Abstract
Humans have five RecQ helicases, whereas simpler organisms have only one. Little is known about whether and how these RecQ helicases co-operate and/or complement each other in response to cellular stress. Here we show that RECQL5 associates longer at laser-induced DNA double-strand breaks in the absence of Werner syndrome (WRN) protein, and that it interacts physically and functionally with WRN both in vivo and in vitro. RECQL5 co-operates with WRN on synthetic stalled replication fork-like structures and stimulates its helicase activity on DNA fork duplexes. Both RECQL5 and WRN re-localize from the nucleolus into the nucleus after replicative stress and significantly associate with each other during S-phase. Further, we show that RECQL5 is essential for cell survival in the absence of WRN. Loss of both RECQL5 and WRN severely compromises DNA replication, accumulates genomic instability and ultimately leads to cell death. Collectively, our results indicate that RECQL5 plays both co-operative and complementary roles with WRN. This is an early demonstration of a significant functional interplay and a novel synthetic lethal interaction among the human RecQ helicases.
INTRODUCTION
RecQ helicases are a highly conserved group of enzymes involved in the maintenance of genomic stability (1,2). Whereas lower organisms such as bacteria and yeast have one RecQ member per species, there are five RecQ helicases in humans. Mutations in three of the five human RecQ helicases are associated with autosomal recessive diseases such as Bloom (BLM), Werner (WRN) and Rothmund–Thomson (RECQL4) syndromes, characterized by premature aging and carcinogenesis (3,4). RECQL1 and RECQL5 are not yet associated with any disease, but are highly abundant and important. RecQ helicases are multifunctional genome caretakers and play significant roles in various metabolic processes such as DNA replication, recombination, transcription, DNA base excision repair and telomere maintenance (5).
BLM plays an important role in suppression of sister chromatid exchanges (SCEs) and is involved in dissolution of double Holliday junctions together with the topoisomerase IIIα /RMI1 (BLAP75) complex (6,7). WRN encodes a multifunctional protein with both DNA helicase and exonuclease activities (8), and coordinated action of both catalytic activities could be involved in processing of replication and recombination intermediates as well as telomere maintenance (9,10). WRN plays an important role in replication fork stabilization and prevents aberrant recombination events at stalled forks. Further, Werner syndrome (WS) cells are highly sensitive to replicative stress (11). Loss of WRN affects cell cycle progression and results in enhanced accumulation of DNA double-strand breaks (DSBs) and instability at common fragile sites in cells experiencing oncogene-induced replication stress (12).
Although much insight has been gained into the functions of BLM and WRN, the role of RECQL5 is still relatively unknown. RECQL5 is abundantly expressed throughout the cell cycle and has been attributed with some important roles. RECQL5 has been implicated in DNA replication (13–15), recombination (16) and transcription (17–19). RECQL5-deficient cells accumulate spontaneous Rad51 and γ-H2AX foci and are prone to gross chromosomal rearrangements in response to replication stress (16). Although there is no human disease yet to be associated with lack of RECQL5, RECQL5 deficiency in mice results in cancer predisposition (16), indicating that RECQL5 functions as a tumor suppressor.
RECQL5 plays a potential role in homologous recombination (HR) by displacing Rad51 nucleoprotein filaments (16) and is proposed to have overlapping functions with that of BLM in suppression of SCEs (20,21). Our present study focuses on understanding the role of RECQL5 and its functional interplay with BLM and WRN. There are only limited studies addressing whether the human RecQ helicases play co-operative or complementary roles with each other in the maintenance of genomic stability. We have previously reported potential physical and functional interactions between BLM and WRN (22) and between RECQL4 and BLM (23). Here, we report a novel co-operation between RECQL5 and WRN in response to replicative stress and demonstrate that RECQL5 is essential for cell survival in the absence of WRN. Our study implies for the first time, a synthetic lethal interaction between RECQL5 and WRN and that RECQL5 could have important backup functions.
MATERIALS AND METHODS
Cell lines and transfection
SV40-transformed normal GM637, AG11395 (WRN-deficient) and GM08505 (BLM-deficient) fibroblasts were obtained from Coriell cell repositories (Camden, NJ, USA) and cultured in complete MEM medium supplemented with 10% FBS, 1% Pen–Strep along with 1X non-essential and essential amino acids, vitamins and 2 mM glutamine. For live-cell micro-irradiation experiments, cells were plated 24 hrs before transfection (with GFP-RECQL5) in 2-cm glass-bottom plates (MatTek Corporation, Ashland, MA, USA) at 60–70% confluency. Lipofectamine LTX was used for transfection, following the manufacturer’s instructions (Invitrogen, Life Technologies, Grand Island, NY, USA).
Laser micro-irradiation and confocal microscopy
Micro-irradiation has been used to induce DNA DSBs. A Nikon Eclipse 2000E spinning disk confocal microscope was used with five laser imaging modules attached with a CCD camera (Hamamatsu, Tokyo, Japan), as previously described (24,25). Localized DNA DSBs were induced using 435-nm SRS NL100 nitrogen laser (Photonics Instruments, St. Charles, IL, USA) with optimization of the laser intensity to 7% (24). Positions internal to the nuclei of live cells transfected with GFP-RECQL5 were targeted using a 40× oil objective lens. Images were captured at various time points and analysed using Volocity 5.0 build 6 (Improvision/PerkinElmer, Coventry, UK). Retention experiments were performed using a specially designed environmental chamber to maintain physiological conditions (37°C, 5% CO2). The recruitment kinetics was calculated by plotting an increase in fluorescence intensity with time. The data points represent the mean of three independent experiments with error bars.
Generation of stable RECQL5 knockdown cells and depletion of other RecQ helicases
Stable RECQL5 lentiviral knockdown cells were generated as previously described (15). The pLKO.1 vector harboring a mission short hairpin RNA (shRNA)-RECQL5 (TRCN0000051415) targeting the coding region of human RECQL5 was obtained from Sigma Aldrich (St. Louis, MO, USA). Plasmid #1864 shRNA construct expressing scrambled sequence (deposited by Sabatini lab) was purchased from Addgene (Cambridge, MA, USA). For lentiviral transduction, 2 × 105 cells were seeded in 10-cm culture plates and transfected the following day with the lentivirus. Cells were split 48 h after transfection and selected in the presence of 2 µg/ml puromycin.
Stable RECQL4, BLM and WRN-depleted cells were also generated by a similar approach using mission shRNA lentiviral constructs, TRCN000000051169 (RECQL4), TRCN00000004906 (BLM) and TRCN00000004902 (WRN) from Sigma Aldrich. Transient depletion of WRN in GM637 fibroblasts was achieved by using small interference RNA (siRNA)–WRN (Dharmacon, Lafayette, CO, USA), as described previously (26). Briefly, cells were plated to 50–60% confluence in 10-cm dishes 24 h before transfection and transfected with siRNA WRN and control siRNA using Lipofectamine 2000 (Invitrogen). A second transfection was performed similarly after 24 h. Seventy-two hours after the initial transfection, cells were harvested for subsequent analysis of growth assays and DNA fiber experiments. The sequence of WRN siRNA was 5′-GTGTATAGTTACGATGCTAGTGATT-3′, and that of the control siRNA was 5′-UUCUCCGACGUGUCACGUUU-3′.
DNA fiber assays
The cells were exposed to 50 µM BrdU for 15 minutes and immediately washed with cold phosphate buffered saline (PBS). Chromosome spreads were prepared as described previously (27,28). DNA spreads were air-dried and fixed in 3:1 methanol/acetic acid. The slides were acid treated with 2.5 M HCl at room temperature for 1 h, neutralized in Tris buffer, pH 7.5 for 10 min and subsequently blocked for 1 h with 5% bovine serum albumin (BSA), 10% goat serum in PBS. Antibodies were diluted in blocking buffer: mouse monoclonal antibody for BrdU (BD Biosciences, San Jose, CA, USA) 1:50; anti-mouse Dylight 488 (Jackson ImmunoResearch, West Grove, PA, USA) 1:1000. Incubations with antibodies were carried out at 37°C for 1 h (for primary antibodies) and 45 min (for secondary antibodies). DNA was stained with SYTOX orange (Invitrogen) for 5 min and washed with PBS. Slides were finally mounted in anti-fade prolong mounting medium (Invitrogen). Microscopy was carried out using a Zeiss Axiovert 200M microscope and analysed using the Axio Vision software packages. The number of fibers were counted and plotted against their length (μm).
Helicase assays
A forked DNA duplex (D50/49, 26 bp, Table 1) was prepared by labeling the D50 oligonucleotide (IDT, Coralville, Iowa, USA) with the T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) and annealed to the complementary oligonucleotide D49. A Holliday junction (HJ) substrate was prepared from X12 oligonucleotides, as described previously (29). Helicase assays were carried out in the buffer containing 20 mM Tris HCl (pH 7.4), 20 mM NaCl, 25 mM KCl, 1 mM DTT, 5% glycerol, 2.5 mM MgCl2, 100 μg/ml BSA and 2.5 mM ATP. Indicated amounts of RECQL5 and WRN were pre-mixed in the helicase buffer, and the reactions were initiated by addition of 0.5 nM substrate, and incubated at 37°C for 20 min. Reactions were then stopped by the addition of stop buffer (35 mM EDTA, 0.9% SDS, 25% glycerol, 0.04% bromophenol blue, 0.04% xylene cyanol), separated on 10% native polyacrylamide gels and analysed by autoradiography.
Table 1.
List of oligonucleotides used in the study
| Oligo name | Sequence 5′-3′ |
|---|---|
| D50 | GGGACGCGTCGGCCTGGCACGTCGGCCGCTGCGGCCAGGCACCCGATGGC |
| D49 | TTTGTTTGTTTGTTTGTTTGTTTGCCGACGTGCCAGGCCGACGCGTCCC |
| X12-1 | GACGCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCCG |
| X12-2 | CGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTC |
| X12-3 | GACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGC |
| X12-4 | GCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAGAATTCGGCAGCGTC |
| Exo 43 | AGTGCAGACTGCTGCTGAACGTACCCTGATCGTCGTCACGTCA |
| Exo 15 | TGACGTGACGACGAT |
| RS1 | ACTATCATTCAGTCATGTAACCTAGTCAATCTGCGAGCTCGAATTCACTGGAGTGACCTC |
| RS2 | ATTGACTAGGTTACATGACTGAATGATAGT |
| RS3 | GAGGTCACTCCAGTGAATTCGAGCTCGCAGCCCCTCTAGGTTACATGACTGAATGATAGT |
Recombinant proteins
Recombinant RECQL5 protein and the ATPase dead K58R RECQL5 were purified from Escherichia coli by overexpression as fusion proteins with a self-cleaving intein–chitin-binding domain in BL21(DE3) CodonPlus RIPL strain (Stratagene, Agilent Technologies, Santa Clara, CA, USA), as previously described (30). Recombinant histidine-tagged WRN protein was purified using a baculovirus/insect cell expression system, as previously described (31). BLM was a kind gift from Ian Hickson, University of Copenhagen, Denmark.
Strand exchange
All the oligonucleotides used were listed in Table 1. Stalled replication fork-like duplex lacking the leading strand (3′-flap duplex) was generated as previously described (14). Homologous arms were designed with a 5-nt region of heterology adjacent to the three-way junction to prevent spontaneous branch migration. Briefly, the oligo RS1 was 5′-end labeled with T4 polynucleotide kinase and annealed with RS2. A three-way flap-like structure was generated by annealing RS1/2 partial duplex with RS3, which is complementary with RS1, except for the five underlined bases (Table 1). This results in the formation of a bubble-like structure as a result of strand exchange. Strand exchange was performed in the buffer containing 20 mM Tris-HCl (pH 7.5), 8 mM DTT, 5 mM MgCl2, 10 mM KCl, 10% glycerol, 80 μg/ml BSA and 5 mM ATP; 2 nM of WRN was incubated with 0.5 nM three-way flap-like duplex and increasing concentrations of RECQL5 at 37°C for 30 min. The reactions were then stopped and analysed on a 10% native PAGE.
Co-immunoprecipiation
Cells were grown in 15-cm glass plates, harvested as indicated and lysed in 1 ml of lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 mM PMSF and 1% Triton X-100), supplemented with protease inhibitor (Roche Applied Sciences, Indianapolis, IN, USA). Lysis was performed in the presence of ethidium bromide (50 μg/ml) by rotating end-over-end for 30 min at 4°C and centrifuged at 14 000 g for 20 min. The supernatants or whole cell extracts were collected and subjected for pre-clearing with Protein A/G beads (Thermo Fisher Scientific, Waltham, MA, USA). The cell extracts were incubated overnight at 4°C with affinity purified anti RECQL5 antibody or with rabbit IgG (Santa Cruz Inc, Santa Cruz, CA, USA). Immune complexes were subsequently incubated with Protein A/G agarose beads for 5 h at 4°C. The beads were then washed 4–5 times with 50 mM Tris pH 7.4, 150 mM NaCl and 0.2% Triton X-100, and finally re-suspended in 20 μl of 2× SDS loading buffer and denatured at 95°C for 5 min to release bound proteins. The immunoprecipitates were analysed by western blotting and probed with custom-made mouse monoclonal antibody for WRN (clone 1B6 (32)).
In vitro immunoprecipitation
Both RECQL5 (1.2 μg) and WRN (1.2 μg) were pre-incubated with either rabbit IgG (Santa Cruz), or anti RECQL5 antibody in 200 μl of helicase buffer with 5 μg/ml BSA and incubated at 4°C for 90 min. Protein A/G beads were subsequently added to the protein mix and incubated at 4°C for 2 h. The beads were then isolated, washed 5 times in 500 μl of helicase buffer containing 150 mM NaCl and 0.1% BSA and processed as described above.
Growth assays
Scrambled and RECQL5-depleted control and WS fibroblasts were counted and plated 96 h after transduction and 48 h post selection into 18 dishes, 104 cells/dish. Three dishes were harvested every 24 h and counted using a Coulter counter. The first set of three harvested on day 1 were used to normalize as plating controls. The data points represent the mean of three independent experiments, with error bars.
Flow cytometry
To analyse the cell cycle distribution, cells were harvested (96 h after transduction and 48 h after selection in puromycin) by trypsinization using the pre-used media to count for floating (mitotic) cells. The cells were then washed twice with PBS, fixed by 70% ice-cold ethanol and stored at −20°C. The cells were then washed twice with ice-cold PBS and resuspended with 10 μg/ml propidium iodide (PI) solution with 1 mg/ml RNAseA. FACS analysis was performed by Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA) and analysed using FlowJO software.
FITC Annexin assays
To analyse cell death/apoptosis, fluorescein isothiocyanate (FITC) Annexin assays were performed, 96 h after transduction and 48 h after selection in puromycin, using BD Pharminogen FITC Annexin V kit, per the manufacturer’s instructions. Flow cytometric analysis was performed using Accuri C6 flow cytometer (BD Biosciences) and analysed by FlowJo software.
Analysis of metaphase chromosomes
Metaphase spreads were prepared from RECQL5-depleted control and WRN-deficient fibroblasts. Cells were then harvested and incubated in 75 mM KCl for 20 min at 37°C, followed by fixation in ice-cold (3:1) methanol and glacial acetic acid. Metaphase spreads were then made by dropping the cells onto a glass slide and stained with DAPI. Images were captured using Cytovision software (Applied Imaging Corp, Grand Rapids, MI, USA).
List of antibodies used
Polyclonal rabbit anti-RECQL5 antibody was generated against the C-terminal portion of RECQL5 (amino acids 813–963) and affinity purified as described previously (15). Mouse monoclonal antibody against WRN (clone 1B6) was generated previously (32). Other antibodies used in this study were: rabbit anti-BLM, rabbit anti-53BP1 and mouse monoclonal anti-actin antibodies purchased from Abcam (Cambridge, MA, USA); rabbit anti-Rad51 and rabbit anti-WRN (H-300) from Santa Cruz Biotechnology and rabbit polyclonal antibody for cleaved caspase-3 (Asp 175) was obtained from Cell Signaling (Danvers, MA, USA).
RESULTS
Retention time of GFP-RECQL5 at the laser-induced DSB sites is increased in the absence of BLM and WRN
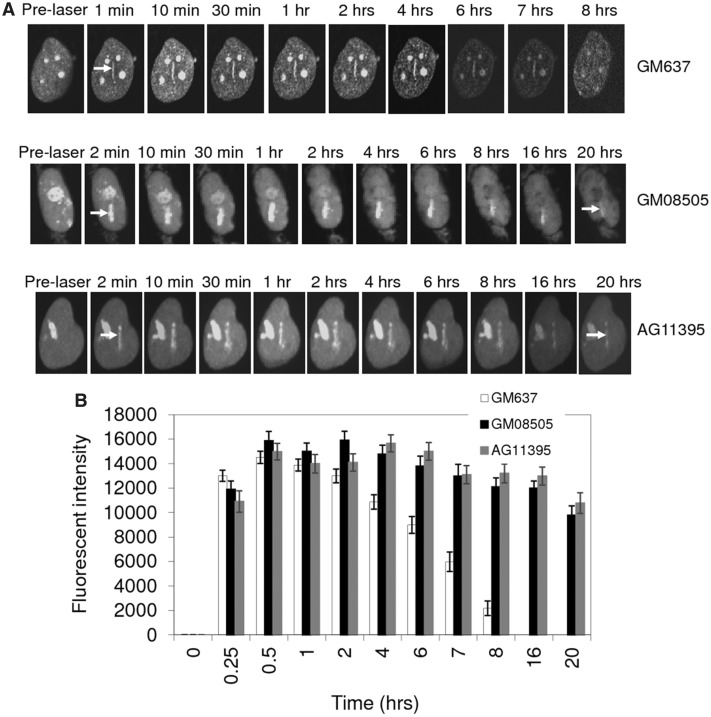
Micro-irradiation is a direct strategy to induce localized DNA DSBs (33–35). We and others have used this approach to study the recruitment of RecQ helicases to the DSB sites (25,36,37). We have recently studied the recruitment dynamics of RECQL5 and shown that the N-terminal helicase and C-terminal KIX domains are essential for the efficient recruitment and stable retention of RECQL5 at DSB sites (24). To study the effect of other human RecQ helicases on RECQL5 function in vivo, similar recruitment and retention studies were performed in WRN-(AG11395) and BLM-(GM08505) deficient fibroblasts. SV40-transformed GM637 were used as control fibroblasts. GFP-RECQL5 associates at the laser-induced DSB sites for 7–8 h in normal (GM637) fibroblasts (24) (Figure 1A). Similar recruitment experiments in GM08505 and AG11395 fibroblasts indicate that the recruitment of RECQL5 is independent of BLM and WRN, but that the dissociation of GFP-RECQL5 from the DSB sites is significantly slower in the absence of BLM and WRN (>20 h, Figure 1A). The plot illustrates the longer retention of GFP-RECQL5 at the damage sites in BLM- and WRN-deficient fibroblasts compared with that of control fibroblasts (Figure 1B).
Figure 1.
Retention of GFP-RECQL5 at laser-induced DSBs is longer in BLM- and WRN-deficient fibroblasts. (A) Retention of GFP-RECQL5 in GM637 (normal), GM08505 (BLM-deficient) and AG11395 (WRN-deficient) fibroblasts. (B) Plot showing longer retention of GFP-RECQL5 in BLM- and WRN-deficient fibroblasts compared with the normal fibroblasts. The increase in the fluorescent intensity was plotted with the time points (in hours).The plot represents mean of three independent experiments (each analysing ∼8–10 cells).
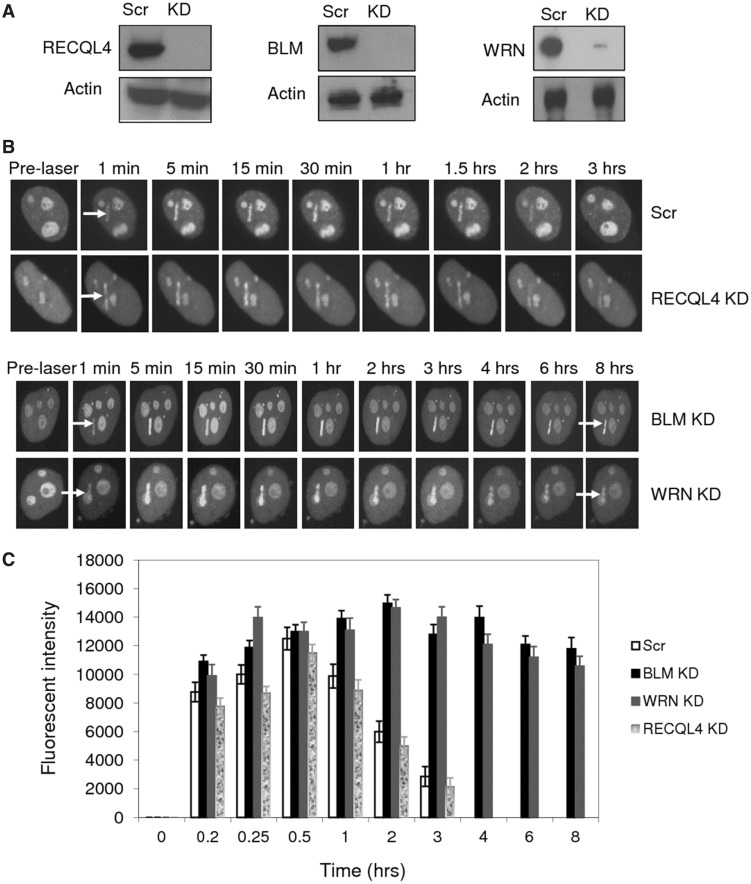
This extended retention of GFP-RECQL5 was further confirmed by performing similar experiments in an isogenic background of HeLa cells, targeted with lentiviral shRNA to stably knockdown BLM, WRN or RECQL4 (Figure 2). Efficient depletion was obtained and the knockdown (KD) efficiency was ∼95% for BLM and RECQL4, ∼85–90% for WRN on average (Figure 2A). Our retention experiments indicate that GFP-RECQL5 dissociated from laser-induced DSBs with similar kinetics in both scrambled and RECQL4-depleted HeLa cells (∼2–3 h) (Figure 2B). However, GFP-RECQL5 dissociated more slowly and was retained longer (>8 h after micro-irradiation) in BLM- or WRN-depleted cells (Figure 2B). The plot compares the retention of GFP-RECQL5 at laser-induced DSB sites in BLM, WRN and RECQL4 stably depleted cells (Figure 2C). Together, these results show that RECQL5 associates longer at the DSB sites in the absence of BLM and WRN, but not in the absence of RECQL4.
Figure 2.
Retention kinetics of GFP-RECQL5 at laser-induced DSBs in the absence of BLM, WRN and RECQL4. (A) Western blots showing stable knockdown of RECQL4, BLM and WRN by lentiviral shRNA in HeLa cells. (B) Retention of GFP-RECQL5 in the absence of BLM, WRN and RECQL4. Stable knockdown cells of RECQL4, BLM and WRN were transiently transfected with GFP-RECQL5 and micro-irradiated at 7% laser intensity, and retention was performed over several hours using the specially designed environmental chamber. (C) Plot comparing the retention kinetics of GFP-RECQL5. The retention was shown up to 8 h (while it retains even longer). The data points represent the mean of three independent experiments (8–10 cells analysed per experiment).
Previous studies have shown accumulation of acute DNA damage in the absence of BLM and WRN (38,39). Further, we confirmed that both BLM- and WRN-deficient fibroblasts have slower DNA repair kinetics and accumulate persistent 53BP1 foci followed by γ-irradiation. In normal GM637 fibroblasts, the 53BP1 foci had almost disappeared within 8 h, but both BLM and WRN fibroblasts accumulate over 5-fold more 53BP1 foci even after 16 h (Supplementary Figure S1A and B). We have previously shown similar accumulation of 53BP1 foci in the absence of RECQL4 (25). Collectively, longer retention of RECQL5 in the absence of BLM or WRN, but not RECQL4, would implicate that RECQL5 could be required to remain bound at these unrepaired lesions in the absence of BLM or WRN, but not in the absence of RECQL4.
RECQL5 physically interacts with WRN both in vivo and in vitro
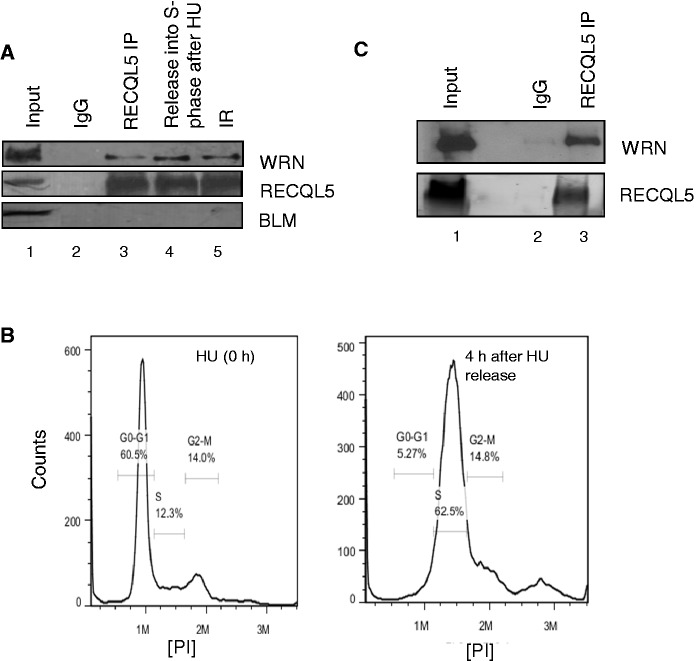
We further investigated whether there were any potential interactions among these RecQ helicases. Co-immunoprecipitation experiments in HeLa cells indicate a physical interaction between RECQL5 and WRN (Figure 3A, lane 3). In contrast, we could not detect any potential interaction of RECQL5 and BLM under similar conditions (Figure 3A). The RECQL5 and WRN association was not affected after γ-irradiation (Figure 3A, lane 5) but increased during S-phase after treatment with hydroxyurea (HU) (Figure 3A, compare lanes 3 and 4). FACS analysis indicates that the HeLa cells were blocked at G1/S after treatment with HU and were in S-phase 4 h after removal of HU (Figure 3B). The higher levels of WRN in the immunoprecipitation indicate that the association could be maximal during S-phase after replicative stress. However, co-immunoprecipitation immediately after HU treatment did not significantly affect their physical association (Supplementary Figure S2A). Previous results indicate that neither RECQL5 nor WRN is cell-cycle regulated, but both are constitutively expressed throughout the cell cycle (22,40,41), suggesting that the higher levels of RECQL5 and WRN immunoprecipitation are not simply owing to the higher protein levels in S-phase. We next explored whether the purified recombinant proteins RECQL5 and WRN interact directly. In vitro immunoprecipitation experiments indicate a physical and direct association of RECQL5 and WRN (Figure 3C).
Figure 3.
RECQL5 physically interacts with WRN both in vivo and in vitro. (A) Co-immunoprecipitation of WRN and RECQL5 from whole-cell extracts of HeLa cells, performed in the presence of ethidium bromide. Lane 1: input; lane 2: immunoprecipitation (IP) using rabbit IgG; lanes 3–5: IP with anti-RECQL5 antibody. Cells were harvested without any treatment (lane 3); 4 h after release from HU treatment (5 mM for 18 h) (lane 4); after γ-irradiation with 10 Gy (lane 5). RECQL5 was confirmed in the pull down using rabbit true blot antibody (central panel); negative IP for BLM was shown in the lower panel. (B) FACS analysis validating the synchronization of HeLa cells in S-phase 4 hrs after removal of HU. (C) In vitro IP was performed using purified recombinant proteins RECQL5 and WRN (1.2 μg each). WRN was pulled down with RECQL5 antibody (lane 3), but not by IgG (lane 2).
WRN relocates from the nucleoli and repositions at stalled replication sites during S-phase after replicative stress (42). We observed that RECQL5 similarly re-localizes outside the nucleolus to the nucleus during S-phase after treatment with HU and that it co-localizes with WRN. A similar re-distribution of both RECQL5 and WRN outside the nucleolus was also observed after treatment with camptothecin. Enlarged images of single representative cells with positive co-localization channels are shown in Supplementary Figure S2B. As we could not observe typical foci formation, this re-distribution and co-localization was validated by calculating the Pearson correlation coefficient using Volocity software 6.0. A Pearson coefficient value between 0.5 and 1 indicates co-localization between the two channels (43). Threshold Pearson coefficient was shown for ∼30 cells and >85–90% of the selected cells had a Pearson coefficient of ∼0.7–0.8, indicating a significant co-localization between RECQL5 and WRN after replicative stress (Supplementary Figure S2C).
Functional co-operation of RECQL5 and WRN
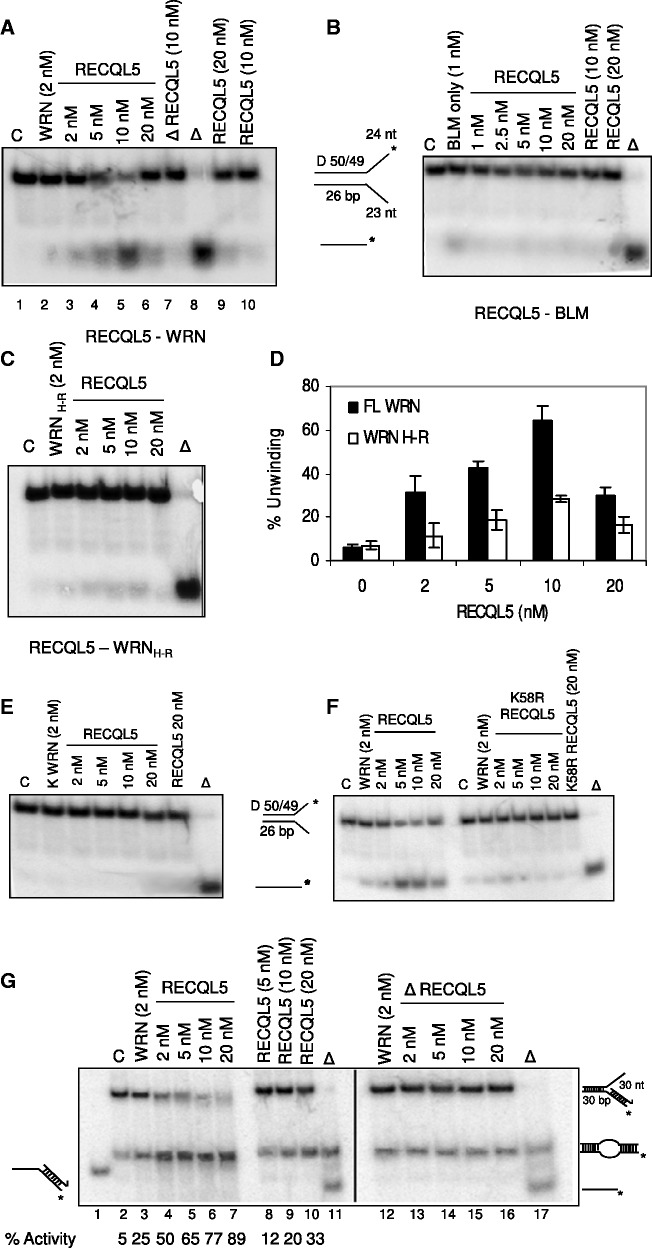
We next analysed the functional consequence of the interaction between RECQL5 and WRN. Our results show that RECQL5 specifically stimulated WRN, but not BLM, on a forked duplex with a 26-bp duplex region (Figure 4A and B). WRN (2 nM) unwound ∼5–10% of the forked DNA duplex (Figure 4A, lane 2); on increasing concentrations of RECQL5 (2–10 nM), there was an increase in WRN helicase activity from ∼10% to almost 70–80% (Figure 4A, compare lanes 3–5). At a molar ratio of 1:1, there was a 2–3-fold increase in the helicase activity of WRN; at 2.5- and 5-fold molar excess of RECQL5, the helicase activity of WRN was increased up to 4- and 6–7-fold, respectively (Figure 4D). However, 10-fold molar excess of RECQL5 (20 nM) resulted in inhibition, possibly owing to competition in binding to the fork substrate (Figure 4A, lane 6). The stimulation observed is more than an additive effect, as RECQL5 is a poorly processive helicase and 20 nM of RECQL5 could only unwind a maximum of 20–25% of the substrate (Figure 4A, lanes 9, 10). GST fragments of WRN protein were previously cloned and purified in our laboratory (44). Interestingly, RECQL5 could also stimulate a WRN fragment containing the helicase and RQC domains (WRN H-R) (Figure 4C), although the stimulation was not as strong as for the full-length WRN (Figure 4D), possibly suggesting the requirement of other domains for a complete functional interaction. This functional interaction between RECQL5 and WRN appears to be more specific, as heat-denatured RECQL5 does not stimulate WRN (Figure 4A, lane 7), nor was the stimulation observed in experiments with an ATPase-dead WRN K577M (45) (Figure 4E). Further, interestingly, ATPase/helicase dead mutant of RECQL5 (K58R RECQL5) did not stimulate WRN under similar conditions (Figure 4F), indicating that the co-operation requires functional RECQL5. Instead, inhibition was observed at higher concentrations, which might be owing to the competition in binding the substrate. Similar co-operation or coupling between two active DNA helicases was previously described by Atkinson et al. (46,47).
Figure 4.
RECQL5 specifically stimulates the helicase activity of WRN but not BLM on fork duplexes. Helicase assays of (A) WRN (2 nM), (B) BLM (1 nM) and (C) GST fragment WRNH−R (2 nM) were performed in the presence of increasing concentrations of RECQL5 (2–20 nM). (D) Quantification of WRN unwinding activity in the presence of increasing concentrations of RECQL5. The plot represents mean of three independent experiments with error bars. (E) Helicase assays of K-WRN (2 nM) on a forked duplex with increasing concentrations of RECQL5. (F) Effect of increasing concentrations of both wt-RECQL5 and K58R RECQL5 on the helicase assays of WRN (2 nM) on a forked duplex. Helicase-dead K58R RECQL5 could not stimulate WRN as wt-RECQL5 under similar conditions. (G) Strand exchange of 2 nM WRN on a 3′-flap duplex with increasing concentrations of RECQL5 (2–20 nM). A synergistic increase in strand exchange of WRN was observed, indicating a functional co-operation of RECQL5 and WRN on synthetic stalled replication forks lacking the leading strand.
Both RECQL5 and WRN were previously shown to perform strand exchange on synthetic stalled replication fork-like structures (14). Our results indicate that RECQL5 can functionally co-operate with WRN by promoting strand exchange on similar stalled replication forks, lacking a leading strand (3′-flap duplex, Figure 4G). Strand exchange is a consequence of both DNA unwinding and strand annealing activities of these RecQ helicases (14) (Supplementary Figure S3A); 2 nM of WRN has ∼20–25% activity (Figure 4G, lane 3); increasing concentrations of RECQL5 (2–20 nM) facilitated strand exchange synergistically to almost 90–95% activity (Figure 4G, lanes 4–7). RECQL5 is poorly processive, as described previously, and 20 nM of RECQL5 yielded ∼35% of activity (lane 10). Heat-denatured RECQL5 did not affect the strand exchange of WRN (lanes 12–16). Collectively, our results indicate that the physical and functional interaction of RECQL5 and WRN could be more significant on replicative stress.
On the other hand, RECQL5 did not affect the exonuclease activity of WRN (Supplementary Figure S3B), nor did it stimulate either BLM or WRN helicase activity on a HJ substrate, an HR intermediate (Supplementary Figure S3C). Instead, inhibition was observed on increasing concentrations of RECQL5, which might be possibly owing to the competition for binding the substrate.
Depletion of RECQL5 in WRN-deficient fibroblasts induces acute DNA damage and severely affects cell viability
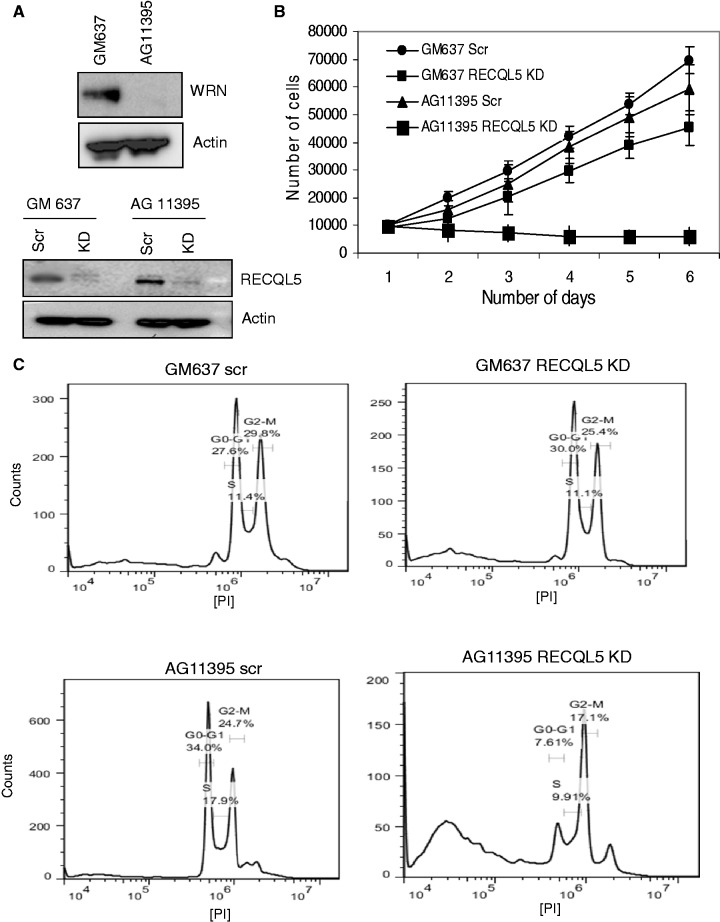
To further explore the functional interaction, depletion of RECQL5 was performed in WRN-deficient fibroblasts (AG11395). We have recently generated stable RECQL5 knockdown cells using lentiviral shRNA (15). Stable depletion of RECQL5 was performed in WRN-deficient fibroblasts using the aforementioned lentiviral shRNA that targets the coding region of RECQL5. GM637 were used as control fibroblasts. AG11395 fibroblasts were confirmed for WRN deficiency (Figure 5A, upper panel), and the level of depletion for RECQL5 was assayed and was ∼90–95% on an average (Figure 5A, lower panel). Growth assays indicated that depletion of RECQL5 in WRN-deficient fibroblasts significantly affected cell survival (Figure 5B). Depletion of RECQL5 in control fibroblasts had a moderate effect on cell proliferation, consistent with our recent observations (15). Interestingly, PI cell-cycle FACS analysis indicated that depletion of RECQL5 in AG11395 fibroblasts resulted in significant cell death (>30% of sub G1 or apoptotic cells), and that most of the remaining cells displayed acute G2/M arrest (Figure 5C). Further, the number of cells in G1 and S-phase were significantly lower than that of the scrambled AG11395 fibroblasts. Surprisingly, an additional peak representing polyploidy (>2 n DNA content) was also observed. Depletion of RECQL5 in control fibroblasts did not show any such major defects. AG11395 fibroblasts show a greater fraction of cells in S-phase, compared with that of the control fibroblasts, and this is consistent with previous observations of delayed S-phase progression in WS fibroblasts (48,49).
Figure 5.
Depletion of RECQL5 in WRN-deficient fibroblasts induces an acute G2/M arrest and severely affects cell survival. (A) Western blot analysis confirming AG11395 fibroblasts are WRN-deficient (upper panel). Depletion of RECQL5 in control (GM637) and WRN-deficient (AG11395) fibroblasts (lower panel). (B) Growth assays of RECQL5-depleted control and WRN-deficient fibroblasts. The number of cells were counted and plotted against number of days. The plot represents the mean of three independent experiments. (C) Depletion of RECQL5 in AG11395 fibroblasts shows prominent cell death and an acute G2/M arrest by FACS analyses.
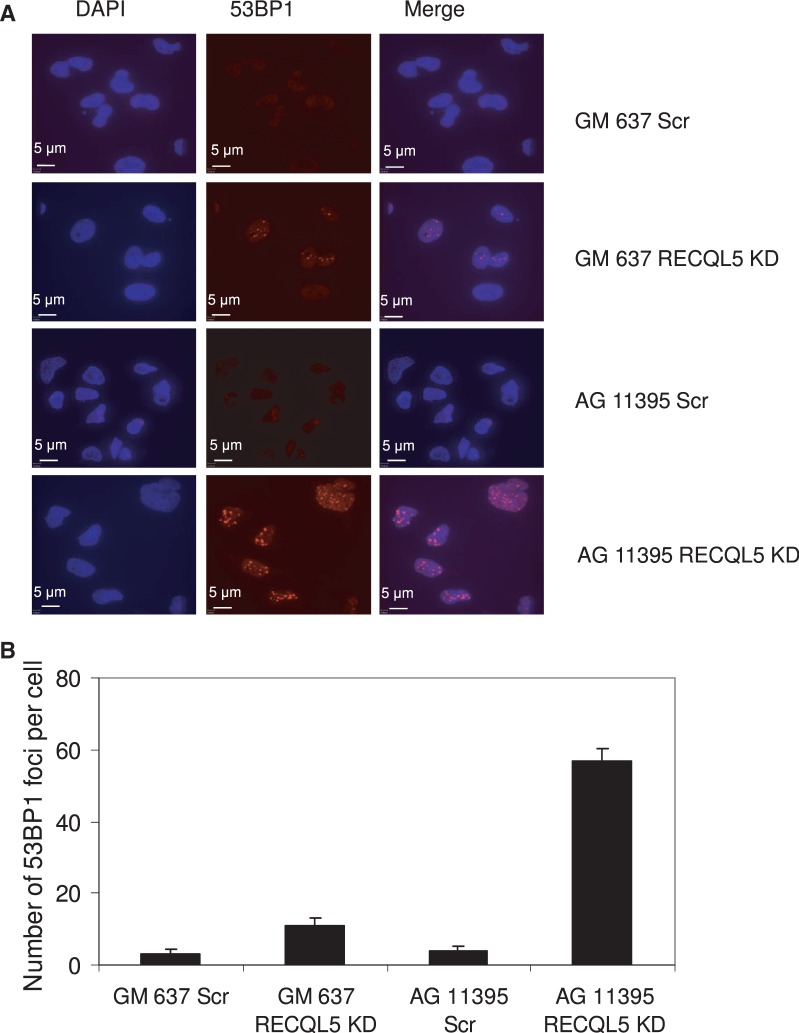
Analysis of 53BP1 foci was performed to assess direct induction of DNA DSBs. Depletion of RECQL5 in WRN-deficient fibroblasts induced over 5–6-fold more 53BP1 foci than in control fibroblasts, indicating the accumulation of acute DNA damage in the absence of both WRN and RECQL5 (Figure 6A and B). Further, MTT cell proliferation assays (Roche Applied Sciences) indicated that depletion of RECQL5 in AG11395 fibroblasts renders them highly sensitive to HU (Supplementary Figure S4). Although the cells die without any treatment, the rate of cell death is significantly higher with very low concentrations of HU such as 0.25 or 0.5 mM.
Figure 6.
Depletion of RECQL5 in WRN-deficient fibroblasts induces acute DNA damage and severe accumulation of DSBs. (A) Spontaneous accumulation of 53BP1 foci in both RECQL5-depleted control and WRN-deficient fibroblasts. (B) Plot showing the average number of 53BP1 foci/cells in RECQL5-depleted control and WRN fibroblasts. The data points represent the mean of three independent experiments.
Depletion of RECQL5 in WS fibroblasts accumulates genomic instability and leads to cell death
To further verify our growth assays and FACS analysis, the extent of cell death or apoptosis was analysed. Annexin V is a marker of apoptotic cell death; therefore, we measured Annexin V staining in our cells. Annexin V assays indicated that depletion of RECQL5 in control fibroblasts showed a mild apoptotic cell death phenotype (Supplementary Figure S5A), and this was consistent with our recent observations (15). However, depletion of RECQL5 in WRN-deficient fibroblasts resulted in significant apoptotic cell death. We observed a 10-fold increase in Annexin V-positive cells, indicating early apoptosis. An ∼14-fold increase in the total number of cells that were positive for both Annexin V and PI indicated late apoptosis or cell death (Supplementary Figure S5A). Consistently, activation of caspase-3 cleavage was observed, further indicating the onset of apoptosis (Supplementary Figure S5B).
We next analysed the cells for accumulation of any genomic instability. Two independent experiments each of ∼25 metaphase spreads (for each cell type) were analysed, and, interestingly, ∼15–18 metaphase spreads of 25 displayed an abnormal phenotype with precocious separation of the two sister chromatids in RECQL5-depleted AG11395 fibroblasts (Supplementary Figure S6A). However, metaphase chromosomes of either RECQL5-depleted control fibroblasts or of scrambled treated AG11395 fibroblasts did not show such an irregular phenotype. The process of sister chromatid pairing (or efficient cohesion of the two sister chromatids) is coupled to DNA replication and is fundamental to proper chromosome segregation and cell viability (50–52). Therefore, we wanted to analyse any replication defects associated with the loss of both RECQL5 and WRN.
Depletion of RECQL5 in WRN-deficient fibroblasts severely impairs DNA replication
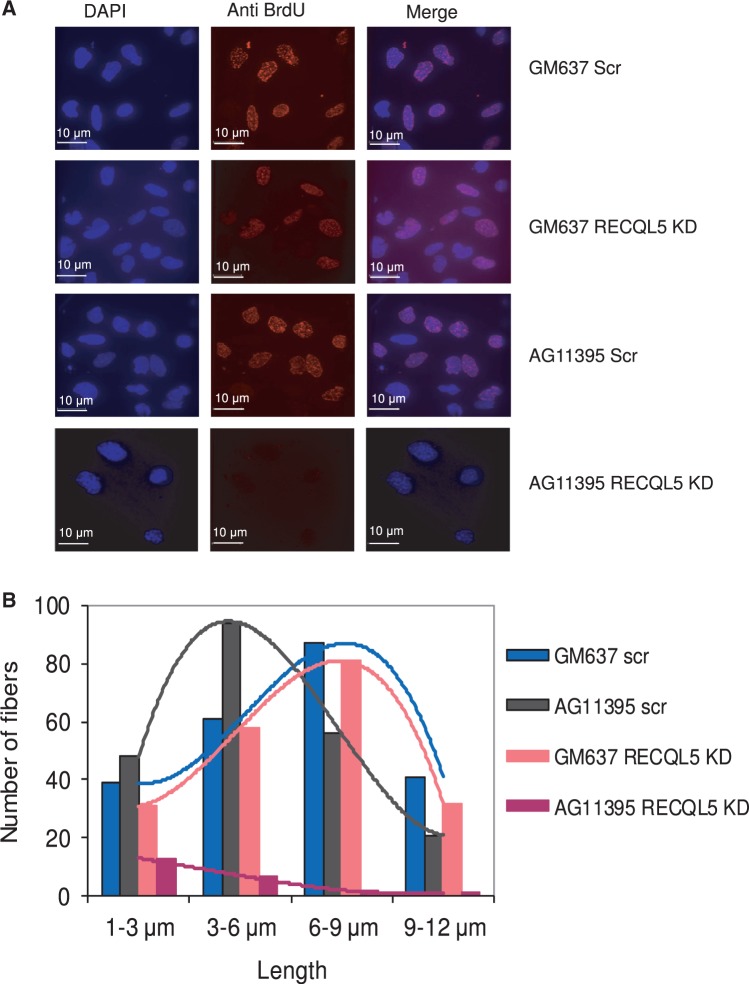
Interestingly, we observed that bromodeoxyuridine (BrdU) intake was severely impaired by depletion of RECQL5 in WRN-deficient fibroblasts compared with RECQL5 depletion in normal fibroblasts (Figure 7A). Replication fork progression was analysed by DNA fiber assays. AG11395 fibroblasts exhibit relatively shorter replication fibers on average (∼3–6 μm, Figure 7B) compared with that of control fibroblasts (∼6–9 μm), and is consistent with defects in replication fork progression reported in WS fibroblasts (49). Depletion of RECQL5 in control fibroblasts did not significantly alter the length of the fibers. However, depletion of RECQL5 in WRN-deficient fibroblasts significantly impaired DNA fiber synthesis (Supplementary Figure S6B). Few fibers were seen, with a very short average length of ∼1–3 μm (Figure 7B), indicating severe defects in DNA replication and accumulation of irreversible stalled or collapsed replication forks. This could be partly due to the fact that relatively low numbers of cells were observed in S-phase; however, the fibers are representative of replication in cells that have lost both RECQL5 and WRN.
Figure 7.
BrdU intake and DNA fiber assays of RECQL5-depleted control and WRN-deficient fibroblasts. (A) Depletion of RECQL5 in AG11395 fibroblasts shows significantly lower BrdU intake. Both RECQL5-depleted control and AG11395 fibroblasts were exposed to a short (30 min) BrdU pulse (20 μM), fixed and immunostained with BrdU antibody. (B) DNA fiber assays of RECQL5-depleted AG11395 fibroblasts show severe defects in DNA fiber synthesis and exhibit very short fibers (average length of 1–3 μm) compared with RECQL5-depleted or scrambled GM637 fibroblasts (average length of 6–9 μm). AG11395 fibroblasts exhibit relatively shorter fibers (average length of 3–6 μm) when compared with GM637 fibroblasts (average length of 6–9 μm). The plot represents the mean of two independent experiments.
We also wanted to analyse possible effects on HR, as defective replication repair can lead to increased recombination at stalled replication forks. Depletion of RECQL5 was previously shown to accumulate spontaneous Rad51 foci (16). WS fibroblasts have relatively few Rad51 foci, consistent with previous observations of attenuated Rad51 foci in the absence of WRN (53,54). Depletion of RECQL5 increased recombination events more in WS fibroblasts than in the control fibroblasts, as indicated by the increase in Rad51 foci (Supplementary Figure S6C).
WS cells are characterized by reduced replicative potential, S-phase defects and hypersensitivity to replication-perturbing agents, all of which are directly related to defects in replication fork progression (55,56). Depletion of RECQL5 induces severe and irreversible replicative blocks, to which WS cells are highly sensitive and this might lead to cell death.
RECQL5 is essential for cell survival and its depletion severely compromises DNA replication in the absence of WRN
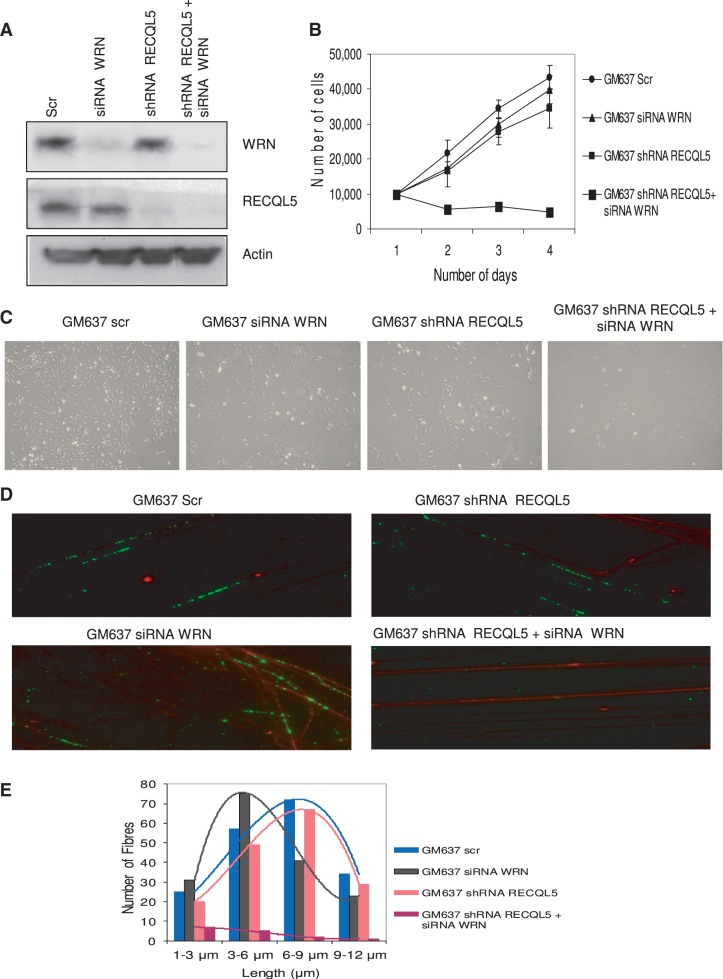
To reproduce the aforementioned results in an isogenic cellular background and to eliminate off-target effects, we generated isogenic double-depleted cells of RECQL5 and WRN in GM637 fibroblasts, by transiently depleting WRN, using siRNA–WRN (26) in RECQL5 stable knockdown cells. The knockdown efficiency was verified by western blot analysis (Figure 8A). Growth assays indicate that RECQL5-WRN double-depleted GM637 fibroblasts could not survive for >2 days (Figure 8B). Bright-field images of the single- and double-depleted cells are shown in Figure 8C.
Figure 8.
Double depletion of RECQL5 and WRN in GM637 fibroblasts. (A) Transient depletion of WRN (using siRNA–WRN) was performed in stable RECQL5 knockdown cells of GM637 fibroblasts. The depletion efficiency was ∼90%. (B) Growth assays indicate poor survival of double-depleted GM637 fibroblasts compared with the single depletions. The number of cells were counted and plotted against number of days. The plot represents the mean of three independent experiments. (C) Bright-field images of single- and double-depleted GM637 fibroblasts were shown. (D) DNA fiber assays were performed with the previously generated single- and double-depleted cells of RECQL5 and WRN. BrdU-labeled fibers were stained in green and DNA in red. (E) Double-depleted GM637 fibroblasts show consistently few and short DNA fibers (in the range 1–3 μm, average length 1.2 μm), compared with the single depletions of RECQL5 (in the range 6–9 μm, average length 7.8 μm) and WRN (in the range 3–6 μm, average length 5.8 μm). Control fibroblasts had fibers in the range 6–9 μm, average length 8.2 μm. The plot represents the mean of two independent experiments.
DNA fiber assays indicate severe replication defects on double depletion of RECQL5 and WRN, compared with single-depleted cells (Figure 8D). WRN–siRNA GM637 fibroblasts show consistently short fibers (average length of ∼5.8 μm, Figure 8E) when compared with the control–siRNA fibroblasts (∼8.2 μm), and is consistent with the previous observations of shorter fiber tracts in WRN-depleted cells (57). RECQL5 depletion alone did not show significant effect (∼7.8 μm), but double depletion of RECQL5 and WRN significantly affected DNA fiber synthesis, as few fibers were seen, and with a very short average length of ∼1.2 μm (Figure 8E).
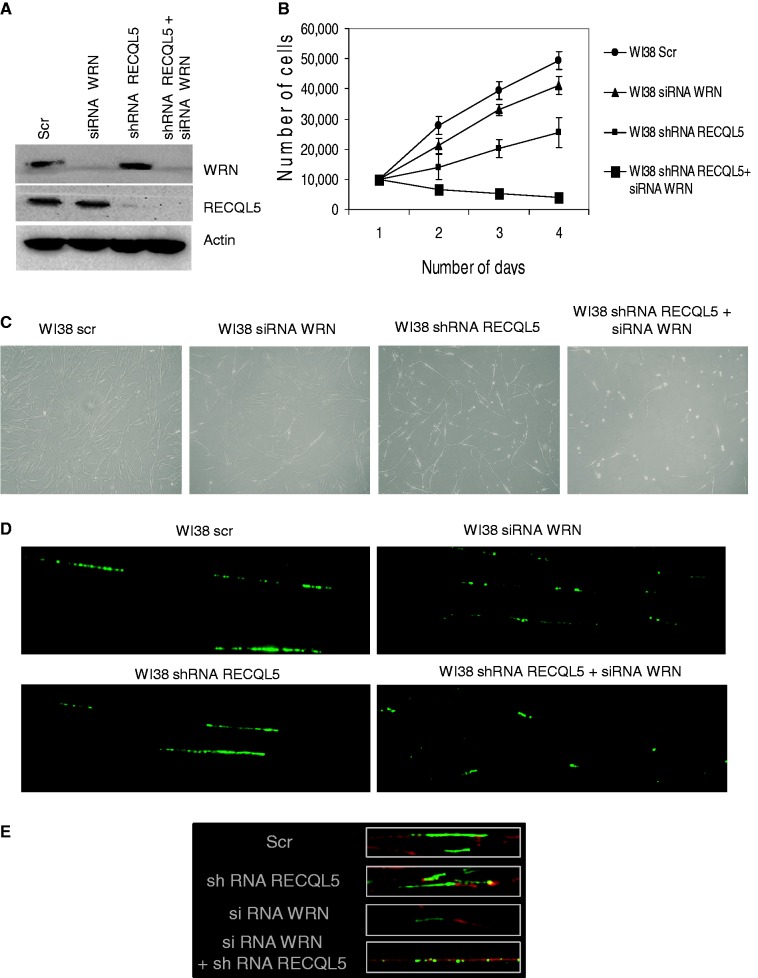
Further, we wanted to validate the effects of double depletion of RECQL5 and WRN using another normal fibroblast cell line WI38. We have previously characterized stable depletion of RECQL5 in WI38 fibroblasts (15). Double depletion of both RECQL5 and WRN was performed as described previously (Figure 9A). Similar results were obtained that the double-depleted (WI38) cells of RECQL5 and WRN were not able to proliferate (Figure 9B), further indicating severe growth deficiency and lethal consequences on the depletion of both RECQL5 and WRN. Bright-field images were shown for both the single- and double-depleted WI38 fibroblasts (Figure 9C). Further, DNA fiber assays were also consistent and the RECQL5–WRN double-depleted WI38 fibroblasts also showed few and significantly short fibers (Figure 9D and E). Collectively, our observations indicate a severe growth deficiency on loss of both RECQL5 and WRN.
Figure 9.
Double depletion of RECQL5 and WRN in WI38 fibroblasts. (A) Transient depletion of WRN (using siRNA–WRN) was performed in stable RECQL5 knockdown cells of WI38 fibroblasts. The depletion efficiency was ∼90%. (B) Growth assays indicate poor survival of double-depleted WI38 fibroblasts compared with the single depletions. The number of cells were counted and plotted against number of days. The plot represents the mean of three independent experiments. (C) Bright-field images of single- and double-depleted WI38 fibroblasts are shown. (D) DNA fiber assays were performed with the previously generated single- and double-depleted cells of RECQL5 and WRN. (E) Enlarged images of DNA fibers of single- and double-depleted cells co-stained with DNA. BrdU-labeled fibers were stained in green and DNA in red.
DISCUSSION
Our study provides novel insight into the recruitment and retention dynamics of RECQL5 at DSB sites and its functional interplay with BLM and WRN. We show that RECQL5 is retained longer at the DSB sites in the absence of BLM and WRN, but not in the absence of RECQL4. Previous studies demonstrate that cells lacking BLM or WRN display defects in both HR and DNA replication, leading to accumulation of stalled or collapsed replication forks (53,58–60). We speculate that RECQL5 could be involved in protecting acute stress or damage in the absence of BLM and WRN. RECQL5 may have roles in vivo that are distinct from, partially overlapping, and also complementary with that of BLM and WRN in the DNA repair process.
RECQL5 plays both non-redundant and overlapping roles with that of BLM (20). Both BLM and RECQL5 were proposed to disrupt Rad51 nucleoprotein/pre-synaptic filaments in HR (16,61). Previous studies by Wang et al. show that deletion of RECQL5 in a BLM−/− background increased the frequency of SCEs, indicating that RECQL5 might suppress SCEs under BLM-deficient conditions (21). Depletion of RECQL5 in BLM−/− mouse ES cells also increased the frequency of SCEs (20). It was suggested that RECQL5, perhaps in conjunction with BLM, provides an initial regulation to prevent the premature or unnecessary engagement of HR (62) and suppresses SCEs. Collectively, these observations indicate that RECQL5 could be involved in protecting the damage sites in the absence of BLM, and that lack of RECQL5 is likely to exacerbate the phenotype resulting from the defects in BLM.
Importantly, our study indicates a previously undisclosed co-operation between RECQL5 and WRN. We show that both RECQL5 and WRN physically interact with each other and that their association is likely enhanced during S-phase under replicative stress. WRN plays a critical role in response to replicative stress and significantly contributes to the recovery of stalled replication forks (53,63,64). WRN re-localizes from the nucleolus to the nucleus after replicative stress and co-localizes with the MRN complex at proliferating cell nuclear antigen (PCNA) sites during S-phase, and it was further shown that this re-localization of WRN requires MRE11 (42,65). Similar co-localization of RECQL5 with the MRN complex and requirement of MRE11 for recruitment of RECQL5 to arrested forks after HU treatment were also reported (66). RECQL5 plays a critical role in stabilization of stalled or collapsed replication forks and associates with PCNA (14); RECQL5-deficient mouse ES cells and primary embryonic fibroblasts are hypersensitive to camptothecin (13). WRN also associates with PCNA (67) and promotes regression of stalled replication forks (68). Consistent with its in vitro substrate preference, WRN has been proposed to reset reversed forks or other replication intermediates and clear the way for replisome progression. Our results indicate that RECQL5 synergistically co-operates with WRN in vitro on synthetic stalled replication fork-like structures and specifically stimulates its helicase activity on a forked duplex. However, RECQL5 does not stimulate WRN on a HJ substrate, possibly suggesting that their co-operation might not be HR mediated. We speculate that although RECQL5 has lower processivity, it co-operates with WRN to facilitate proper replication fork progression in vivo and could be more significant under replicative stress, and that this functional co-operation could also be enhanced by their interactions with other major players such as the MRN complex or PCNA.
Although the retention of RECQL5 at DSBs is extended in the absence of BLM, we did not detect co-operation between RECQL5 and BLM, as we did for WRN. RECQL5 does not interact with BLM, nor does it stimulate BLM helicase activity on fork duplexes or HJ substrates. This finding may have implications for the divergent and complementary roles that these RecQ helicases play in the cell. We speculate that RECQL5 responds to stress regardless of the source of the stress. In BLM-deficient cells, the stress likely arises from elevated SCEs and deposition of RAD51 nucleoprotein filaments to which RECQL5 gets associated. In WRN-deficient cells and in normal cells after replication-blocking DNA damage, RECQL5 is probably responding to stalled or collapsed replication forks; in the later case, WRN’s catalytic activities may be insufficient to handle the total cellular burden and therefore RECQL5 may provide assistance and stimulation. Although there is little insight into why any of the RecQ helicases interact or stimulate one another, we have observed some important scenarios of this interplay (22,23). As humans have five RecQ helicases, it would suggest that the activity of just one RecQ helicase might be insufficient to meet the demands of normal cellular metabolism.
Multiple mechanisms likely conspire and lead to the synthetic lethal interaction between RECQL5 and WRN. Specifically, replication dysfunction, altered checkpoint signaling and dysregulation of topoisomerases could all contribute to the observed phenotype. There could also be a partial defect in replication origin firing, as it was shown that both WRN and RECQL5 associate at the origin of replication in response to replicative stress (69). Loss of WRN is known to induce replicative stress, and we speculate that RECQL5 could be important both at origin firing and replication fork progression. WRN-depleted cells accumulate DNA breaks if challenged with replication-perturbing agents, an indication of incorrect handling of stalled replication forks (53). However, WS cells are able to recover from DNA synthesis perturbation, suggesting that loss of WRN is compensated for by other proteins. Here, we show that RECQL5 can support ongoing DNA replication and cell survival in the absence of WRN. We confirm this by two independent observations: depletion of RECQL5 in WS fibroblasts and generation of isogenic double-depleted cells for both RECQL5 and WRN using two independent fibroblast cell lines.
Interestingly, our results indicate that loss of both RECQL5 and WRN results in abnormal or defective separation of the two sister chromatids. Efficient sister chromatid pairing is established during DNA replication and is fundamental for cell viability (50,52). Defects in DNA replication can also trigger mitotic arrest, with onset of apoptosis and polyploidy being hallmarks of mitotic cell death (51,70). On the other hand, loss of checkpoint (CHK1) activation and cells entering mitosis with defective DNA replication can also result in loss of cell viability (70,71). Our previous studies indicate that WRN plays upstream roles to both ATM and ATR. In WRN-depleted cells, there is a defect in the activation of pATM, pATR and pCHK1, specifically in response to replicative stress (32,72). WRN-depleted cells also progress through S-phase at a faster rate in response to replicative stress, suggesting a defect in the intra S-phase checkpoint (32). On depletion of RECQL5 in the absence of WRN, we consistently observed that most of the cells were arrested at G2/M, with relatively few cells in S-phase. We speculate that loss of both RECQL5 and WRN could result in a form of mitotic cell death, which is consistent with the onset of apoptosis, observation of polyploidy and significant accumulation of cells at G2/M and their subsequent cell death (indicated by accumulation of sub G1 cells by FACS analysis).
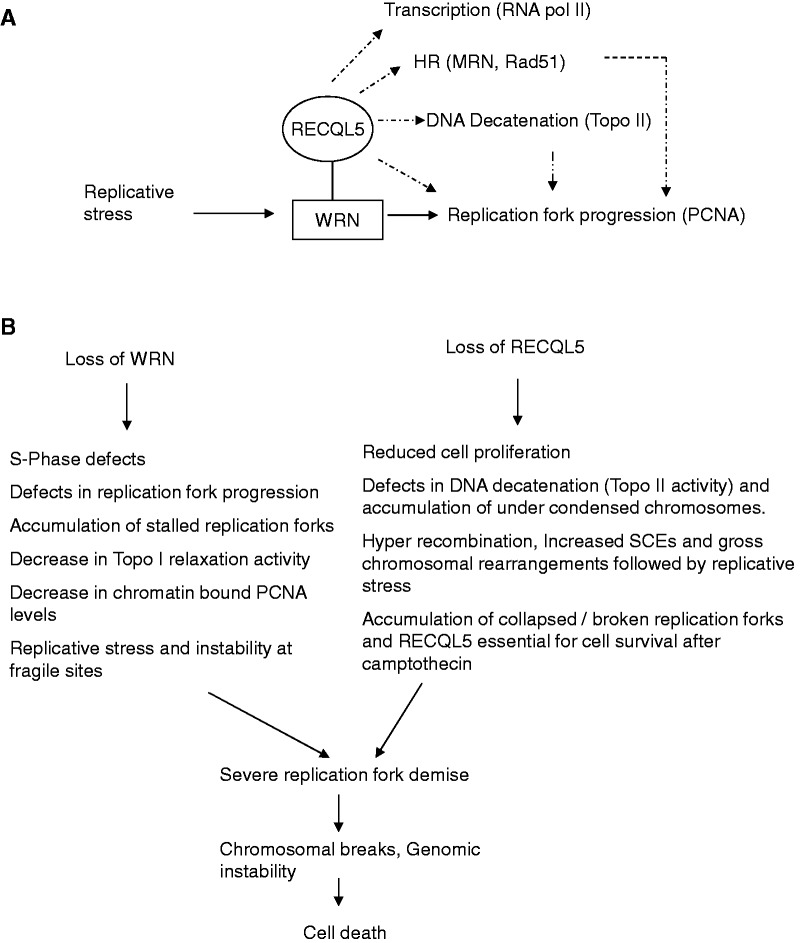
We propose that RECQL5 has both distinct and co-operative roles with that of WRN (Figure 10A). For example, WRN was shown to interact with topoisomerase I (Topo I), stimulating its re-ligation step of the relaxation process (73) and shown to have a potential role in combating Topo I lesions (74). Cell extracts from WS fibroblasts display a marked decrease in Topo I relaxation activity of negatively supercoiled DNA (73). Interestingly, RECQL5-deficient mouse embryonic fibroblasts were shown hypersensitive to camptothecin, a drug that inhibits Topo I (13). Additionally, our recent studies indicate that RECQL5 facilitates DNA decatenation and that depletion of RECQL5, but not WRN, induces defects in Topo II-mediated DNA decatenation (15). Therefore, loss of RECQL5 and WRN could impact both Topo I and Topo IIα (Figure 10B), and this could induce severe defects in DNA decatenation and chromosomal segregation and may in turn lead to polyploidy.
Figure 10.
(A) Proposed roles of RECQL5: The dotted lines represent previously proposed and distinct roles of RECQL5. The major reported interacting partners are shown in the parenthesis. The solid lines indicate our proposed co-operation of RECQL5 and WRN. (B) A model reasoning the cell death with the loss of both RECQL5 and WRN. Both distinct and overlapping consequences were listed.
Our observation of massive cell death by depletion of RECQL5 and WRN is, to our knowledge, the first reported synthetic lethal interaction among RecQ helicases and appears to be specific, as we and others have shown that co-depletions of other human RecQ helicases like BLM and RECQL4 (23,75), WRN and BLM (76) and BLM and RECQL5 (20) resulted in viable cells, exhibiting mostly additive phenotypes compared with the single depletions. We propose that RECQL5 may also have a potential backup function (in addition to its specialized roles) and that depletion of RECQL5 in a WRN-defective background results in a severe replicative block that eventually leads to cell death. Future studies are important to determine whether loss of RECQL5 function is selectively lethal for WRN-deficient tumor cells and exhibits minimal toxicity on normal cells. Additional studies on the complex interactions and functional interplay between the human RecQ helicases will provide insight into the complicated mechanisms required to maintain genomic stability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Intramural Research Program of the National Institute on Aging. Funding for open access charge: National Institutes of Health [AG000726-20].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. Haritha Vallabhaneni and Manikandan Paramasivam for critical reading of the manuscript. They thank Kent Horvath, Drs Haritha Vallabhaneni, Yie Liu and Michael S. Seidman for their timely help and discussions. They thank Joshua A. Sommers for the purification of recombinant WRN. They also thank Dr Pavel Janscak (University of Zurich) for providing the expression vector for the ATPase/helicase dead K58R RECQL5. BLM was a kind gift from Ian Hickson, University of Copenhagen, Denmark.
REFERENCES
- 1.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 3.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 4.Lindor NM, Furuichi Y, Kitao S, Shimamoto A, Arndt C, Jalal S. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet. 2000;90:223–228. doi: 10.1002/(sici)1096-8628(20000131)90:3<223::aid-ajmg7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 5.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 6.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J. Biol. Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 9.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl Acad. Sci. USA. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murfuni I, Nicolai S, Baldari S, Crescenzi M, Bignami M, Franchitto A, Pichierri P. The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene. 2012 doi: 10.1038/onc.2012.80. March 12 (doi:10.1038/onc.2012.80; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Lu X, Zhou G, Barnes EL, Luo G. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol. Biol. Cell. 2009;20:114–123. doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramamoorthy M, Tadokoro T, Rybanska I, Ghosh AK, Wersto R, May A, Kulikowicz T, Sykora P, Croteau DL, Bohr VA. RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res. 2012;40:1621–1635. doi: 10.1093/nar/gkr844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J. Biol. Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanagaraj R, Huehn D, MacKellar A, Menigatti M, Zheng L, Urban V, Shevelev I, Greenleaf AL, Janscak P. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 2010;38:8131–8140. doi: 10.1093/nar/gkq697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, et al. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von KC, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 23.Kumar SD, Popuri V, Kulikowicz T, Shevelev I, Ghosh AK, Ramamoorthy M, Rossi ML, Janscak P, Croteau DL, Bohr VA. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012;40:6632–6648. doi: 10.1093/nar/gks349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popuri V, Ramamoorthy M, Tadokoro T, Singh DK, Karmakar P, Croteau DL, Bohr VA. Recruitment and retention dynamics of RECQL5 at DNA double strand break sites. DNA Repair (Amst) 2012;11:624–635. doi: 10.1016/j.dnarep.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL, Burks L, Plon SE, Bohr VA. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl Acad. Sci. USA. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 28.Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 29.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 30.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 31.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Cheng WH, Muftic D, Muftuoglu M, Dawut L, Morris C, Helleday T, Shiloh Y, Bohr VA. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol. Biol. Cell. 2008;19:3923–3933. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 36.Karmakar P, Seki M, Kanamori M, Hashiguchi K, Ohtsuki M, Murata E, Inoue E, Tada S, Lan L, Yasui A, et al. BLM is an early responder to DNA double-strand breaks. Biochem. Biophys. Res. Commun. 2006;348:62–69. doi: 10.1016/j.bbrc.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 37.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, et al. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 2004;166:801–813. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szekely AM, Bleichert F, Numann A, Van KS, Manasanch E, Ben NA, Canaan A, Weissman SM. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol. Cell Biol. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 41.Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 42.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinchuk V, Zinchuk O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr. Protoc. Cell Biol. 2008;4 doi: 10.1002/0471143030.cb0419s39. 4.19. [DOI] [PubMed] [Google Scholar]
- 44.Doherty KM, Sommers JA, Gray MD, Lee JW, von KC, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J. Biol. Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 45.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 46.Atkinson J, Guy CP, Cadman CJ, Moolenaar GF, Goosen N, McGlynn P. Stimulation of UvrD helicase by UvrAB. J. Biol. Chem. 2009;284:9612–9623. doi: 10.1074/jbc.M808030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atkinson J, Gupta MK, McGlynn P. Interaction of Rep and DnaB on DNA. Nucleic Acids Res. 2011;39:1351–1359. doi: 10.1093/nar/gkq975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 50.Carson DR, Christman MF. Evidence that replication fork components catalyze establishment of cohesion between sister chromatids. Proc. Natl Acad. Sci. USA. 2001;98:8270–8275. doi: 10.1073/pnas.131022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- 52.Skibbens RV. Establishment of sister chromatid cohesion. Curr. Biol. 2009;19:1126–1132. doi: 10.1016/j.cub.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zecevic A, Menard H, Gurel V, Hagan E, DeCaro R, Zhitkovich A. WRN helicase promotes repair of DNA double-strand breaks caused by aberrant mismatch repair of chromium-DNA adducts. Cell Cycle. 2009;8:2769–2778. doi: 10.4161/cc.8.17.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brosh RM, Jr, Bohr VA. Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp. Gerontol. 2002;37:491–506. doi: 10.1016/s0531-5565(01)00227-3. [DOI] [PubMed] [Google Scholar]
- 56.Shen J, Loeb LA. Unwinding the molecular basis of the Werner syndrome. Mech. Ageing Dev. 2001;122:921–944. doi: 10.1016/s0047-6374(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 57.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imamura O, Fujita K, Itoh C, Takeda S, Furuichi Y, Matsumoto T. Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene. 2002;21:954–963. doi: 10.1038/sj.onc.1205143. [DOI] [PubMed] [Google Scholar]
- 59.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70:6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao ZH, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- 61.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu X, Lou H, Luo G. A Blm-Recql5 partnership in replication stress response. J. Mol. Cell Biol. 2011;3:31–38. doi: 10.1093/jmcb/mjq056. [DOI] [PubMed] [Google Scholar]
- 63.Sharma S, Sommers JA, Brosh RM., Jr In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication. Hum. Mol. Genet. 2004;13:2247–2261. doi: 10.1093/hmg/ddh234. [DOI] [PubMed] [Google Scholar]
- 64.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3:1331–1339. doi: 10.4161/cc.3.10.1185. [DOI] [PubMed] [Google Scholar]
- 66.Zheng L, Kanagaraj R, Mihaljevic B, Schwendener S, Sartori AA, Gerrits B, Shevelev I, Janscak P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez-Lopez AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech. Ageing Dev. 2003;124:167–174. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- 68.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 71.Yin L, Locovei AM, D'Urso G. Activation of the DNA damage checkpoint in mutants defective in DNA replication initiation. Mol. Biol. Cell. 2008;19:4374–4382. doi: 10.1091/mbc.E08-01-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patro BS, Frohlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J. Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laine JP, Opresko PL, Indig FE, Harrigan JA, von KC, Bohr VA. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 74.Christmann M, Tomicic MT, Gestrich C, Roos WP, Bohr VA, Kaina B. WRN protects against topo I but not topo II inhibitors by preventing DNA break formation. DNA Repair (Amst) 2008;7:1999–2009. doi: 10.1016/j.dnarep.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kohzaki M, Chiourea M, Versini G, Adachi N, Takeda S, Gagos S, Halazonetis TD. The helicase domain and C-terminus of human RecQL4 facilitate replication elongation on DNA templates damaged by ionizing radiation. Carcinogenesis. 2012;33:1203–1210. doi: 10.1093/carcin/bgs149. [DOI] [PubMed] [Google Scholar]
- 76.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70:6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.