Abstract
Dan is a transcription factor that regulates the ttd operon encoding tartrate dehydratase. During anaerobic conditions, its copy number increases by 100-fold, making Dan an abundant nucleoid-associated protein. However, little is known about the mode of Dan–DNA interaction. To understand its cellular functions, we used single-molecule manipulation and imaging techniques to show that Dan binds cooperatively along DNA, resulting in formation of a rigid periodic nucleoprotein filament that strongly restricts accessibility to DNA. Furthermore, in the presence of physiologic levels of magnesium, these filaments interact with each other to cause global DNA condensation. Overall, these results shed light on the architectural role of Dan in the compaction of Escherichia coli chromosomal DNA under anaerobic conditions. Formation of the nucleoprotein filament provides a basis in understanding how Dan may play roles in both chromosomal DNA protection and gene regulation.
INTRODUCTION
The chromosome of Escherichia coli is a large circular molecule (∼4.65 megabases) that is compacted with the aid of nucleoid-associated proteins (NAPs), which form the nucleoid (1,2). NAPs are involved in both gene regulation and chromosome compaction (3–5). Escherichia coli survives under various stressful environments. For instance, it can switch from aerobic to anaerobic respiration in an environment such as the human gut, where oxygen is deprived. This switch occurs through global changes in transcriptional activity that shift genome expression and change NAP composition. Indeed, NAPs that are specific to growth conditions have been identified, such as Fis in rapidly growing cells and Dps in stationary-phase cells (1,2,6). Recently, it was shown that TtdR, a positive regulator of the ttdA-ttdB-ygjE operon (previously known as the transcription factor YgiP), is also a growth condition–specific NAP that is up-regulated under anaerobic conditions (7,8). It was renamed Dan for DNA-binding proteins under anaerobic conditions (7,8). It is likely that Dan is also involved in uncharacterized anaerobic activities.
To understand the biological role of Dan as a growth phase–specific NAP, one must first understand its DNA-binding properties and subsequent effects on DNA organization. Here, a combination of single-molecule force manipulation and imaging techniques were used to study Dan–DNA interactions under various physiological conditions. An important finding is that Dan binds cooperatively along DNA to form a rigid periodic nucleoprotein filament that restricts accessibility of other proteins to DNA; other NAPs including HU, H-NS and StpA act similarly (9–12). Furthermore, in the presence of magnesium, the Dan nucleoprotein filaments interact with each other, leading to condensation of DNA. Overall, these results suggest that, similar to many other NAPs, Dan is a multi-functional NAP involved in physical organization and protection of chromosomal DNA and, possibly, regulation of gene expression.
MATERIALS AND METHODS
Over-expression and purification of Dan
A plasmid containing the Dan gene (expressing a C-terminus 6xHis-tagged protein) was constructed as described previously (7) and transformed into BL21 (DE3) E. coli cells using a standard heat-shock method. The transformed cells were grown to an OD600 of ≈0.6–0.7 in lysogeny broth (LB) media containing ampicillin at 37°C before isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce Dan expression. After induction, the cells were harvested and lysed using sonication. DNase (RQ1 DNase, Promega, USA) was added to the cell lysate, which was then incubated in ice for at least 3 h to digest any remaining chromosomal DNA fragments. The lysate was then centrifuged, and the supernatant was collected. The supernatant solution was adjusted to contain a final concentration of 1 M NaCl and 20 mM imidazole. Dan was purified using gravity-flow immobilized metal affinity chromatography. Nickel-charged resin (Ni-NTA Agarose, Qiagen, Singapore) was added to bind to the 6xHis-tagged Dan. The sample was placed in a filter column and washed with 10 ml of washing buffer (20 mM imidazole in a 50 mM phosphate, 500 mM NaCl buffer), followed by 1 ml of pre-elution buffer (100 mM imidazole). The protein was then eluted in 2.5 ml of elution buffer (250 mM imidazole) before dialyzed against storage buffer (10 mM Tris–HCl, 200 mM KCl, 5 mM MgCl2, pH 7.6 and 50% glycerol) to remove residual imidazole. Sodium dodecyl sulfate polyacrylamide gel electrophoresis was run to determine the purity and molecular weight of the protein, and the protein concentration was measured using Nanodrop ND1000 (Wilmington, USA). Mass spectroscopy was done to ensure the desired protein was obtained. Glycerol was added up to 50% before storage at −20°C.
Electrophoresis mobility shift assay and DNase I digestion assay
For Dan–DNA electrophoresis mobility shift assay (EMSA), 576 bp DNA was used as the DNA template. The 576 bp DNA was amplified from the 992–1152 region of λ-DNA and purified before EMSA application. Typically, 40 ng of DNA was incubated with indicated concentrations of Dan protein for 10 min at room temperature before agarose gel electrophoresis. DNase I digestion assay was performed by incubating 1 U DNase I (Promega) with Dan–DNA complexes for 10 min, followed by addition of ethylenediaminetetraacetic acid to quench the digestion before agarose gel electrophoresis.
Atomic force microscopy imaging and data analysis
Glutaraldehyde-coated mica surfaces allow protein–DNA complexes to be deposited on the surface in any buffer conditions and preserve conformation of the complexes. This has been shown using nucleosome arrays, integration host factor (IHF)–DNA complexes and H-NS–DNA complexes (10,13,14). In this study, DNA was incubated with Dan and deposited on a glutaraldehyde-coated mica surface for atomic force microscopy (AFM) imaging. The mica was prepared by depositing 0.1% APTES solution on a 0.5 cm × 0.5 cm piece of mica for 15 min. The mica was then rinsed with deionized water, dried with nitrogen gas and incubated in a desiccator for at least 2 h. Next, 1% glutaraldehyde solution was deposited on the mica for 15 min to allow glutaraldehyde molecules to bind covalently to the (3-aminopropyl)triethoxysilane (APTES)–mica surface before the mica was again rigorously washed and dried. A solution of 0.2 ng/µl linearized φX174 DNA (5386 bp) was mixed with an appropriate concentration of Dan (300 nM for a 1:1 monomer:base pair ratio) and incubated for 20 min. The solution was then deposited on the mica for 20 min, and then the mica was washed with 3 ml of deionized water and dried with nitrogen gas. When 576 bp DNA was used, the same protocol was used except that the Dan–DNA reaction mix was diluted once before depositing it on the mica. Imaging was done using an AFM (5500 AFM, Agilent Technologies, Singapore) in Acoustic AC mode. Sample imaging was done at scan size of 1–4 µm per square length, resolution of 512 × 512 or 1024 × 1024 points and scan speed of 1 line per second. AFM imaging data was processed using Gwyddion software (http://gwyddion.net/) before measuring DNA contour length and end-to-end distance using homebrew software.
Magnetic tweezers experiments
The setup was similar to that used in previous magnetic tweezers experiments (10,15). Biotin-labelled oligonucleotides were attached to both ends of λ-DNA molecules, forming B-λ-B DNA molecules. A low concentration (∼300 pg/µl) of these molecules was added to the reaction channel to allow only one end of the DNA to attach to the streptavidin-functionalized surface. Streptavidin-coupled paramagnetic beads (Dynabeads M-280 Streptavidin, Invitrogen, Singapore) were then added to the channel to attach to the other end of the B-λ-B DNA molecule, forming a DNA tether for the single-DNA stretching experiments.
RESULTS
Dan proteins cooperatively bind to DNA and cause DNA stiffening
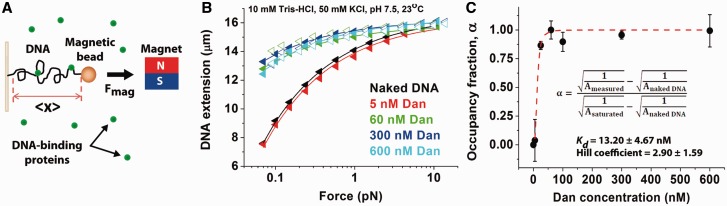
To study the DNA-binding properties of Dan, we first performed single-DNA stretching experiments using transverse magnetic tweezers (Figure 1A), as previously described (10). Single-DNA stretching allows identification of changes in DNA elasticity through measurement of DNA contour length and persistence length (16). Force-extension (FE) curves of λ DNA (NEB, 48 582 bp) were obtained by incubating λ DNA with increasing concentrations of Dan in 10 mM Tris–HCl, 50 mM KCl, pH 7.5 buffer conditions (Figure 1B). At 5 nM Dan concentration, the DNA extension is similar to that of the naked DNA, indicating no or minimal Dan binding to the force-extended λ DNA. At Dan concentrations above 60 nM at ∼10 pN, DNA extension is significantly longer than that of naked DNA during the low DNA-stretching force regime (<1 pN). This extension indicates an apparent increase in DNA rigidity. Increasing the Dan concentration to 600 nM slightly increased DNA extension, and the DNA stiffening effect was largely saturated at Dan concentrations above 60 nM.
Figure 1.
Dan causes DNA stiffening through cooperative DNA-binding. (A) An illustration of the transverse magnetic tweezers setup for the single-DNA stretching experiment. (B) Force-extension curves of single-DNA stretching experiments were performed using increasing concentrations of Dan. DNA stiffening is saturated by 60 nM Dan concentration. Solid curves are fitting curves according to the WLC model. The apparent persistence length and contour length of Dan nucleoprotein filament is 488 ± 290 and 16 002 ± 320 nm. (C) The occupancy fraction of Dan–DNA binding was calculated from the measured apparent persistence length (inserted equation) and plotted with respect to the Dan concentration. Fitting with Hill equation (solid line) suggests that Dan has DNA-binding Kd of 13.20 ± 4.67 nM and Hill coefficient of 2.90 ± 1.59, indicating strong cooperative DNA binding by Dan.
The level of DNA stiffening was quantified by fitting the Dan–DNA FE curves with Marko–Siggia worm-like chain (WLC) formula (Supplementary Methods: FE curves WLC model fitting). The two-parameter fitting allowed semi-empirical determination of DNA contour length and persistence length (17). At the saturated Dan concentration of 600 nM, the WLC model fittings showed that the apparent DNA contour length decreased from 16 490 ± 26 nm to 16 002 ± 320 nm, whereas the DNA persistence length significantly increased from 51 ± 1 to 488 ± 290 nm (Supplementary Figure S1). The fitting values were obtained from the average of three independent experiments. Because the persistence length of naked DNA is ∼50 nm (18), saturated Dan–DNA binding resulted in an increase of >8-fold in apparent DNA rigidity.
The dissociation constant, Kd, of Dan–DNA complexes has been measured in bulk, but the binding cooperativity has not been reported (7). Here, we examined the DNA-binding kinetics of Dan by using the fitted DNA persistence length values to calculate Dan DNA occupancy fraction (Figure 1C and Supplementary Methods: Occupancy fraction calculation). The dissociation constant Kd and the Hill coefficient, n, of Dan–DNA interaction were semi-empirically calculated by fitting the Hill equation to data in Figure 1C. The Kd and n were calculated to be 13.20 ± 4.67 nM and 2.90 ± 1.59, respectively. These values were confirmed using the conventional EMSA, which suggested Kd and n values of 66.8 ± 6.4 nM and 2.8 ± 0.5, respectively (Supplementary Figure S2). The Kd measured in our experiments is consistent with the previously reported value of 10–100 nM (7). In addition, the Hill coefficient of >1 indicates that Dan binds to DNA in a positive cooperative manner.
Dan proteins form rigid periodic filaments along DNA
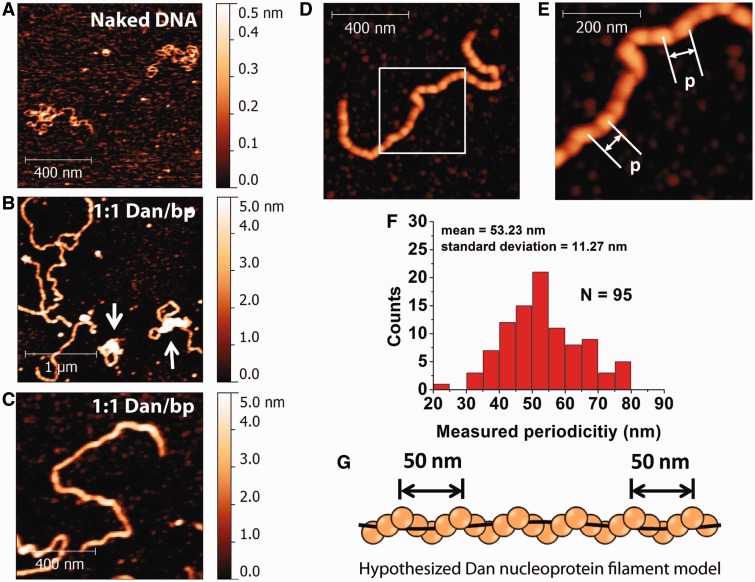
AFM imaging was used to study Dan–DNA organization visually and to complement the single-DNA stretching experiment results. In our experiments, Dan associated with linearized double-stranded φX174 DNA (NEB, 5386 bp) was trapped on glutaraldehyde-modified mica surfaces, which are particularly useful for imaging large DNA–protein complexes under various buffer conditions (10,12,13,19). It should be noted that the glutaraldehyde molecules are covalently bound to the surface. Any unbound molecules are removed by rinsing the mica with large volume of deionized water. Thus, we do not expect significant amount of glutaraldehyde molecules to diffuse into the solution to cause potential non-specific aggregation of the DNA and DNA–protein complexes. This allowed us to study the effects of buffer conditions on Dan–DNA organization. Under the same buffer condition used for single-DNA stretching experiments, naked DNA is randomly coiled (Figure 2A). At a 1:1 ratio of Dan to base pairs, the majority of Dan–DNA complexes are extended (Figure 2B) in contrast to the coiled conformation of naked DNA (Figure 2A). Although some of the DNA–Dan complexes show partial DNA compaction, the majority show extended DNA conformations that are consistent with DNA stiffening observed in the single-DNA stretching experiments. In addition, the evenly distributed height and width of the Dan–DNA complex indicate that the Dan nucleoprotein complex has a regular filamentous structure (Figure 2C). Cooperative binding of Dan to DNA suggests that the nucleoprotein filament is likely formed by Dan polymerizing along DNA.
Figure 2.
Visualization of the Dan nucleoprotein filaments. (A) AFM image of linearized φX174 DNA molecules in the absence of Dan protein showed relaxed conformation. (B) Incubation with Dan protein at a 1:1 ratio of Dan to base pairs forms elongated DNA conformation, indicating that DNA stiffens on Dan binding. In addition, some of the DNA is condensed (white arrows). (C) An example of a monomeric rigid nucleoprotein filament at a 1:1 ratio of Dan to base pairs (300 nM Dan, 10 ng DNA) suggests that Dan is evenly coated along the entire DNA length. (D) High-resolution AFM images show that the Dan nucleoprotein adopts a periodic filament structure. (E) Close-up image of the white box in (D) indicates the periodic feature spacing, p. (F) Semi-empirical measurements of the feature spacing, p, suggests there is a regular spacing of ∼50 nm (N = 95). (G) The hypothesized helical structure of the Dan nucleoprotein filament with a periodic protein filament (orange spheres) wrapping along DNA (black line).
The regular Dan nucleoprotein filament is particularly interesting because similar filaments were observed as a conserved nucleoprotein structure formed by the gene-silencing family of H-NS proteins (10,12,20,21). To identify the basic structure of this nucleoprotein filament, the Dan nucleoprotein filaments were imaged at higher resolution using a larger set of data points and slower scan speed (Figure 2D and Supplementary Figure S3). Periodic features are visible along the contour of Dan nucleoprotein filament. Feature spacing, p, is shown to illustrate the periodic structure of Dan nucleoprotein filament (Figure 2E). A semi-automatic measurement of Dan nucleoprotein filaments reveals that p = 53.23 ± 11.27 nm (Figure 2F).
On the basis of observed periodic feature (Figure 2F) and the small reduction in DNA contour length after nucleoprotein filament formation (Supplementary Figure S1), we hypothesize that it is likely that the nucleoprotein filament is composed of a Dan filament wrapping around the DNA in a helical fashion (Figure 2G), whereby the DNA is threaded through the periodic helical protein filament structure and does not cause significant extension–reduction due to DNA distortion. In summary, at saturated DNA binding, Dan binds cooperatively along DNA to form a periodic rigid filament and causing DNA to stiffen. At unsaturated DNA-binding conditions, Dan can cause local DNA compaction (Supplementary Figure S4), which is consistent with previous AFM studies (7).
Dan filament restricts DNA accessibility
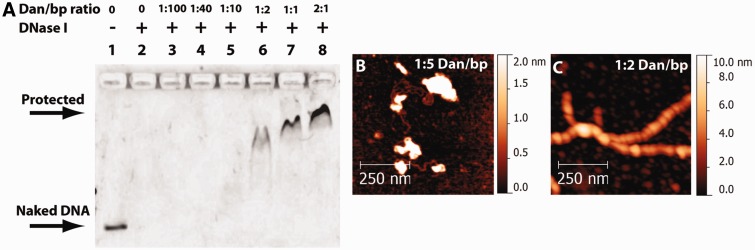
The formation of a protein filament on DNA could form a continuous physical barrier to restrict DNA accessibility. To test this hypothesis, DNase I assay was conducted by incubating 40 ng of linear 576 bp DNA at various ratios of Dan to base pairs in standard buffer followed by digestion with 2 U of DNase I (Promega) (Figure 3A). At ratio of Dan to base pairs ≥1:2, DNA was not degraded in the presence of DNase I, indicating that DNA was protected by Dan. In contrast, DNA incubated without Dan was digested over the same time scale (Figure 3A).
Figure 3.
DNase I DNA protection assay in the presence of varying Dan concentrations. (A) DNA was incubated with varying Dan to base pair ratios was treated with DNase I to determine the protection ability of Dan. At ≥1:2 Dan:base pair ratio, DNA showed signs of DNase I protection, whereas lower ratios resulted in complete DNA digestion by DNase I. (B) AFM images of DNA–Dan complexes at a 1:5 ratio of Dan to base pairs suggests incomplete DNA coverage by Dan, causing DNA compaction, whereas the naked DNA portion will be prone to DNase I digestion. The height scale (0–2 nm) is used to facilitate visualization of the naked DNA (∼0.4–0.6 nm) compared with the white spots that are height-saturated compacted Dan-bound DNA portions (∼20–30 nm). (C) At a of 1:2 ratio of Dan to base pairs, DNA is saturated by Dan, resulting in rigid nucleoprotein filament formation that protects DNA from DNase I digestion. A bigger height scale (0–10 nm) is used due to Dan nucleoprotein filament higher height.
The appearance of protected DNA at 1:2 ratio of Dan to base pairs (Figure 3A) suggests that coating of DNA by the Dan filament is nearly saturated. Indeed at a ratio of 1:5, AFM image analysis shows unsaturated DNA binding, which results in DNA compaction (Figure 3B), whereas at 1:2, Dan nucleoprotein filaments formed (Figure 3C). These data show that complete coating of DNA by the Dan filament can protect DNA from nuclease digestion by physically blocking nuclease access to DNA.
Dan nucleoprotein filaments mediate DNA condensation in the presence of physiological MgCl2 concentrations
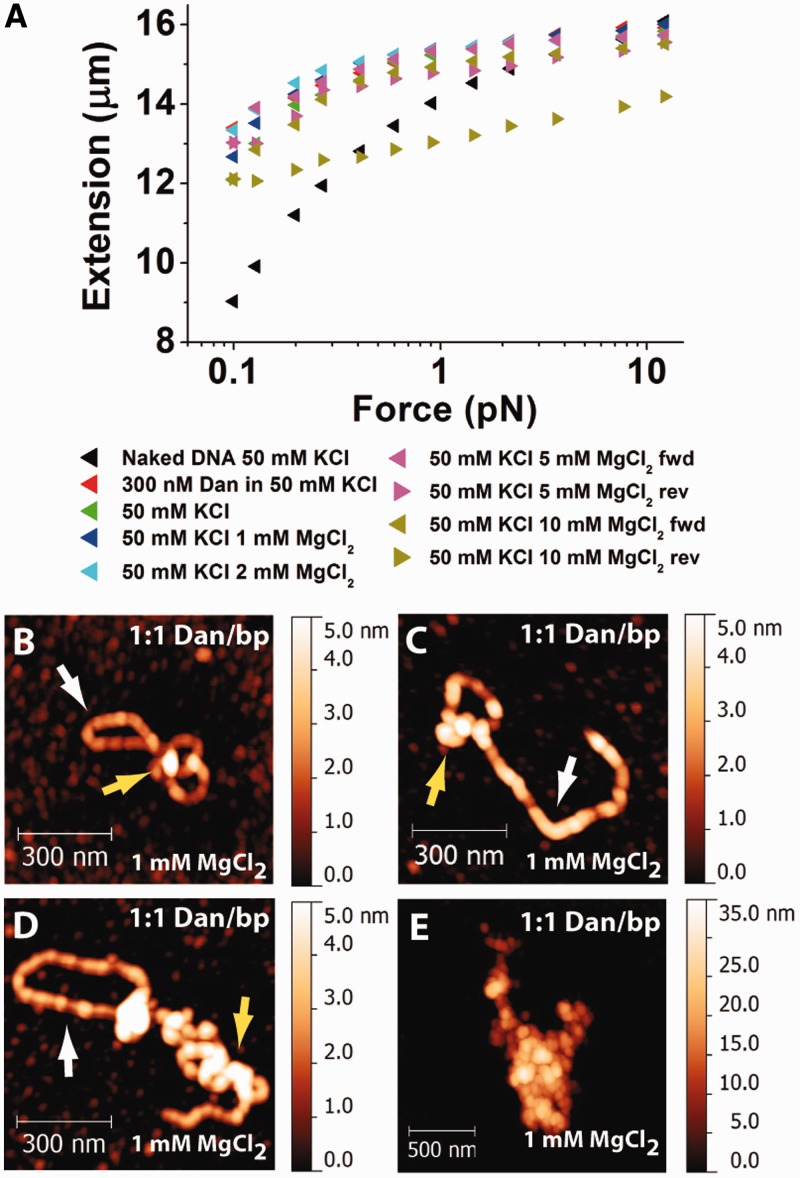
Magnesium is essential for many enzymatic reactions in bacteria, and its concentration is 2–4 mM in vivo (22,23). In addition, it has been demonstrated that magnesium affects the DNA-binding properties of H-NS proteins (10,12). Therefore, we investigated whether the Dan–DNA interaction is also sensitive to physiological concentrations of magnesium. We found that MgCl2 induces interactions between Dan nucleoprotein filaments and causes DNA compaction into higher-order structures (Figure 4). A pre-formed Dan nucleoprotein filament in the absence of free proteins and magnesium can be folded by addition of buffers containing ≥5 mM MgCl2, indicated by the hysteresis between the respective forward and reverse FE curves (Figure 4A). The presence of both DNA stiffening and FE curve hysteresis suggests that Dan nucleoprotein filaments interact with each other at high MgCl2 concentrations.
Figure 4.
Magnesium causes higher-order DNA compaction mediated by inter-filament interactions of Dan. (A) Single-DNA stretching experiments show that the Dan nucleoprotein filament is relatively stable up to 10 mM MgCl2. At 10 mM MgCl2, hysteresis between DNA relaxation and stretching suggests DNA folding. (B–E) AFM images of DNA–Dan complexes incubated at a 1:1 ratio in the presence of 1 mM MgCl2 suggests that magnesium causes Dan filaments to interact with each other, resulting in a higher-order DNA compaction. Dan nucleoprotein filament is still observed at 1 mM MgCl2 (white arrows), and DNA compaction is also observed (yellow arrows). (E) Occasionally, large-scale DNA aggregation is also observed. For this image, different scales are used to accommodate the large DNA–Dan complex.
To complement the single-DNA stretching experiments and obtain a visual understanding of Dan–DNA interaction in the presence of magnesium, Dan–DNA complexes were incubated in the presence of 1 mM MgCl2 and imaged using AFM (Figure 4B–E). Dan is still able to form rigid nucleoprotein filaments in the presence of magnesium (Figure 4B–D). In addition, nucleoprotein filaments compaction was also observed (Figure 4B–E). DNA compaction/folding in 1 mM MgCl2 buffer conditions was probably not observed in our single-DNA stretching experiments because the stretching force applied to the DNA likely increased the folding energy barrier and may have also obscured weaker interactions between filaments.
Dan nucleoprotein filaments can form linear concatemers or circles through end-to-end interactions
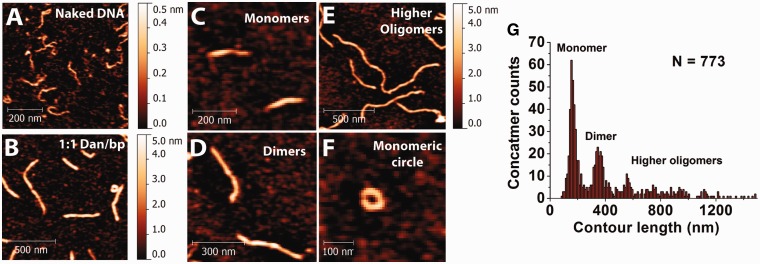
Another surprising observation in our study is that Dan nucleoprotein filaments form both rigid linear concatemers and circles, depending on the filament density (Figure 5). It should be noted that this was done in the absence of any ligase. AFM imaging of short, linear 576 bp DNA incubated in a 1:1 ratio of Dan to base pairs shows various lengths of Dan nucleoprotein filaments (Figure 5A and B). The DNA contour length of 576 bp DNA is ∼200 nm, but most of the Dan nucleoprotein filaments are much longer. Figure 5C–E show the monomeric, dimeric and higher oligomeric forms of the nucleoprotein filament, respectively. This suggests that Dan nucleoprotein filament is able to form concatemers. Occasionally, circularized nucleoprotein filaments were also observed (Figure 5F). To analyze the formation of concatemers quantitatively, the linear nucleoprotein filament contour length was measured (N = 773), and the histogram was plotted (Figure 5G). The contour lengths are quantized at approximately 200 nm, suggesting that Dan nucleoprotein filaments form linear concatemers via end-to-end connection. Due to the low probability of the circular concatemers, we did not perform similar contour length analysis.
Figure 5.
End-joining of Dan nucleoprotein filaments to DNA concatemers and circular DNA. (A) Naked DNA control. (B) DNA–Dan complexes at 1:1 ratio show multiple DNA conformations. (C and D) Dan mediates formation of linear DNA concatemers. (F) Occasionally, circular DNA conformation of a monomeric DNA was also observed. (G) Histogram of DNA contour length determined from AFM analysis shows that Dan causes the formation of linear DNA concatemers, likely caused by ends joining.
DISCUSSION
In this study, we show that Dan can form a rigid nucleoprotein filament and induce DNA compaction in the presence of ≥1 mM MgCl2. Interestingly, the nucleoprotein filament formation has been reported as a conserved nucleoprotein structure formed by silencing of genes encoding the H-NS proteins (9,10,12) and is shown to be crucial to H-NS gene-silencing and antagonizing functions (20,24). In addition, nucleoprotein filaments were also reported for E. coli HU and eukaryotic HMGB, two well-known DNA-bending proteins, in high protein concentration and low ionic strength (11,25). However, the relevancy of the filaments formation to the in vivo functions of these proteins is not clear. The ability of Dan to form a continuous nucleoprotein filament that restricts DNA accessibility and organizes DNA into different buffer-dependent conformations demonstrates certain level of homology between the DNA-binding properties of Dan and H-NS proteins.
The rigid nucleoprotein filament formation by Dan occurs through a cooperative binding process, which is also a similar property observed in H-NS proteins (9,10,12). The cooperative polymerization of H-NS proteins along DNA is believed to be related to their capability to form higher-ordered oligomerization states in solution (26–28). Interestingly, Dan can also form higher-ordered oligomerization states in solution (Supplementary Figure S5). These similarities between Dan and the H-NS proteins suggest that the Dan protein–protein interactions may also contribute to the cooperative binding of Dan along DNA. The finding of concatemers formed by the Dan nucleoprotein filaments through end-to-end interactions is unexpected and interesting, as it has not been observed in other proteins that form rigid nucleoprotein filaments on double-stranded DNA. The mechanism behind concatemer formation is not clear and requires further study.
During anaerobic condition, Dan protein copy number is up-regulated to approximately 7000–9000 (7), which translates to in vivo Dan concentration in the order of micromolar. Based on our work here, this range of concentration favours Dan nucleoprotein filament formation and thus suggests the filament relevancy in vivo. Deletion of dan from the E. coli chromosome slows cell growth during anaerobic conditions (7,8,29), indicating that Dan is important for proper anaerobic function. However, the precise roles of Dan during anaerobiosis are unclear. Genomic SELEX screening identified a total of 688 Dan-binding sites along the E. coli genome (7), implying that Dan targets regulation sites in addition to the ttd operon in E. coli. From this genome-wide screening assay, a high-affinity consensus DNA sequence was also found for Dan upstream of its regulated genes (7). Although Dan positively regulates the ttdA-ttdB-ygjE operon and also its own expression (8,29), the impact of Dan on the global gene expression of E. coli under anaerobic conditions has not been investigated. For comparison, the E. coli H-NS proteins, which also forms the protective rigid nucleoprotein filaments, causes global gene silencing despite being involved in the up-regulation of some genes (30). The similarity of the DNA-binding properties between Dan and H-NS suggests that Dan might play a role in global gene silencing under anaerobic conditions. This warrants future studies of Dan gene regulation at a global level during anaerobic stress growth conditions.
Other than gene regulatory function, Dan may also contribute to chromosomal DNA packaging and protection. The addition of this new nucleoid protein Dan during anaerobic growth condition is expected to alter the overall nucleoid structure by binding to naked DNA regions or replacing pre-existing nucleoid proteins. In addition, the in vivo magnesium concentration was estimated to be around 2–4 mM (22,31). According to our results, Dan causes higher-order DNA compaction via protein filament interactions in the presence of magnesium in this concentration range. Taking all of the above together, Dan may aid chromosome packaging and DNA protection. The idea that Dan is involved in chromosomal protection is consistent with a previous finding that Dan is generally localized at the surface of nucleoid (7). Similarly, the stress-response E. coli NAP DNA-binding proteins from starved cells are abundant during starvation stress conditions and are involved in chromosomal DNA compaction and protect DNA from oxidative stress damage (32–34). In summary, this work shows that the Dan protein is able to form rigid nucleoprotein filament, which provides a potential mechanistic platform in understanding Dan roles as a NAP during E. coli anaerobic growth conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
Ministry of Education of Singapore [MOE2008-T2-1-096 to J.Y.]; Mechanobiology Institute at National University of Singapore (to J.Y.); Grants-in-Aid for Scientific Research (A) from Ministry of Education, Culture, Sports, Science and Technology of Japan [21241047 to A.I.]. Funding for open access charge: the Mechanobiology Institute at National University of Singapore (to J.Y.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Adam Yuen and the Mechanobiology Institute, Singapore Protein Expression Facility for performing protein expression and purification services. We also thank Dr Chen Hu for setting up the magnetic tweezers. C.J.L., S.Y.L. and J.T. performed the experiments. A.I. and J.Y. conceived the research. C.J.L, S.Y.L., A.I. and J.Y. designed the experiments and interpreted the data. C.J.L., S.Y.L., J.T., A.I. and J.Y. wrote the article.
REFERENCES
- 1.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli—sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 2.Ishihama A. The Nucleoid: An Overview. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 2009. [Google Scholar]
- 3.Bertin P, Hommais F, Krin E, Soutourina O, Tendeng C, Derzelle S, Danchin A. H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie. 2001;83:235–241. doi: 10.1016/s0300-9084(01)01247-0. [DOI] [PubMed] [Google Scholar]
- 4.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 5.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 6.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teramoto J, Yoshimura SH, Takeyasu K, Ishihama A. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res. 2010;38:3605–3618. doi: 10.1093/nar/gkq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshima T, Biville F. Functional identification of ygiP as a positive regulator of the ttdA-ttdB-ygjE operon. Microbiology. 2006;152:2129–2135. doi: 10.1099/mic.0.28753-0. [DOI] [PubMed] [Google Scholar]
- 9.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim CJ, Whang YR, Kenney LJ, Yan J. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 2012;40:3316–3328. doi: 10.1093/nar/gkr1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HD, Bash R, Yodh JG, Hager GL, Lohr D, Lindsay SM. Glutaraldehyde modified mica: a new surface for atomic force microscopy of chromatin. Biophys J. 2002;83:3619–3625. doi: 10.1016/S0006-3495(02)75362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Droge P, Bao QY, Chen H, Liu YJ, Yan J, Davey CA. A divalent metal-mediated switch controlling protein-induced DNA bending. J. Mol. Biol. 2007;367:731–740. doi: 10.1016/j.jmb.2006.09.082. [DOI] [PubMed] [Google Scholar]
- 15.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 16.Yan J, Marko JF. Effects of DNA-distorting proteins on DNA elastic response. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;68:011905. doi: 10.1103/PhysRevE.68.011905. [DOI] [PubMed] [Google Scholar]
- 17.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 18.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 19.Fu HX, Freedman BS, Lim CT, Heald R, Yan J. Atomic force microscope imaging of chromatin assembled in Xenopus laevis egg extract. Chromosoma. 2011;120:245–254. doi: 10.1007/s00412-010-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim CJ, Lee SY, Kenney LJ, Yan J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci. Rep. 2012;2:509. doi: 10.1038/srep00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winardhi RS, Fu W, Castang S, Li Y, Dove SL, Yan J. Higher order oligomerization is required for H-NS family member MvaT to form gene-silencing nucleoprotein filament. Nucleic Acids Res. 2012;40:8942–8952. doi: 10.1093/nar/gks669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lusk JE, Williams RJP, Kennedy EP. Magnesium and growth of Escherichia coli. J. Biol. Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 23.Hurwitz C, Rosano CL. The intracellular concentration of bound and unbound magnesium ions in Escherichia coli. J. Biol. Chem. 1967;242:3719–3722. [PubMed] [Google Scholar]
- 24.Walthers D, Li Y, Liu YJ, Anand G, Yan J, Kenney LJ. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCauley M, Hardwidge PR, Maher LJ, III, Williams MC. Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys. J. 2005;89:353–364. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badaut C, Williams R, Arluison V, Bouffartigues E, Robert B, Buc H, Rimsky S. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–41666. doi: 10.1074/jbc.M206037200. [DOI] [PubMed] [Google Scholar]
- 27.Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 29.Kim OB, Reimann J, Lukas H, Schumacher U, Grimpo J, Dunnwald P, Unden G. Regulation of tartrate metabolism by TtdR and relation to the DcuS-DcuR-regulated C4-dicarboxylate metabolism of Escherichia coli. Microbiology. 2009;155:3632–3640. doi: 10.1099/mic.0.031401-0. [DOI] [PubMed] [Google Scholar]
- 30.Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J. Bacteriol. 2002;184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paymaster NJ. Magnesium-metabolism—brief review. Ann. Roy. Coll. Surg. 1976;58:309–314. [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiancone E, Ceci P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Yoshimura SH, Hizume K, Ohniwa RL, Ishihama A, Takeyasu K. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 2004;32:1982–1992. doi: 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.