Abstract
MicroRNAs (miRNAs) play important roles in biological development and disease. Much less is known about their role in normal adult physiology. The proximal convoluted tubule (PCT) and the medullary thick ascending limb (mTAL) in the kidney consist of epithelial cells with different transport activities. We identified 55 possible miRNA-target pairs of which the miRNAs and their predicted target proteins, many of which are involved in epithelial transport, were inversely enriched in PCT and mTAL. Some miRNAs appeared to have synergistic effects on shared targets. miR-192 and its predicted target the β-1 subunit of Na+/K+-ATPase (Atp1b1), an enzyme providing the driving force for tubular transport, were inversely enriched in kidney regions. In mice, knockdown of miR-192 led to up-regulation of Atp1b1 protein. When mice were fed with a high-salt diet, knockdown of miR-192 blunted the adaptational increase of urine output. Interestingly, miR-192 appeared to target Atp1b1 through the 5′-, rather than 3′-untranslated region. The study suggests a novel physiological mechanism in which miR-192 suppresses Na+/K+-ATPase and contributes to renal handling of fluid balance. It supports an important role of miRNAs in determining cellular characteristics that may appear subtle yet are physiologically critical.
INTRODUCTION
MicroRNAs (miRNAs) are a class of endogenous and conserved small RNA molecules that regulate gene expression. They primarily act by binding to the 3′-untranslated region (3′-UTR) of their target mRNAs to decrease protein abundance (1,2). Mechanisms of action other than the 3′-UTR interaction have been reported, most of them not yet well understood (3–7). Computational predictions based on sequence characteristics suggest the presence of multiple target genes for any given miRNA (8,9). Many miRNAs have been shown to play important roles in regulating developmental and pathological processes (10,11).
Much less, however, is known about the role of miRNAs in normal, adult physiology. An exciting possibility is that miRNAs might have a critical role in determining or maintaining cell-type-specific physiological characteristics (i.e. cell identity) in a fully developed organism. The mechanism could work in coordination with transcriptional controls. For example, miRNAs can repress leaky transcripts or adjust the abundance of expressed genes (12). While many miRNAs are clearly expressed in a tissue-specific manner, direct evidence for a functional role of miRNAs in maintaining cell-type-specific physiological characteristics has been scarce.
Nephron segments in the kidney, such as the proximal convoluted tubule (PCT) and the medullary thick ascending limb (mTAL), provide an excellent model for studying the role of miRNAs in the regulation of cell-type-specific physiological characteristics. Both PCT and mTAL consist of a homogeneous population of epithelial cells whose primary function is vectorial transport. Cells in both segments are derived from the same origin, the metanephric mesenchyme. However, PCT and mTAL differ in specific transport activities, which are critical for normal kidney physiology including the regulation of whole body fluid and solute homeostasis. It is unknown whether miRNAs play a role in regulating physiological characteristics of specific nephron segments including PCT or mTAL.
We report here evidence suggesting that miRNAs might be involved in maintaining numerous physiological characteristics that are specific to PCT or mTAL. Further experiments showed that miR-192 regulates Na+/K+-ATPase in human renal epithelial cells and in vivo in animal models and revealed a novel physiological mechanism in which miR-192 suppressed Na+/K+-ATPase and contributed to renal handling of fluid balance on a high-salt diet.
MATERIALS AND METHODS
Animals
For the miRNA profiling study, male Sprague–Dawley rats weighting 290–330 g were used and maintained on the AIN-76A diet containing 0.4% NaCl (Dyets). To study the effect of salt intake on miR-192 expression, Sprague–Dawley rats were fed with the Basal Diet 5755 (0.24% sodium; TestDiet) and then switched to Low Sodium Diet 5881 (0.03% sodium; TestDiet). For the miR-192 knockdown studies, we used CD-1 mice, maintained on the 0.4% NaCl diet or a 4% NaCl diet (Dyets). For chronic monitoring of urine output and water intake, mice were housed in metabolic cages (Lab Products Inc.). Animal protocols were approved by the Institutional Animal Care and Use Committee.
Isolation of nephron segments
Left kidney was perfused with cold dissection solution, followed by digestion solution. The kidney cortex and outer medulla were separated and incubated in digestion solution. Digested tissue was microdissected under a stereomicroscope. See Supplementary Data for more details.
miRNA isolation
Isolated nephron segments from three rats were pooled together and miRNAs were isolated using the RT2 qPCR-Grade miRNA Isolation Kit (SA Biosciences).
Real-time PCR miRNA array
We used RT2 miRNA PCR Array System (SA Biosciences) and followed the manufacturer’s protocol (13). Three arrays were run for every nephron segment, each containing pooled samples from three rats (nine rats in total). Relative expression results were normalized across plates according to the total expression of all miRNAs on each plate.
Taqman real-time PCR
Expression levels of several miRNA and mRNA were measured using real-time PCR with Taqman chemistry (Applied Biosystems) as described previously (13–15).
Protein database
The database of proteins that are segment-specific was compiled from original articles and two textbooks (Brenner and Rector: The Kidney, Saunders, 8th edition; Koeppen and Stanton: Renal Physiology, Elsevier, 4th edition).
Selection of possible miRNA-target pairs
miRNA-target pairs were selected based on two criteria: (i) sequence characteristics (based on miRNA target prediction tools) and (ii) reciprocal expression of a miRNA and its predicted target protein in two nephron segments (16). Three prediction tools were used: TargetScan v5.1 (http://www.targetscan.org/), MicroCosm Targets v5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) and microRNA.org (http://www.microrna.org/microrna/home.do).
Western blot
Western blot was performed using primary antibodies obtained from Santa Cruz Biotechnology (17,18).
Na+/K+-ATPase activity assay in crude membrane fractions
The assay was performed as described previously (19,20). See Supplementary Data for more details.
3′-UTR and P+5′-UTR reporter constructs
Reporter gene vectors, containing the 3′-UTR or the promoter region and 5′-UTR (P+5′-UTR) of a miRNA target gene, were constructed as previously described (13–15). The UTR of the gene of interest was identified and amplified from rat genomic DNA. Primer sequences are listed in Supplementary Data. The PCR product was inserted into pMIR-REPORT vector (Ambion), adjacent to the 3′-end or 5′-end of the luciferase reporter gene for the 3′-UTR or P+5′-UTR assay, respectively.
3′-UTR-mutation, P+5′-UTR-mutation and P+5′-UTR-deletion constructs
Site-directed mutagenesis was performed with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene), following the protocol suggested by the company (21). Primers used for introducing mutations/deletions are listed in Supplementary Data. The 3′-UTR-mutation construct had mutations introduced at nucleotides +143 to +145 relative to the first nucleotide in the 3′-UTR. The P+5′-UTR construct included the sequence ∼1 kb upstream of the transcription start site and the subsequent 5′-UTR of the Atp1b1 gene. Deletion and mutation were introduced at nucleotides −386 to −391 and −387 to −389, respectively, where +1 is the translation initiation site.
UTR reporter assay
The assay was performed as described previously (13–15). HeLa or 3T3-L1 cells cultured in 96-well plates were co-transfected with the UTR reporter construct (100 ng per well), a pMIR-REPORT β-gal plasmid (50 ng per well) and pre-miR or control oligonucleotides (10 pmol per well). Twenty-four hours after transfection, luminescence from luciferase and β-galactosidase were measured. β-Galactosidase activity was used to normalize luciferase signals. See Supplementary Data for more details.
Atp1b1 promoter construct and promoter activity assay
We followed the previously described approach (21). The 1-kb sequence upstream of the transcription start site of the Atp1b1 gene was inserted into the pGL4.81 vector containing Renilla luciferase gene (Promega). Human renal epithelial cells cultured in 96-well plates were transfected with the Atp1b1 promoter-pGL4.81 vector, the pGL2-control vector and pre-miR oligonucleotides, as described earlier. Twenty-four hours after transfection, luciferase activity was measured. pGL2 control containing the firefly luciferase gene was used to normalize the Renilla luciferase. See Supplementary Data for more details.
In vitro and in vivo use of oligonucleotides
Anti-miR and pre-miR oligonucleotides from Ambion were used for suppression and over-expression of miRNAs in vitro, respectively. Cells were transfected with pre-miR-192, anti-miR-192 or other oligonucleotides and their respective controls (at 100 nM unless otherwise indicated), using Lipofectamine 2000 (Invitrogen). After 24 or 48 h, cells were collected for analysis. Locked nucleic acid (LNA)-modified anti-miR oligonucleotides from Exiqon were used for in vivo suppression and delivered by intraperitoneal (i.p.) injection (10 mg/kg body weight) (14).
Statistics
miRNA expression profiles in glomeruli, PCT and mTAL were analyzed using one-way ANOVA. t-Test was used to compare expressions in PCT and mTAL and analyze the UTR assays, the miR-192 suppression/over-expression experiments and the miR-192 and Atp1b1 expressions in the in vivo studies. Two-way ANOVA was used for comparison of urine output and water intake in mouse studies. P < 0.05 was considered significant. Data are presented as mean ± SEM.
RESULTS
miRNA expression profiles in glomeruli, PCT and mTAL
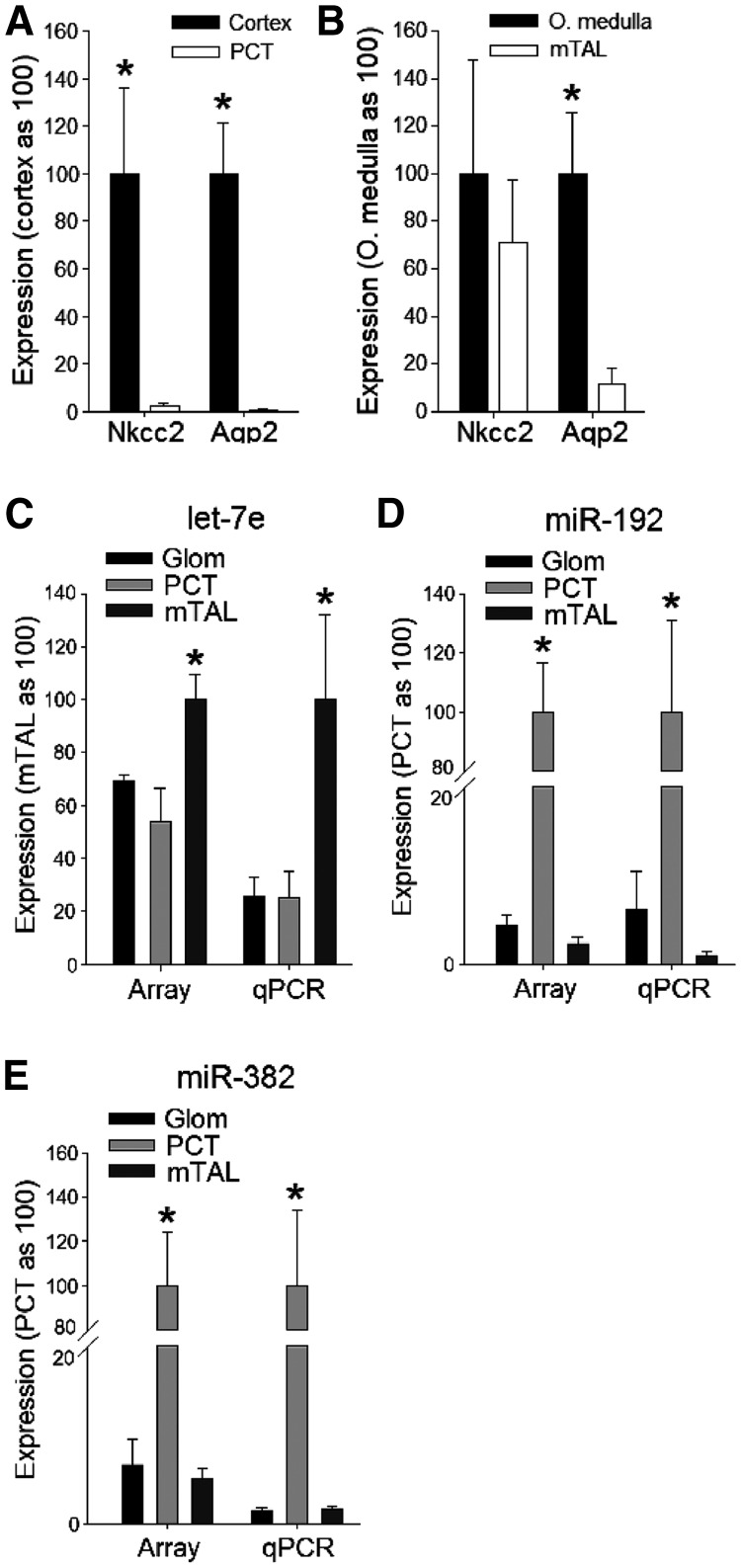
The three nephron segments were microdissected from rat kidneys and the sample purity of PCT and mTAL validated by determining expression of segment-specific marker genes (Figure 1A and B). Abundance of 118 miRNAs in glomeruli, PCTs and mTALs was determined with miRNA PCR arrays, and the obtained expression levels are shown in Supplementary Table S1. Among the analyzed miRNAs expressed in glomeruli, the most abundant were miR-126, miR-23b, miR-23a, miR-26a and let-7c. In PCTs, we found miR-16, miR-21, miR-192, miR-194, miR-30c and let-7c to have the highest expressions. miRNAs that were most abundant in mTALs were miR-100, let-7c, miR-30c, let-7b, miR-26a, miR-23a and miR-30a.
Figure 1.
Validation of purity of microdissected PCT and mTAL samples and the miRNA expression profiles obtained by PCR array. To exclude possible contamination of isolated nephron segments, we determined mRNA expressions of Na+/K+/2Cl− cotransporter (Nkcc2) and aquaporin 2 (Aqp2) in the isolated nephron segments and in homogenates of cortex and outer medulla (O. medulla). Nkcc2 is selectively expressed in cortical and medullary thick ascending limbs. Aqp2 is selectively expressed in cortical and medullary collecting ducts. Both Nkcc2 and Aqp2 would be expected to be detectable in homogenates of the renal cortex and medulla. (A) In isolated PCT samples, expression of Nkcc2 and Aqp2 was nearly undetectable, excluding contamination by other tubular segments present in kidney cortex that express those genes (i.e. the cortical thick ascending limb and cortical collecting duct). *P < 0.05 versus the isolated segments. (B) In isolated mTAL samples, Nkcc2 was highly expressed whereas Aqp2 was largely depleted compared to the whole renal outer medulla, excluding significant contamination by the medullary collecting duct. *P < 0.05 versus the isolated segments. Relative expression levels of let-7e (C), miR-192 (D) and miR-382 (E) in glomeruli (Glom), PCT and mTAL obtained by PCR array using Sybr Green chemistry (Array) were confirmed by individual real-time PCR using Taqman chemistry (qPCR). n = 3, with each sample consisting of tissues obtained from three rats. *P < 0.05 versus the other segments.
We selected three miRNAs with various degrees of differential expression and measured their abundance by performing individual real-time PCRs based on Taqman chemistry. The Taqman analysis confirmed the expression patterns shown by the PCR array based on the SYBR Green method (Figure 1C–E).
Differential miRNA expression between PCT and mTAL
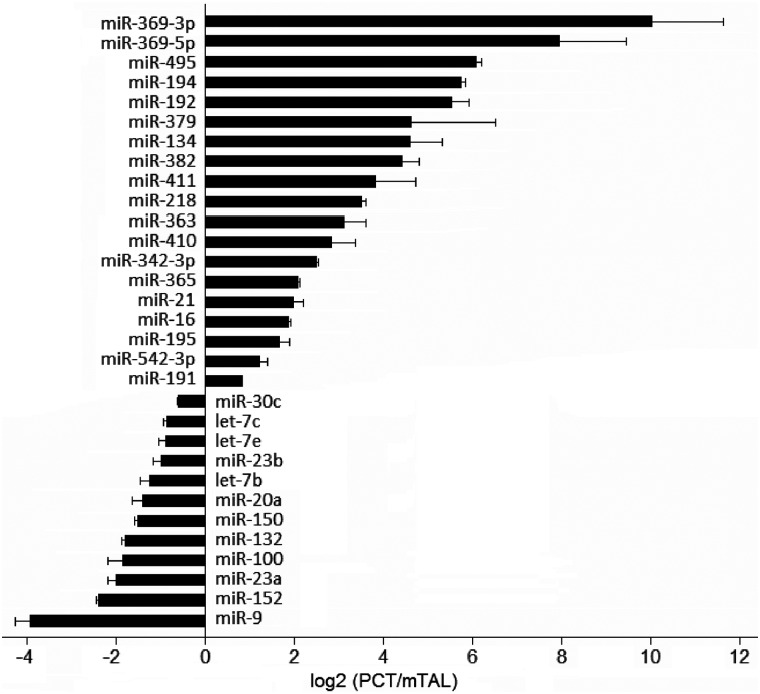
PCTs and mTALs consist of epithelial cells that are generally similar and derived from the same developmental origin, yet significantly different in their transport activities. Thus, they provide a good model for identifying miRNAs involved in determining cellular differences that are subtle yet important. Figure 2 shows 31 miRNAs that were differentially expressed in these two nephron segments. Nineteen of those miRNAs had significantly higher abundance in PCT compared with mTAL, and 12 miRNAs were more highly expressed in the mTAL segment.
Figure 2.
miRNAs differentially expressed in the PCT and the mTAL. Relative expression difference is presented as fold difference (PCT/mTAL) on logarithmic scale. n = 3, P < 0.05.
Segment-specific protein expression database
By performing an extensive literature search, we generated a list of genes known to be differentially expressed in PCT and mTAL at the level of protein abundance (Supplementary Table S2). We identified 23 proteins, and the large majority (21 of 23) was involved in tubular transport processes.
miRNA-target pairs in PCT and mTAL
miRNAs bind to target sequences with imperfect complementarity, making it highly challenging to predict miRNA-target pairs. Only a small fraction of computationally predicted miRNA-target interactions has been experimentally confirmed. We reasoned that reciprocal expression of a miRNA and a predicted target would support the presence and relevance of a miRNA-target protein pair (16). Reciprocal expression was defined as a miRNA being up-regulated and a protein being down-regulated, or the other way around. We examined miRNAs and proteins that were found to be differentially expressed in PCT and mTAL (shown in Figure 2 and Supplementary Table S2), which formed 620 possible pairs. Of the 620 pairs, 55 were predicted to be high-probability miRNA-target pairs based on the following: (i) their sequence characteristics, as determined by TargetScan v5.1, MicroCosm Targets v5 or microRNA.org and (ii) reciprocal expression as defined earlier. The 55 pairs involved 26 different miRNAs and 17 different proteins (Table 1).
Table 1.
Possible miRNA-target pairs in PCT and mTAL
| miRNA | Higher expression | Target symbol | Higher expression | Target common name |
|---|---|---|---|---|
| rno-let-7b | mTAL | Slc4a4 | PCT | Na(+)/HCO3(−) cotransporter |
| rno-let-7b | mTAL | Slc22a1 | PCT | Organic cation transporter 1 |
| rno-let-7c | mTAL | Slc4a4 | PCT | Na(+)/HCO3(−) cotransporter |
| rno-let-7e | mTAL | Aqp11 | PCT | Aquaporin 11 |
| rno-let-7e | mTAL | Slc4a4 | PCT | Na(+)/HCO3(−) cotransporter |
| rno-let-7e* | mTAL | Aqp11 | PCT | Aquaporin 11 |
| rno-miR-100 | mTAL | Slc9a1 | PCT | Na(+)/H(+) exchanger 1 |
| rno-miR-100 | mTAL | Slc22a6 | PCT | Organic anion transporter 1 |
| rno-miR-132 | mTAL | Aqp1 | PCT | Aquaporin 1 |
| rno-miR-132 | mTAL | Aqp8 | PCT | Aquaporin 8 |
| rno-miR-134 | PCT | Romk2 | mTAL | ATP-sensitive inward rectifier K channel 1.1b |
| rno-miR-150 | mTAL | Lrp2 | PCT | Megalin |
| rno-miR-150 | mTAL | Slc4a4 | PCT | Na(+)/HCO3(−) cotransporter |
| rno-miR-150 | mTAL | Slc22a2 | PCT | Organic cation transporter 2 |
| rno-miR-152 | mTAL | Slc9a1 | PCT | Na(+)/H(+) exchanger 1 |
| rno-miR-152 | mTAL | Slc22a2 | PCT | Organic cation transporter 2 |
| rno-miR-16 | PCT | Romk2 | mTAL | ATP-sensitive inward rectifier K channel 1.1b |
| rno-miR-16 | PCT | Umod | mTAL | Uromodulin (Tamm–Horsfall glycoprotein) |
| rno-miR-16 | PCT | Slc12a1 | mTAL | Na(+)-K(+)-2Cl(−) cotransporter |
| rno-miR-16 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-192 | PCT | Atp1b1 | mTAL | Na(+)-K(+)-ATPase beta 1 |
| rno-miR-195 | PCT | Romk2 | mTAL | ATP-sensitive inward rectifier K channel 1.1b |
| rno-miR-195 | PCT | Umod | mTAL | Uromodulin (Tamm–Horsfall glycoprotein) |
| rno-miR-195 | PCT | Slc12a1 | mTAL | Na(+)-K(+)-2Cl(−) cotransporter |
| rno-miR-195 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-20a | mTAL | Aqp1 | PCT | Aquaporin 1 |
| rno-miR-20a | mTAL | Aqp8 | PCT | Aquaporin 8 |
| rno-miR-21 | PCT | Atp1a1 | mTAL | Na(+)-K(+)-ATPase alpha 1 |
| rno-miR-21 | PCT | Umod | mTAL | Uromodulin (Tamm–Horsfall glycoprotein) |
| rno-miR-21 | PCT | Slc12a1 | mTAL | Na(+)-K(+)-2Cl(−) cotransporter |
| rno-miR-21 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-218 | PCT | Atp1b1 | mTAL | Na(+)-K(+)-ATPase beta 1 |
| rno-miR-218 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-23a | mTAL | Slc9a1 | PCT | Na(+)/H(+) exchanger 1 |
| rno-miR-23a | mTAL | Slc4a4 | PCT | Na(+)/HCO3(−) cotransporter |
| rno-miR-23b | mTAL | Slc9a1 | PCT | Na(+)/H(+) exchanger 1 |
| rno-miR-23b | mTAL | Slc4a4 | PCT | Na(+)/HCO3(−) cotransporter |
| rno-miR-30c | mTAL | Slc9a1 | PCT | Na(+)/H(+) exchanger 1 |
| rno-miR-342-3p | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-363 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-363* | PCT | Slc4a2 | mTAL | Anion exchange protein 2 |
| rno-miR-369-3p | PCT | Atp1a1 | mTAL | Na(+)-K(+)-ATPase alpha 1 |
| rno-miR-369-3p | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-382* | PCT | Romk2 | mTAL | ATP-sensitive inward rectifier K channel 1.1b |
| rno-miR-410 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-411 | PCT | Slc12a1 | mTAL | Na(+)-K(+)-2Cl(−) cotransporter |
| rno-miR-411 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-495 | PCT | Romk2 | mTAL | ATP-sensitive inward rectifier K channel 1.1b |
| rno-miR-495 | PCT | Slc4a7 | mTAL | Na(+)-HCO3(−) cotransporter 3 |
| rno-miR-542-3p | PCT | Atp1b1 | mTAL | Na(+)-K(+)-ATPase beta 1 |
| rno-miR-9 | mTAL | Aqp1 | PCT | Aquaporin 1 |
| rno-miR-9 | mTAL | Aqp8 | PCT | Aquaporin 8 |
| rno-miR-9 | mTAL | Aqp11 | PCT | Aquaporin 11 |
| rno-miR-9 | mTAL | Cubn | PCT | Cubilin |
| rno-miR-9 | mTAL | Slc9a1 | PCT | Na(+)/H(+) exchanger 1 |
On the left side are miRNAs and the nephron segment in which they were more highly expressed. On the right side are their predicted protein targets and the nephron segment in which the proteins were more highly expressed. See text for details.
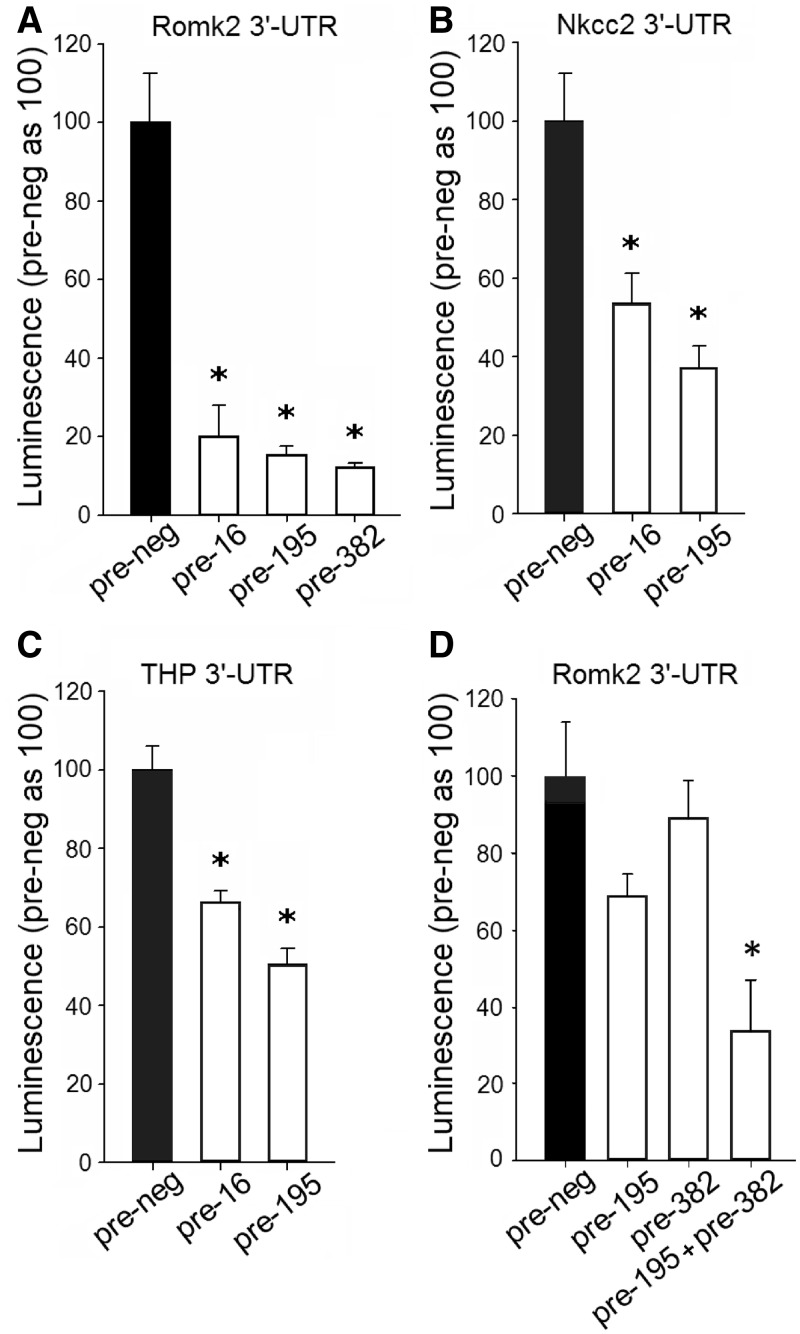
Interaction between selected miRNAs and their predicted targets was tested with the 3′-UTR reporter assay. We selected miRNAs that were highly abundant and that were predicted to have multiple targets relevant in kidney physiological processes. miRNA mimics, compared with control oligonucleotides, significantly reduced luciferase activity of the reporter vectors containing their target 3′-UTR sequence, which supported the presence of interaction between miR-16 and ATP-sensitive inward rectifier K+ channel (Romk2), Na+/K+/2Cl− cotransporter (Nkcc2 or Slc12a1) and Uromodulin or Tamm–Horsfall glycoprotein (Umod); miR-195 and Romk2, Nkcc2 and Umod; and miR-382 and Romk2 (Figure 3A–C).
Figure 3.
Experimental confirmation of interaction between selected miRNAs and their putative 3′-UTR targets and evidence for apparent synergistic effects of miR-195 and miR-382. HeLa cells were transfected with luciferase reporter constructs containing the following 3′-UTR segments: ATP-sensitive inward rectifier K+ channel (Romk2), Na+/K+/2Cl− cotransporter (Nkcc2 or Slc12a1) or Uromodulin or Tamm–Horsfall glycoprotein (Umod or THP). Cells were also transfected with 100 nM of the following oligonucleotides: pre-miR-16 (pre-16), pre-miR-195 (pre-195) and pre-miR-382 (pre-382) or control oligonucleotides (pre-neg). (A) n = 5–10, *P < 0.05 versus control. (B) n = 5–10, *P < 0.05 versus control. (C) n = 13–14, *P < 0.05 versus control. (D) HeLa cells were transfected with luciferase reporter constructs containing the 3′-UTR of Romk2. Cells were also transfected with the following miRNA mimics: pre-195 (30 nM), pre-382 (30 nM), pre-195 (15 nM) + pre-382 (15 nM) or pre-neg control (30 nM) oligonucleotides. In all treatment conditions, the total concentration of miRNA mimics was equal (30 nM). The extent of luciferase suppression was greater with simultaneous treatment with miR-195 and miR-382 mimics, compared with the sum of the effect of each miRNA applied individually. n = 5, *P < 0.05 versus the other groups.
miR-16 and miR-195 have the same seed region sequence important for target recognition. It is, therefore, not surprising that they share the same targets. miR-195 (or miR-16) and miR-382 have different seed region sequences and are predicted to bind to different sites in the 3′-UTR of Romk2. We tested the effect of simultaneous treatment with miR-195 and miR-382 mimics at low concentrations on the 3′-UTR of Romk2. When applied individually at 15 nM, miR-195 or miR-382 mimic did not significantly suppress luciferase activity (71 ± 11% and 87 ± 15% of control, respectively, n = 6, P > 0.05 versus negative control mimic). When applied jointly at the same concentration (15 nM each), miR-195 and miR-382 substantially suppressed luciferase activity (Figure 3D). This was not because the total concentration of the mimics was greater with the combined treatment (30 nM in total) because the effect of the combined treatment was significantly larger than when each mimic was applied at 30 nM individually (Figure 3D). In addition, the effect size of the combined treatment (15 nM each for a total of 30 nM) was larger than the sum of the effects of each miRNA mimic applied individually at either 15 nM or 30 nM. The result indicated possible synergistic effects of miR-195 and miR-382 on the 3′-UTR of Romk2.
Several combinations of three of the five miRNAs showing the largest fold enrichment in PCT or mTAL had one or more shared predicted target genes (Supplementary Table S3).
Na+/K+-ATPase β1 subunit (Atp1b1) and miR-192 were reciprocally expressed in the kidney
In rat nephron segments, we found high miR-192 expression in PCT and low expression in mTAL (Figures 1 and 2; Supplementary Table S1). Consistent with these data, we previously showed high miR-192 expression in the rat renal cortex (where PCT are abundant) and low expression in the medulla (where mTAL are abundant) (16). In fact, miR-192 is one of the most abundant miRNAs in the kidney cortex and the PCT.
As shown in Table 1, miR-192 was predicted to target the Na+/K+-ATPase β1 subunit (Atp1b1). McDonough et al. (22) previously analyzed Atp1b1 protein in nephron segments and determined lower expression in PCT and higher expression in mTAL. Consistently, in our study we found Atp1b1 protein to be expressed at lower levels in rat kidney cortex compared with the medulla, whereas there was no significant difference in the alpha subunit expression. Moreover, the enzymatic activity of Na+/K+-ATPase in the membrane fraction was significantly lower in the renal cortex compared with the medulla (42 ± 7 versus 88 ± 14 nmol/mg/min; n = 6–7, P < 0.05).
miR-192 contributes to renal handling of fluid balance
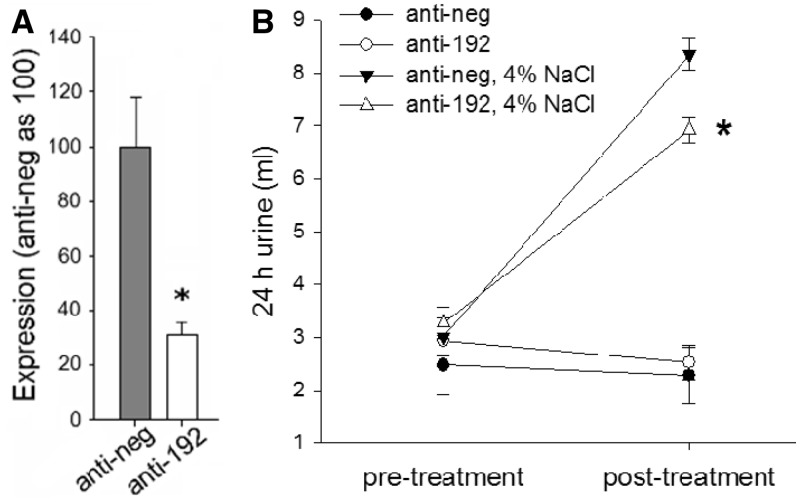
We examined the in vivo functional importance of miR-192 in conditions of increased sodium and water load. Mice were housed in metabolic cages, with urine output and water intake monitored over 24-h periods. miR-192 suppression was achieved by LNA-modified anti-miR-192 delivered as i.p. injections (10 mg/kg). Anti-miR-192 substantially suppressed miR-192 in kidney cortex 2 days after the injection (Figure 4A).
Figure 4.
Knockdown of miR-192 blunted the increase of urine output in condition of high salt intake in mice. (A) LNA-modified anti-miR-192 decreased miR-192 expression in kidney cortex in mice. Anti-miR-192 (anti-192) or anti-scrambled control (anti-neg) oligonucleotides were delivered to mice by intraperitoneal injection (10 mg/kg body weight). Forty-eight hours after injection, miR-192 expression was suppressed to ∼30% of control levels. n = 6, *P < 0.05 versus anti-neg. (B) Knockdown of miR-192 significantly attenuated the increase of urine output when mice were switched from a 0.4% NaCl diet to a 4% NaCl diet. All animals were fed with a 0.4% NaCl diet after receipt. After 4 days of acclimatization, 24-h urine collection was started and urine output on the 0.4% NaCl diet was measured for 5 days to ensure a stable baseline. Mice were then divided into four groups based on oligonucleotide and dietary treatment: (i) remaining on the 0.4% NaCl diet and receiving scrambled anti-miR (anti-neg) (n = 6); (ii) remaining on the 0.4% NaCl diet and receiving anti-miR-192 (anti-192) (n = 6); (iii) switched to a 4% NaCl diet and receiving anti-neg (n = 8) and (iv) switched to a 4% NaCl diet and receiving anti-192 (n = 8). *P < 0.05 versus the ‘anti-neg, 4% NaCl’ group.
Suppression of miR-192 did not significantly affect urine output in mice that were fed with a 0.4% NaCl diet (Figure 4B). However, when mice were fed with a high-salt (4%) diet, the anti-miR-192 injection blunted the high salt-induced increase of urine output by ∼30% in the first 24 h of high salt intake (Figure 4B). Water intake was not statistically different between the animal groups.
Suppression of miR-192 causes up-regulation of Atp1b1 that correlates with a decrease in urine output
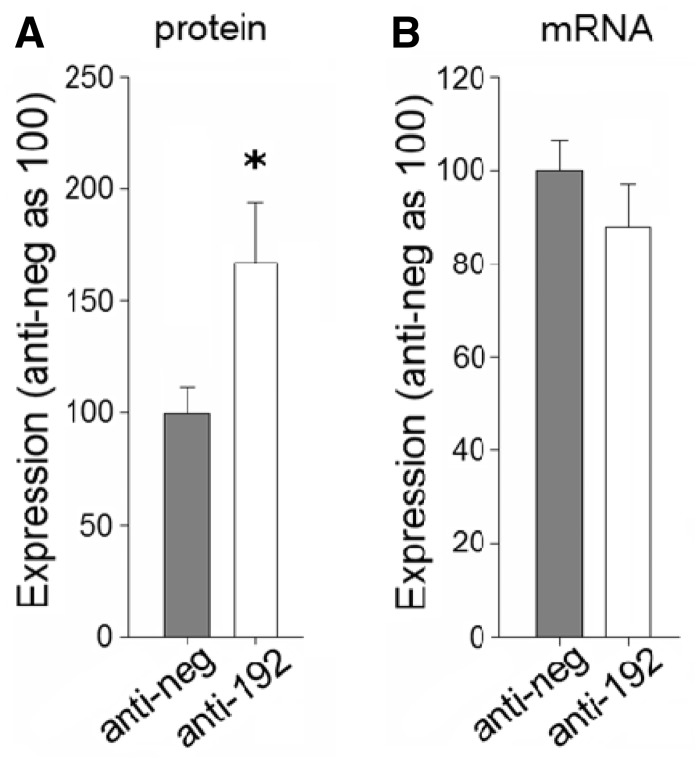
Mice injected with anti-miR-192 showed significant up-regulation of Atp1b1 protein to ∼160% of control levels, measured in kidney cortex 48 hours after injection (Figure 5). Atp1b1 abundance appeared to correlate with miR-192 effects on urine output. By day 7 after the anti-miR injection, Atp1b1 protein was no longer up-regulated and urine output was not different in mice treated with anti-miR-192 compared with mice treated with scrambled anti-miR, even though miR-192 was still suppressed.
Figure 5.
Knockdown of miR-192 causes up-regulation of Atp1b1 in kidney cortex in mice. Anti-miR-192 or anti-scrambled control (anti-neg) oligonucleotides were delivered to mice by intraperitoneal injection (10 mg/kg). Expression of Atp1b1 was assessed 48 h after knocking down miR-192. (A) Protein abundance of Atp1b1. n = 6, *P < 0.05 versus control. (B) mRNA levels of Atp1b1. n = 6.
Sodium depletion results in suppression of miR-192 and is accompanied by a trend of increased Atp1b1 abundance in rat kidney
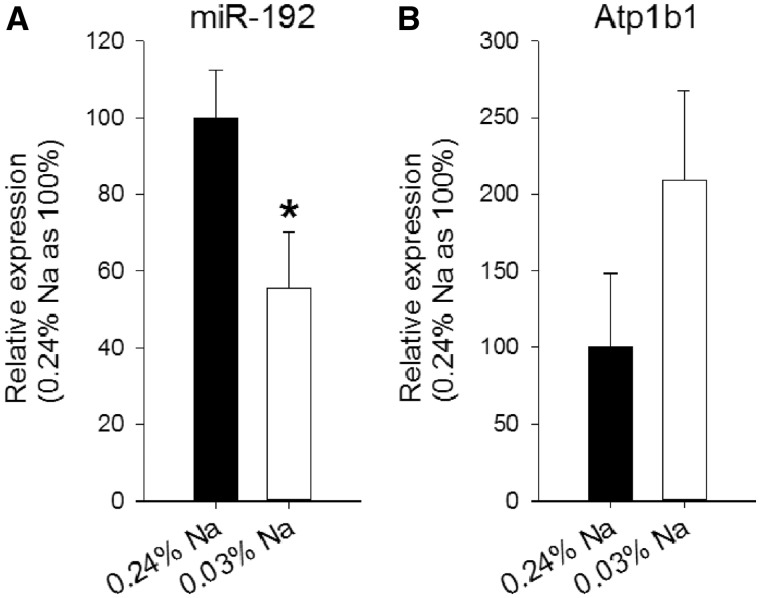
To further examine endogenously regulated changes in miR-192 and Atp1b1 expression, we fed rats with a control diet containing 0.24% sodium and then with a diet with only 0.03% sodium. Forty-eight hours after starting the low-salt diet, miR-192 was significantly suppressed to ∼55% of the basal level, whereas Atp1b1 abundance tended to increase (Figure 6).
Figure 6.
miR-192 and Atp1b1 abundance in rat kidney cortex in condition of sodium depletion. Rats were fed with a basal diet containing 0.24% sodium (control). One group of animals was then switched to a diet containing 0.03% sodium and maintained on that diet for 48 h. (A) Relative miR-192 expression in kidney cortex, in the two animal groups. After 48 h of low sodium diet, miR-192 was significantly suppressed. n = 5–6/group, *P < 0.05 versus control. (B) Relative Atp1b1 expression in kidney cortex, in the two animal groups. At 48 h of low-salt diet Atp1b1 protein abundance was approximately doubled, although the change did not reach statistical significance. n = 5–6/group.
miR-192 regulates Atp1b1 expression and Na+/K+-ATPase activity in human kidney cells
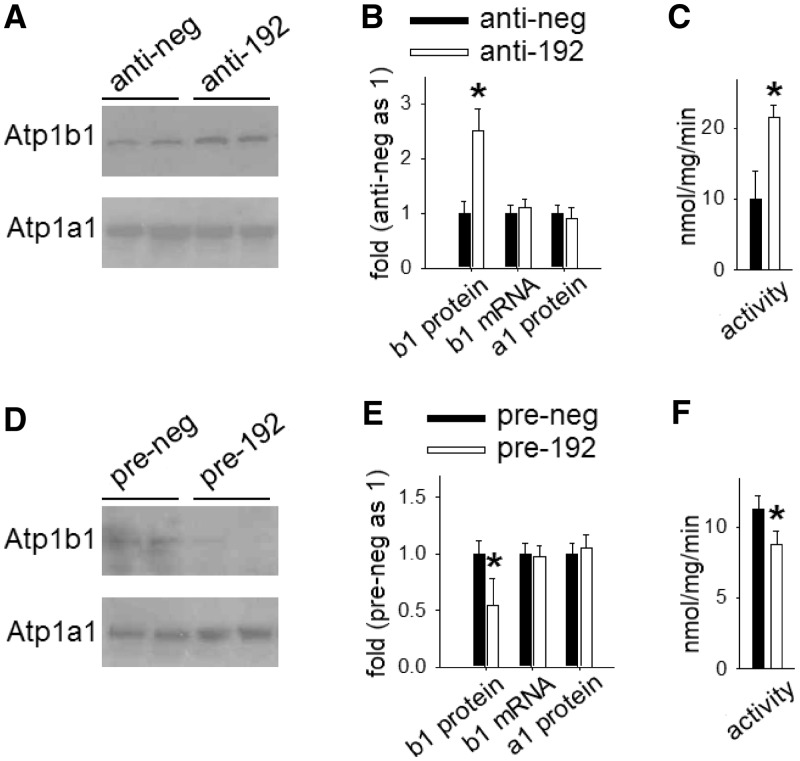
In cultured human kidney epithelial cells, suppression of miR-192 up-regulated Atp1b1 protein and membrane-bound Na+/K+-ATPase enzymatic activity to ∼250 and ∼210% of control values, respectively (Figure 7A–C). Conversely, a miR-192 mimic (pre-miR-192) decreased Atp1b1 protein abundance and Na+/K+-ATPase enzymatic activity to ∼50 and ∼75% of control levels, respectively (Figure 7D–F). Importantly, the α1 subunit of Na+/K+-ATPase (Atp1a1) was not affected, suggesting that Atp1b1 modulation caused the changes in membrane-bound Na+/K+-ATPase activity. Also, miR-192 expression did not affect Atp1b1 mRNA levels, which suggests regulation at translational level.
Figure 7.
Atp1b1 expression and membrane-bound Na+/K+-ATPase activity in human kidney cells is regulated by miR-192. Cultured human renal epithelial cells were transfected with 100 nM anti-miR-192 oligonucleotides for miR-192 knockdown and pre-miR-192 oligonucleotide for miR-192 over-expression and their respective scrambled control oligonucleotides (anti-neg and pre-neg). Cells were harvested after 48-h incubation period. Images on the left (A and D) show western blots for Atp1b1 and Atp1a1; graphs on the right represent quantification of Atp1b1 and Atp1a1 protein or mRNA expression and Na+/K+-ATPase activity. (A–C) Knocking down miR-192 resulted in up-regulation of Atp1b1 protein as well as increased membrane-bound Na+/K+-ATPase. mRNA levels of Atp1b1 and protein levels of Atp1a1 were not affected. n = 6–7, *P < 0.05 versus control. (D–F) Over-expressing miR-192 resulted in down-regulation of Atp1b1 protein as well as decreased membrane-bound Na+/K+-ATPase activity, without affecting Atp1b1 mRNA or Atp1a1 protein expressions. n = 6–7, *P < 0.05 versus control.
miR-192 interacts with the 5′-UTR of Atp1b1
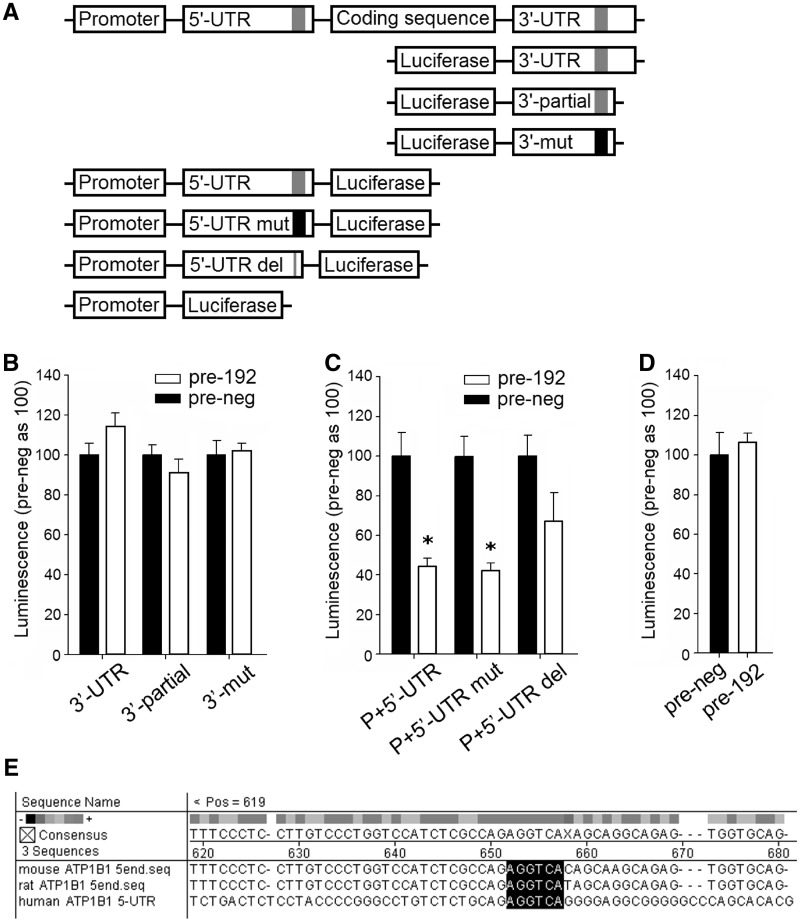
To test direct interaction between miR-192 and Atp1b1 3′-UTR, we performed the luciferase reporter assay. We tested constructs containing the whole 3′-UTR sequence, a 3′-UTR segment containing the predicted miR-192 binding site and the segment with introduced mutation at the predicted binding site (Figure 8A). The result indicated there was no direct interaction between miR-192 and Atp1b1 3′-UTR (Figure 8B).
Figure 8.
Analysis of miR-192 interaction with 3′-UTR, 5′-UTR and promoter of rat Atp1b1. (A) Schematic description of the constructs used in the analysis. The top scheme represents the Atp1b1 gene structure including the promoter, 5′-UTR, coding sequence and 3′-UTR. The gray segment represents the putative miR-192 binding region and the black segment represents mutation of the same region. (B) 3′-UTR reporter analysis did not confirm interaction between miR-192 and 3′-UTR of Atp1b1. 3T3-L1 cells were co-transfected with pre-miR-192 or pre-neg (control oligonucleotides) and the luciferase reporter constructs containing one of the following: 3′-UTR (the whole 3′-UTR of Atp1b1), 3′-UTR-partial (a segment of the 3′-UTR containing the putative miR-192 binding site) or 3′-mut (the 3′-UTR segment with mutated putative miR-192 binding site, at nucleotides +143 to +145, where +1 is the first nucleotide in the 3′-UTR of Atp1b1). n = 8–19/group. (C) 5′-UTR reporter analysis confirmed interaction between miR-192 and 5′-UTR of Atp1b1. 3T3-L1 cells were co-transfected with pre-miR-192 or pre-neg and the luciferase reporter construct containing one of the following: P+5′-UTR (promoter region and 5′-UTR of Atp1b1), P+5′-UTR-mut (the promoter region and 5′-UTR with mutated putative miR-192 binding site, at nucleotides −387 to −389, where +1 is the translation initiation site) or P+5′-UTR-del (the promoter region and 5′-UTR with deletion of 6 bp containing the putative miR-192 binding site, at nucleotides −386 to −391, where +1 is the translation initiation site). Pre-miR-192 suppressed luciferase activity of the P+5′-UTR and P+5′-UTR-mut constructs, whereas suppression of the P+5′-UTR-del construct did not reach statistical significance. n = 4/group, *P < 0.05 versus control. (D) miR-192 did not affect Atp1b1 promoter activity. Human renal epithelial cells were co-transfected with pre-miR-192 or pre-neg and the luciferase expression vector driven by the Atp1b1 promoter. n = 8. (E) Alignment of a segment of the 5′-UTR of Atp1b1 in mouse, rat and human containing a single, conserved predicted miR-192 binding site (marked in black). To predict the miR-192 binding site in the Atp1b1 5′-UTR, we screened the 5′-UTR for sequence complementarity with the miR-192 seed region sequence (nucleotides 2–7 of the miR-192). We allowed for wobble base pairs (G–U).
We examined the possibility that miR-192 interacted with the Atp1b1 5′-UTR (5–7,23). Alignment of the entire 5′-UTR sequence of Atp1b1 with the seed region sequence of miR-192 identified a single binding site at nucleotides −386 to −391, where +1 is the translation initiation site (Figure 8E). When cells were treated with pre-miR-192, luciferase activity was significantly reduced in the construct containing both the Atp1b1 promoter and 5′-UTR, which indicated that miR-192 interacted with Atp1b1 promoter and/or 5′-UTR (Figure 8C). The inclusion of the Atp1b1 promoter (the 1-kb sequence upstream of the transcription start site) in the construct was necessary for driving luciferase expression at a physiologically relevant level. Introducing mutations of 3 nt at the putative miR-192 binding site within the 5′-UTR did not affect luciferase activity. However, deleting all 6 nt at the binding site partially blunted the miR-192 effect (Figure 8C). miR-192 did not suppress luciferase signal when the construct contained only the Atp1b1 promoter but not the 5′-UTR, excluding the possibility of transcriptional suppression mediated through the Atp1b1 promoter (Figure 8D). Overall, these results suggested miR-192 interaction with Atp1b1 5′-UTR as the possible mechanism by which miR-192 reduces Atp1b1 abundance.
DISCUSSION
The present study provided novel evidence for an important and broad role of miRNAs in maintaining cell-specific characteristics in PCT and mTAL that may appear subtle but are physiologically critical. We demonstrated a direct role that miR-192 had in the regulation of a major transporter in renal epithelial cells, Na+/K+-ATPase. In vivo studies suggest a novel physiological mechanism in which miR-192 suppresses Atp1b1, which could contribute to kidney handling of solutes and fluids. We also showed that miR-192 interacted with Atp1b1 in an unconventional manner involving the 5′-UTR.
miRNAs have been shown to be involved in a large number of developmental and pathological processes in different organs and tissues (10,11). However, the role of miRNAs in adult physiology is much less understood. Several studies have demonstrated the significance of miRNAs in determining cell-type-specific characteristics that are functionally important (24–26). For example, Van Rooij et al. demonstrated that miR-208b and miR-499 knockout mice had substantial loss of slow (Type I) myofibers and increased abundance of fast (Type II) myosin isoforms. Conversely, over-expression of miR-499 induced complete conversion of fast into slow myofibers (26).
In this study, we identified 55 miRNA-target pairs that consisted of segment-specific miRNAs and their predicted segment-specific protein targets, mainly transporters and channels, in PCT and mTAL. These miRNAs might contribute to determining segment-specific molecular characteristics of PCT and mTAL. miRNAs that are equally expressed across the segments may still be important in regulating transport activities; however, they are less likely to be involved in maintaining the specificity or differences between the segments. It would be important to confirm direct interactions between miRNAs and target genes in any future studies that intend to further examine the significance of the high-probability miRNA-target pairs that we identified. Moreover, it would be important to understand the molecular mechanisms underlying the differential enrichment of miRNAs in different nephron segments. The abundance of a miRNA, similar to mRNA, can be regulated at transcriptional and post-transcriptional levels.
miRNAs have been shown to be important in renal development and renal injury. Experiments in animals with cell-specific loss of dicer, an enzyme important for miRNA maturation, supported a role of miRNA in development and function of glomeruli, proximal tubules and renin-producing juxtaglomerular cells (27–30). miRNAs are involved in pathological molecular pathways in the kidney including epithelial–mesenchymal transition, fibrosis, diabetic or hypertensive injury, ischemic preconditioning and possibly hypertension (13–15,31–35). The role of miRNAs in renal physiology, however, is only beginning to be studied. Lin et al. (36) showed that miR-802 mediates the stimulatory effects of potassium on ROMK channel activity in renal cells possibly by targeting and suppressing caveolin-1 expression. Another study demonstrated that several miRNAs participate in cellular responses to osmotic stress in kidney cells (37). The findings of the current study suggest that miRNAs could have wide-spread effects on renal physiology.
As a specific example, we demonstrated regulation of Na+/K+-ATPase activity by miR-192 in kidney cells. Na+/K+-ATPase provides the primary driving force for transport of nearly all solutes and fluids in the kidney. Suppression of miR-192 significantly blunted the adaptational increase in urine output, indicating the importance of maintaining high miR-192 levels during increased sodium/water intake. This result seems to be in agreement with the study by Elvira-Matelot et al. (38) that detected decreased kidney miR-192 levels in condition of sodium depletion in mice, an effect confirmed in rats in the present study. It would be valuable to determine in future studies whether miR-192 influences sodium excretion as suggested by the urine output data and which nephron segments mediate any such effect of miR-192.
Although miR-192 remained suppressed 7 days after the anti-miR injection, urine output was decreased only during the first 24 h following treatment. Changes in urine output correlated with expression of Atp1b1, which was only transiently up-regulated during the first 48 h after miR-192 suppression. Effects of miR-192 knockdown on Atp1b1 expression and urine output are modest and transient, which is likely due to the robust control of Na+/K+-ATPase and urine output that involves a complex regulatory network. In normal physiological conditions, Na+/K+-ATPase activity is regulated by multiple factors including angiotensin II, norepinephrine and alpha agonists, dopamine, endothelin, nitric oxide, prostaglandins and others (39–43). In addition to Na+/K+-ATPase-mediated control, there are other mechanisms contributing to maintaining fluid balance, which is critical for whole body homeostasis and must be tightly controlled. It is the integrated response of multiple regulatory systems that determines Na+/K+-ATPase activity and fluid balance under various physiological challenges. The robust regulatory network would be expected to make the effect of altering any one component (such as miR-192 and Na+/K+-ATPase) modest and/or transient.
Knockdown of miR-192 in mouse kidney was achieved by i.p. injections of anti-miR-192 in the present study. Despite systemic administration, direct effects of anti-miR-192 were likely to be localized, as miR-192 is expressed at substantial levels only in kidney and gastrointestinal tract (44,45). Within the kidney, miR-192 is predominantly expressed in the cortex (16).
The Na+/K+-ATPase transporter is composed of at least two subunits, alpha and beta. Our in vitro studies demonstrated that miR-192-mediated changes in Atp1b1 expression could modulate membrane-bound Na+/K+-ATPase enzymatic activity. Although alpha subunit contains the catalytic site, beta subunit is required for normal Na+/K+-ATPase activity, as it is involved in maturation and cellular localization of Na+/K+-ATPase and has a stabilizing effect on the alpha subunit (46–49). Experiments in Xenopus oocytes showed that pre-existing beta subunit could assemble with alpha subunit expressed later, resulting in functional Na+/K+-ATPase. On the other hand, functional Na+/K+-ATPase was not formed when the alpha subunit was expressed first, followed by beta subunit expression later (50). Further, beta subunit has a longer half-life than unassembled alpha subunit. These studies indicate that the beta subunit is a modulator with an important fine-tuning role in the enzyme’s activity.
The majority of reported miRNA-target interactions occur through binding of miRNAs to 3′-UTR of target mRNAs. However, alternative mechanisms, such as interaction between miRNAs and 5′-UTR, have been shown in a small number of cases (5–7,23). Moretti et al. (6) demonstrated that miR-2 could specifically regulate translational process in vitro and in vivo, not only through interaction with the 3′-UTR but also equally effectively through interaction with the 5′-UTR or the open reading fragment (ORF). The 5′-UTR (and the ORF) can function as miRNA binding sites with mechanistically similar interactions as the 3′-UTR. It was suggested that miRNA-5′-UTR interaction obstructs translational initiation and also interferes with translating ribosomes (6,51). In our study, we have experimentally demonstrated inhibitory effect of miR-192 on the 5′-UTR of Atp1b1. The interaction appeared to depend in part on nucleotides 385–390 in the 5′-UTR. However, we cannot rule out the possibility that additional binding sites with a lower degree of homology may contribute to the effect of miR-192. miR-192 could also influence the reporter gene activity through indirect mechanisms. For example, miR-192 could potentially affect a protein that binds to the −386 to −391 region in the 5′-UTR sequence of Atp1b1.
The kidney regulates whole body fluid and electrolyte homeostasis, which is mediated by filtration and transport activities in distinct nephron segments. The findings of the current study suggest that dozens or more miRNA-target interactions might be involved in determining transport and other physiological characteristics of PCT and mTAL and that a novel physiological mechanism exists in which miR-192 suppresses Na+/K+-ATPase and contributes to renal handling of fluid balance on a high-salt diet.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Materials and Methods.
FUNDING
National Institutes of Health (NIH) [HL085267, DK084405, HL077263, HL082798, HL029587, HL111580 to M.L.]; American Heart Association Pre-Doctoral Fellowship (to D.M.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai NP, Lin YL, Wei LN. MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem. J. 2009;424:411–418. doi: 10.1042/BJ20090915. [DOI] [PubMed] [Google Scholar]
- 6.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′UTR. Proc. Natl Acad. Sci. USA. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 11.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38(Suppl.):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 13.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382. Nucleic Acids Res. 2010;38:8338–8347. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kriegel AJ, Liu Y, Cohen B, Usa K, Liu Y, Liang M. MiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosis. Physiol. Genomics. 2012;44:259–267. doi: 10.1152/physiolgenomics.00173.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang M, Pietrusz JL. Thiol-related genes in diabetic complications: a novel protective role for endogenous thioredoxin 2. Arterioscler. Thromb. Vasc. Biol. 2007;27:77–83. doi: 10.1161/01.ATV.0000251006.54632.bb. [DOI] [PubMed] [Google Scholar]
- 18.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW, Jr, Liang M. Novel role of fumarate metabolism in dahl-salt sensitive hypertension. Hypertension. 2009;54:255–260. doi: 10.1161/HYPERTENSIONAHA.109.129528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang M, Knox FG. Nitric oxide reduces the molecular activity of Na+,K+-ATPase in opossum kidney cells. Kidney Int. 1999;56:627–634. doi: 10.1046/j.1523-1755.1999.00583.x. [DOI] [PubMed] [Google Scholar]
- 20.Liang M, Knox FG. Nitric oxide activates PKCalpha and inhibits Na+-K+-ATPase in opossum kidney cells. Am. J. Physiol. 1999;277:F859–F865. doi: 10.1152/ajprenal.1999.277.6.F859. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Mladinov D, Pietrusz JL, Usa K, Liang M. Glucocorticoid response elements and 11 beta-hydroxysteroid dehydrogenases in the regulation of endothelial nitric oxide synthase expression. Cardiovasc. Res. 2009;81:140–147. doi: 10.1093/cvr/cvn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonough AA, Magyar CE, Komatsu Y. Expression of Na(+)-K(+)-ATPase alpha- and beta-subunits along rat nephron: isoform specificity and response to hypokalemia. Am. J. Physiol. 1994;267:C901–C908. doi: 10.1152/ajpcell.1994.267.4.C901. [DOI] [PubMed] [Google Scholar]
- 23.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Quiat D, Voelker KA, Pei J, Grishin NV, Grange RW, Bassel-Duby R, Olson EN. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc. Natl Acad. Sci. USA. 2011;108:10196–10201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J. Am. Soc. Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J. Am. Soc. Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2010;21:756–761. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, Carretero O, Sigmund CD, Gomez RA. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J. Am. Soc. Nephrol. 2010;21:460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl Acad. Sci. USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J. Am. Soc. Nephrol. 2011;22:1668–1681. doi: 10.1681/ASN.2010111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Kriegel AJ, Liu Y, Usa K, Mladinov D, Liu H, Fang Y, Ding X, Liang M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012;82:1167–1175. doi: 10.1038/ki.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marques FZ, Campain AE, Tomaszewski M, Zukowska-Szczechowska E, Yang YH, Charchar FJ, Morris BJ. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011;58:1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 36.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J. Am. Soc. Nephrol. 2011;22:1087–1098. doi: 10.1681/ASN.2010090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Liu H, Wang T, Zhang T, Kuang J, Luo Y, Chung SS, Yuan L, Yang JY. Tonicity-responsive microRNAs contribute to the maximal induction of osmoregulatory transcription factor OREBP in response to high-NaCl hypertonicity. Nucleic Acids Res. 2011;39:475–485. doi: 10.1093/nar/gkq818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elvira-Matelot E, Zhou XO, Farman N, Beaurain G, Henrion-Caude A, Hadchouel J, Jeunemaitre X. Regulation of WNK1 expression by miR-192 and aldosterone. J. Am. Soc. Nephrol. 2010;21:1724–1731. doi: 10.1681/ASN.2009111186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banday AA, Lokhandwala MF. Loss of biphasic effect on Na/K-ATPase activity by angiotensin II involves defective angiotensin type 1 receptor-nitric oxide signaling. Hypertension. 2008;52:1099–1105. doi: 10.1161/HYPERTENSIONAHA.108.117911. [DOI] [PubMed] [Google Scholar]
- 40.Bertorello AM, Katz AI. Short-term regulation of renal Na-K-ATPase activity: physiological relevance and cellular mechanisms. Am. J. Physiol. 1993;265:F743–F755. doi: 10.1152/ajprenal.1993.265.6.F743. [DOI] [PubMed] [Google Scholar]
- 41.Bertorello A, Hokfelt T, Goldstein M, Aperia A. Proximal tubule Na+-K+-ATPase activity is inhibited during high salt diet: evidence for DA-mediated effect. Am. J. Physiol. 1988;254:F795–F801. doi: 10.1152/ajprenal.1988.254.6.F795. [DOI] [PubMed] [Google Scholar]
- 42.Guzman NJ, Fang MZ, Tang SS, Ingelfinger JR, Garg LC. Autocrine inhibition of Na+/K(+)-ATPase by nitric oxide in mouse proximal tubule epithelial cells. J. Clin. Invest. 1995;95:2083–2088. doi: 10.1172/JCI117895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herman MB, Rajkhowa T, Cutuli F, Springate JE, Taub ML. Regulation of renal proximal tubule Na,K-ATPase by prostaglandins. Am. J. Physiol. Renal Physiol. 2010;298:F1222–F1234. doi: 10.1152/ajprenal.00467.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jorgensen PL, Hakansson KO, Karlish SJ. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. [DOI] [PubMed] [Google Scholar]
- 47.Geering K. The functional role of the beta-subunit in the maturation and intracellular transport of Na,K-ATPase. FEBS Lett. 1991;285:189–193. doi: 10.1016/0014-5793(91)80801-9. [DOI] [PubMed] [Google Scholar]
- 48.Geering K. The functional role of beta subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- 49.Béguin P, Hasler U, Staub O, Geering K. Endoplasmic reticulum quality control of oligomeric membrane proteins: topogenic determinants involved in the degradation of the unassembled Na,K-ATPase alpha subunit and in its stabilization by beta subunit assembly. Mol. Biol. Cell. 2000;11:1657–1672. doi: 10.1091/mbc.11.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noguchi S, Higashi K, Kawamura M. Assembly of the α-subunit of Torpedo californica Na+/K+-ATPase with its pre-existing β-subunit in Xenopus oocytes. Biochim. Biophys. Acta. 1990;1023:247–253. doi: 10.1016/0005-2736(90)90420-s. [DOI] [PubMed] [Google Scholar]
- 51.Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.