appeared elongated and in close contact, and it was frequently impossible to distinguish the individual vRNPs (Figure 11 and Supplementary Figure S4). In this region, EN virions differ from MO and WSN virions, in which individual vRNPs can be observed (36,43). Considering that vRNPs adopt a panhandle structure in virions (7,8,50), the numerous contacts that we have observed in the central and lower part of EN virions (Figure 11 and Supplementary Figure S4), but not MO virions (36), are consistent with interactions involving sequences distributed all along the EN vRNAs, as we observed in vitro (Figure 5).
Our in vitro studies suggest that different vRNA/vRNA interactions might underlie the selective packaging of vRNAs in different IAVs. In the present study, we identified five intermolecular interactions between EN vRNAs at the nucleotide level, and sequence comparison shows that they are not conserved in the MO and PR8 IAV strains (Supplementary Figure S5). This idea might seem contradictory with the observation that conserved codons tend to accumulate in the regions of the vRNAs known to contain packaging and with the fact that mutations of some of these codons affect vRNA packaging (33–35). However, some of these conserved codons appeared to be important for vRNA packaging in WSN, but not PR8 (35), indicating that some packaging signals are not conserved, even among human IAVs of the same serotype. The fact that the sequences involved in the interactions between the EN vRNAs are not conserved in the MO and PR8 IAV strains is consistent with the observation that EN and MO vRNAs establish different networks of intermolecular interactions in vitro. This lack of conservation could have major consequences for genetic reassortment by creating genomic incompatibilities between vRNAs from different IAVs. Indeed, by studying genetic reassortment between EN and MO viruses, we recently demonstrated that reassortment between these two viruses is restricted at the level of the vRNAs themselves, and not only at the protein level (Essere, B. Yver, M. et al., submitted for publication).
The ultimate step of our strategy will be to demonstrate that at least some of the interactions we observed in vitro do exist in virions or/and in infected cells and to test their importance in the selective packaging of the interacting vRNAs. An important step towards this goal is the precise identification of the nucleotides involved in five vRNA/vRNA interactions (Figures 5–10). In addition, for two interactions, we confirmed the existence of the base pairing using compensatory mutations in the two interacting vRNAs (Figure 9 and 10). As the interacting sequences we identified are all located in coding regions (in antisense orientation) (Figure 5), testing the existence and function of these interactions in cell culture will ideally involve using silent compensatory mutations. However, as usually only the third base of the codons can be mutated without affecting the protein sequence, it is generally not possible to design such mutants. For instance, the mutations engineered in vRNA 3 to test the interaction with vRNA 8 are silent, but those in vRNA 8 are not (Figure 9E). However, the mutations introduced in vRNAs 1 and 2 to test their interaction are both silent (Figure 10D). This pair of mutants, or similar ones, opens up the possibility to study the existence and the functional role of some of the vRNA/vRNA interactions we detected in vitro and to test the hypothesis that vRNA/vRNA interactions might drive the selective incorporation of vRNPs into IAV budding particles (3,26). Indeed, work in progress in our laboratories indicated that the interaction between vRNAs 2 and 8 does take place in vivo and participates in the selective packaging of these vRNAs (Gavazzi, C. et al., in preparation).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Funding for open access charge: Centre National de la Recherche Scientifique (CNRS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Dr Redmond Smyth is acknowledged for critical reading of the manuscript.
REFERENCES
- 1.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Micro. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 2.Palese P, Shaw M. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott. Williams and Wilkins, Philadelphia; 2006. pp. 1647–1689. [Google Scholar]
- 3.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J. Gen. Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 4.Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 5.Neumann G, Hobom G. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 1995;76:1709–1717. doi: 10.1099/0022-1317-76-7-1709. [DOI] [PubMed] [Google Scholar]
- 6.Neumann G, Zobel A, Hobom G. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology. 1994;202:477–479. doi: 10.1006/viro.1994.1365. [DOI] [PubMed] [Google Scholar]
- 7.Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortin J, Martin-Benito J. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad. Sci. USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou YY, Vafabakhsh R, Doganay S, Gao Q, Ha T, Palese P. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl Acad. Sci. USA. 2012;109:9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laver WG, Downie JC. Influenza virus recombination: I. Matrix protein markers and segregation during mixed infections. Virology. 1976;70:105–117. doi: 10.1016/0042-6822(76)90240-3. [DOI] [PubMed] [Google Scholar]
- 13.Lubeck M, Palese P, Schulman J. Nonrandom association of parental genes in influenza A virus recombinants. Virology. 1979;95:269–274. doi: 10.1016/0042-6822(79)90430-6. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima K, Sugiura A. Three-factor cross of influenza virus. Virology. 1977;81:486–489. doi: 10.1016/0042-6822(77)90165-9. [DOI] [PubMed] [Google Scholar]
- 15.Chambers TM, Webster RG. Defective interfering virus associated with A/Chicken/Pennsylvania/83 influenza virus. J. Virol. 1987;61:1517–1523. doi: 10.1128/jvi.61.5.1517-1523.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis AR, Nayak DP. Sequence relationships among defective interfering influenza viral RNAs. Proc. Natl Acad. Sci. USA. 1979;76:3092–3096. doi: 10.1073/pnas.76.7.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duhaut SD, Dimmock NJ. Heterologous protection of mice from a lethal human H1N1 influenza A virus infection by H3N8 equine defective interfering virus: comparison of defective RNA sequences isolated from the DI inoculum and mouse lung. Virology. 1998;248:241–253. doi: 10.1006/viro.1998.9267. [DOI] [PubMed] [Google Scholar]
- 18.Duhaut SD, Dimmock NJ. Defective segment 1 RNAs that interfere with production of infectious influenza A virus require at least 150 nucleotides of 5′ sequence: evidence from a plasmid-driven system. J. Gen. Virol. 2002;83:403–411. doi: 10.1099/0022-1317-83-2-403. [DOI] [PubMed] [Google Scholar]
- 19.Duhaut SD, McCauley JW. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology. 1996;216:326–337. doi: 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 20.Jennings P, Finch J, Robertson J. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983;34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima K, Ueda M, Sugiura A. Origin of small RNA in von Magnus particles of influenza virus. J. Virol. 1979;29:1142–1148. doi: 10.1128/jvi.29.3.1142-1148.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nayak DP, Sivasubramanian N, Davis AR, Cortini R, Sung J. Complete sequence analyses show that two defective interfering influenza viral RNAs contain a single internal deletion of a polymerase gene. Proc. Natl Acad. Sci. USA. 1982;79:2216–2220. doi: 10.1073/pnas.79.7.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odagiri T, Tashiro M. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J. Virol. 1997;71:2138–2145. doi: 10.1128/jvi.71.3.2138-2145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivasubramanian N, Nayak D. Defective interfering influenza RNAs of polymerase 3 gene contain single as well as multiple internal deletions. Virology. 1983;124:232–237. doi: 10.1016/0042-6822(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 25.Fujii K, Ozawa M, Iwatsuki-Horimoto K, Horimoto T, Kawaoka Y. Incorporation of influenza A virus genome segments does not absolutely require wild-type sequences. J. Gen. Virol. 2009;90:1734–1740. doi: 10.1099/vir.0.010355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl Acad. Sci. USA. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Q, Chou YY, Doganay S, Vafabakhsh R, Ha T, Palese P. The Influenza A Virus PB2, PA, NP, and M Segments Play a Pivotal Role during Genome Packaging. J. Virol. 2012;86:7043–7051. doi: 10.1128/JVI.00662-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y, Huang T, Ly H, Parslow TG, Liang Y. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 2008;82:229–236. doi: 10.1128/JVI.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozawa M, Maeda J, Iwatsuki-Horimoto K, Watanabe S, Goto H, Horimoto T, Kawaoka Y. Nucleotide sequence requirements at the 5′ end of the influenza A virus M RNA segment for efficient virus replication. J. Virol. 2009;83:3384–3388. doi: 10.1128/JVI.02513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gog JR, Afonso EDS, Dalton RM, Leclercq I, Tiley L, Elton D, von Kirchbach JC, Naffakh N, Escriou N, Digard P. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh GA, Rabadán R, Levine AJ, Palese P. Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 2008;82:2295–2304. doi: 10.1128/JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fournier E, Moules V, Essere B, Paillart JC, Sirbat JD, Isel C, Cavalier A, Rolland JP, Thomas D, Lina B, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 2012;40:2197–2209. doi: 10.1093/nar/gkr985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brule F, Marquet R, Rong L, Wainberg MA, Roques BP, Le Grice SF, Ehresmann B, Ehresmann C. Structural and functional properties of the HIV-1 RNA-tRNA(Lys)3 primer complex annealed by the nucleocapsid protein: comparison with the heat-annealed complex. RNA. 2002;8:8–15. doi: 10.1017/s1355838202010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moules V, Ferraris O, Terrier O, Giudice E, Yver M, Rolland J, Bouscambert-Duchamp M, Bergeron C, Ottmann M, Fournier E, et al. In vitro characterization of naturally occurring influenza H3NA- viruses lacking the NA gene segment: toward a new mechanism of resistance. Virology. 2010;404:215–224. doi: 10.1016/j.virol.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 39.Marquet R, Baudin F, Gabus C, Darlix JL, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tounekti N, Mougel M, Roy C, Marquet R, Darlix JL, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J. Mol. Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 41.Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 2005;79:3766–3774. doi: 10.1128/JVI.79.6.3766-3774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 43.Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, Sagara H, Kawaoka Y. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 2012;3:639. doi: 10.1038/ncomms1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polge E, Darlix JL, Paoletti J, Fosse P. Characterization of loose and tight dimer forms of avian leukosis virus RNA. J. Mol. Biol. 2000;300:41–56. doi: 10.1006/jmbi.2000.3832. [DOI] [PubMed] [Google Scholar]
- 45.Ng AKL, Zhang H, Tan K, Li Z, Liu JH, Chan PKS, Li S-M, Chan WY, Au SWN, Joachimiak A, et al. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22:3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 47.Baudin F, Bach C, Cusack S, Ruigrok RW. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc. Natl Acad. Sci. USA. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elton D, Digard P, Tiley L, Ortin J. In: Influenza Virology: Current Topics. Kawaoka Y, editor. Caister Academic Press; 2006. Wymondham, Norfolk NR18 0JA, England, pp. 1–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.