Abstract
Bacterial RNase J and eukaryal cleavage and polyadenylation specificity factor (CPSF-73) are members of the β-CASP family of ribonucleases involved in mRNA processing and degradation. Here we report an in-depth phylogenomic analysis that delineates aRNase J and archaeal CPSF (aCPSF) as distinct orthologous groups and establishes their repartition in 110 archaeal genomes. The aCPSF1 subgroup, which has been inherited vertically and is strictly conserved, is characterized by an N-terminal extension with two K homology (KH) domains and a C-terminal motif involved in dimerization of the holoenzyme. Pab-aCPSF1 (Pyrococcus abyssi homolog) has an endoribonucleolytic activity that preferentially cleaves at single-stranded CA dinucleotides and a 5′–3′ exoribonucleolytic activity that acts on 5′ monophosphate substrates. These activities are the same as described for the eukaryotic cleavage and polyadenylation factor, CPSF-73, when engaged in the CPSF complex. The N-terminal KH domains are important for endoribonucleolytic cleavage at certain specific sites and the formation of stable high molecular weight ribonucleoprotein complexes. Dimerization of Pab-aCPSF is important for exoribonucleolytic activity and RNA binding. Altogether, our results suggest that aCPSF1 performs an essential function and that an enzyme with similar activities was present in the last common ancestor of Archaea and Eukarya.
INTRODUCTION
RNA processing and degradation are critical to the survival of all cells and acknowledged as a means of regulating gene expression. In particular, the nature of RNA 5′ and 3′-ends is known to have major impact because they control the entry and directionality of endo- and exoribonucleases involved in these processes. In Archaea, exploration of RNA processing and degradation pathways is still in its early stages. Because easy genetic approaches are not readily available, functional studies in the Archaea are often based on genomic and proteomic analyses that are interpreted in light of our understanding of RNA metabolism in Bacteria and Eukarya. Unlike its eukaryal counterpart, archaeal mRNA is not capped at its 5′-end nor is it polyadenylated at its 3′-end [(1) for review]. Transcription in the Archaea is performed by a eukaryal-like RNA polymerase that initiates at ‘TATA-boxes’ [for review see (2)]. However, little is known about mRNA 3′-end maturation and transcription termination.
Processing of the 3′-end is an essential step in converting eukaryotic pre-mRNAs to mature polyadenylated mRNAs [for review see (3)]. This process is executed by the cleavage and polyadenylation macromolecular complex, which is well-described in both yeast and mammals (4–9). Among other proteins, this machinery includes the cleavage and polyadenylation specificity factor (CPSF-73), a 73 kDa subunit that carries out the endonucleolytic cleavage at a CA motif 20–30 nt downstream of the AAUAAA consensus sequence before polyadenylation (10,11). In the maturation of metazoan histone pre-mRNA, CPSF-73 has also been shown to act as a 5′–3′ exoribonuclease in the degradation of the transcript downstream of the cleavage site (12). Based on sequence similarity, a gene encoding a homolog of the eukaryotic CPSF-73 has been reported to be prevalent in archaeal genomes (13) raising the question of its role in archaeal RNA metabolism. Recently, CPSF-73 homologs in Methanocaldococcus jannashii and Methanothermobacter thermautotrophicus have been shown to have nuclease activity (14–16). However, the enzymatic properties and specificities of archaeal CPSF(aCPSF)-73 homologs remain to be clearly delineated.
The eukaryotic CPSF-73 is a member of the β-CASP family of metallo-β-lactamases (17), which includes ribonucleases important in RNA metabolism that are widespread in all three domains of life (18). The β-CASP proteins have the first four signature motifs of the metallo-β-lactamase superfamily followed by a distinct region [β-CASP domain, (19)] that is characterized by three short conserved motifs A (Asp or Glu), B (His) and C (His). These enzymes use a zinc-dependent mechanism in catalysis and act as 5′–3′ exonucleases and/or endonucleases (19,20). Examples of β-CASP nucleases include RNase J1, a key player of Bacillus subtilis RNA metabolism that functions as an endonuclease and a 5′–3′ exonuclease (21–23). We have shown recently that orthologs of bacterial RNase J with 5′–3′ exonuclease activity are widespread in the Euryarchaea (24). Similar activity has been described for a closely related β-CASP protein of the crenarchaeon Sulfolobus solfataricus (25). Moreover, several crystal structures of aCPSF-73 homologs in M. thermoautotrophicus, Pyrococcus horikoshii and Methanosarcina mazei have been solved. These proteins are dimeric (14,26) and have a tripartite architecture consisting of a N-terminal region with two K homology (KH) RNA-binding motifs, a central β-metallo-lactamase domain and a C-termimal β-CASP domain (14,16,26). Altogether, identification of homologs of the eukaryotic CPSF-73 and of bacterial RNase J in the Archaea raises the question of their role in RNA metabolism and their evolutionary origin.
The metallo β-lactamase protein superfamily is highly represented in Archaea (26). Among them, the archaeal β-CASP family members were proposed to be RNA hydrolases as their sequences are more closely related to bacterial RNase J and to eukaryotic CPSF-73 than to β-CASP proteins involved in DNA repair and recombination (19). Here we report the inventory, classification and phylogenetic analysis of the archaeal β-CASP proteins, allowing us to identify seven β-CASP clusters. Among them, one is clearly related to bacterial RNase J [archaeal RNase (aRNase J)] and three are related to CPSF-73. Members of the aCPSF1 group are present in all Archaea whose genomes have been sequenced. We show that Pab-aCPSF1 from the thermococcal archaeon Pyrococcus abyssi has the same activities and specificity as its eukaryotic counterpart, CPSF-73. Thus, aCPSF1 family members are authentic orthologs of CPSF-73.
MATERIALS AND METHODS
Collection of archaeal β-CASP proteins
Genome entries of the complete archaeal and bacterial genomes were retrieved from EMBL (http://www.ebi.ac.uk/genomes/) and processed by a set of perl programs into a mySQL database. We used the RPS-Blast program to annotate protein sequences according to the conserved domain database available at the NCBI (http://www.ncbi.nih.gov/Structure/cdd/cdd.shtml) and computed pairs of one-to-one ortholog genes with BlastP as follows: two genes a and b from genomes A and B, are considered to be orthologs if a is the best hit of b in genome A and reciprocally, and if a (or b) has a paralogous gene named c then the score of a versus b should be greater than the score of a (or b) versus c. The proteins of the β-CASP family have the first four signature motifs of the metallo-β-lactamase superfamily followed by a distinct globular domain [named β-CASP domain, (19)] that is used to identify new members of the family. Callebaut et al. (19) identified a list of β-CASP family members in eukaryotes, bacteria and archaea. The archaeal list included only 14 species and 40 β-CASP candidate proteins. To update the annotation of the β-CASP proteins in the 110 complete archaeal genomes, we used the β-CASP domain of each candidate sequence as query in a Psi-Blast search against the protein sequences of complete archaeal genomes. To maximize the sensibility of the prediction, we set the maximal E-value <1e-05 and the maximal number of iterations to 20 to be able to recover all putative candidates in each individual Psi-Blast search. This resulted in an initial collection of 375 proteins.
Protein classification
For each protein of the initial collection, we retrieved orthologs in each archaeal genome to obtain orthologous proteins pairs. Among these, 13 proteins were not identified by the PsiBlast search. A graph was produced where vertices correspond to proteins and edges to orthologous relationships. This graph included six connected components. The application of a partition algorithm [MCL with an inflation operator setup to 1.2 (27)] revealed nine well-defined groups of proteins (orthologous groups, OGs). The protein members of two groups (GloB, COG0491, Zn-dependent hydrolases including glyoxylases and MtrA, COG4063, tetrahydromethanopterin S-methyltransferase, subunit A) do not have the A, B and C β-CASP signature motifs. Hence, they were false positives and were discarded from further analysis. The other OG proteins possess the signature of the β-CASP family and fall in three related COGs: COG1782 (predicted metal-dependent RNase, consisting of a metallo-beta-lactamase domain and an RNA-binding KH domain), COG1236 (predicted exonuclease with beta-lactamase fold involved in RNA processing) and COG0595 (predicted hydrolase of the metallo-beta-lactamase superfamily). As the different protein groups were not found in all archaeal genomes, we systematically searched for putatively unannotated genes (missed by annotation or pseudogenes) with the TblastN program. The strategy used to compute the clusters of orthologous proteins was validated by the very low level of paralogy observed in each group (less than three paralogs per genome).
Alignments and trees computation
The archaeal phylogeny was inferred from a set of 53 ribosomal proteins using the super matrix approach (28) with only one strain per species. Sequences were aligned using the MUSCLE program (29). The alignments were inspected and manually refined using the SEAVIEW sequence editor (30) and trimmed using trimA1 (31). These parsed alignments were concatenated to produce a single alignment of 6938 residues. The maximum likelihood trees were computed with PhyML (32) using the LG model of sequence evolution. The gamma-distributed substitution rate variation was approximated by four discrete categories with shape parameter and proportion of invariant sites estimated from the data. Non-parametric bootstrap values were computed (100 replications of the original dataset) using the same parameters. Trees were visualized and annotated with TreeDyn (33).
Construction of vectors for the expression of Pab-CPSF1 and variants
The supplementary Table S1 summarizes oligonucleotides used in this study. The plasmid used for expression of HIS-tagged Pab-CPSF1 was pET15b. The coding sequence (PAB1868) was amplified by PCR from P. abyssi genomic DNA and cloned as an XhoI-BamHI PCR fragment using OLC5 and OLC3 oligonucleotides to give the plasmid pEC-Pab-CPSF1. The ΔKH and ΔCter variants were constructed using OLΔKHC5/OLC3 and OLC5/OLΔC3 oligonucleotide pairs. The H261A and H594A variants were generated by site-directed mutagenesis of pEC-Pab-CPSF1 with the appropriate oligonucleotides (OLIA261/OL2A261; OL1A594/OL2A594, respectively) and the QuikChange II XL Kit (Stratagene).
Overexpression and purification of wild type Pab-aCPSF1 and variants
The BL21-CodonPlus (DE3) Escherichia coli strain carrying pEC-Pab-aCPSF1 or plasmids bearing mutations was induced at an OD600 of 0.6 by addition of 0.1 mM IPTG, and incubated 3 h at 30°C. A cell extract was heated to 70°C for 10 min and clarified by low-speed centrifugation. TALON Metal resin (Clontech) was used for IMAC purification of the HIS-tagged protein. Proteins (∼10 µg/µl) were dialysed against 20 mM Hepes pH 7.5, 300 mM NaCl, 1 mM EDTA, 1 mM DTT and 1% glycerol and stored at 4°C. The His-tag was removed by treatment with thrombin (0.5 u/µl) for 2 h at room temperature, before incubation at 70°C for 15 min and centrifugation to recover the supernatant. An aliquot of the purified protein was analysed by Coomassie stained 10% SDS-PAGE and migrated at 72 kDa in agreement with the predicted molecular mass of 72 kDa for the wild type protein and at 55 kDa for the ΔKH variant.
RNA synthesis and labeling
In vitro transcription with T7 RNA polymerase was performed as described by the manufacturer (Promega) using PCR fragments as templates. The sR47 and sRkB templates were prepared as described previously (34,35) and the sR47MutCG and sR47Mut21U using oligonucleotides OLsR47MutCG5/OLsR47MutCG3 and OLsR47Mut21U5/OLsR47Mut21U3, respectively (Supplementary Table S1). [α−32P] UTP and [γ−32P] GTP were added to the in vitro transcription mix to synthesize uniformly labeled transcripts and 5′-end triphosphorylated labeled RNAs (p*pp RNA), respectively. 5′-end monophosphate RNA labeling was performed on dephosphorylated RNA or synthetic RNA with T4 polynucleotide kinase in the presence of [γ−32P] ATP. 3′-end labeling was carried out with T4 RNA ligase in presence of [5′-32P] pCp and DMSO. All labeled RNAs were purified on denaturing 8 or 10% PAGE.
RNase assay
A typical enzyme excess reaction in a final volume of 15 µl contained 5 nM 32P-RNA, 6 µM wild type Pab-aCPSF1 or variants, 20 mM Hepes, pH 7.5, 100 mM KCl, 1.5 mM MgCl. Reactions were started by addition of the enzyme and incubated at 65°C and repeated in at least three independent experiments. Samples of 4 µl were withdrawn at the indicated times and the reactions were stopped by incubation with proteinase K (20 u) for 10 min at 37°C before addition of formamide-containing dye supplemented with 10 mM EDTA or spotted directly on thin layer chromatography (TLC) plates (PEI-cellulose, Nagel). The samples and T1/OH ladders were denatured for 1 min at 95°C before separation on 10% PAGE/8 M urea sequencing gels. TLC plates were developed with 0.25 M KH2PO4 and gels were dried before analysis using PhosphoImager and MultiGauge software.
Electrophoretic mobility shift assay
EMSA was performed as previously described (34). RNA and ribonucleoprotein (RNP) complexes were separated on a native 5% (19:1) polyacrylamide gel containing 0.5× TBE and 5% glycerol. Electrophoresis was performed at room temperature at 250 V in 0.5× TBE running buffer containing 5% glycerol. The gels were dried and visualized using a Fuji-Bas 1000 phosphorImager.
Size exclusion chromatography
After IMAC purification, Pab-aCPSF1 and its variants were concentrated by ultrafiltration (Millipore Anicon Ultra 30 K) and loaded onto a Superdex 200 10/300GL gel filtration column (GE Healthcare), pre-equilibrated in 20 mM HEPES (pH 7.5), 300 mM NaCl, 1 mM DTT, 1 mM EDTA and 1% glycerol. The protein standard kit (GE-healthcare) containing ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa) and ribonuclease A (13.7 kDa) was used to estimate the molecular mass of the native protein. The flow rate was fixed at 0.5 ml/min and elution of the protein was monitored by absorbance at 280 nm.
RESULTS
Groups of orthologous β-CASP proteins in the Archaea.
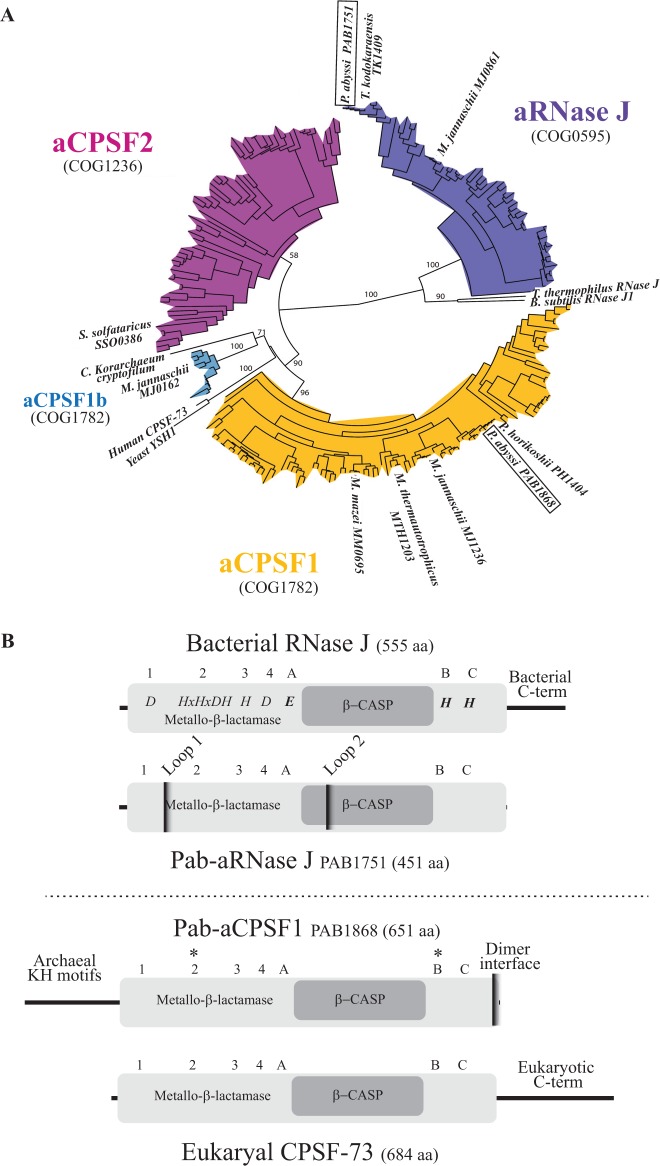
We undertook a detailed analysis of all archaeal β-CASP members to define OGs and to elucidate their evolutionary relationships. We collected β-CASP sequences from 110 complete archaeal genomes and classified them based on sequence conservation as described in Methods. Of the nine clusters that emerged from this analysis (Supplementary Figure S1A), members of the GloB and MtrA clusters did not have the conserved A, B and C motifs characteristic of β-CASP proteins; members of the αβCx cluster had non-canonical spacing between the A and B sequence motifs (Supplementary Figure S1B); members of the αβCy and αβCz clusters were not monophyletic, suggesting complex evolution that might include horizontal gene transfers with bacteria. These OGs were not further considered here. Using the remaining four groups of orthologous β-CASP proteins, we constructed a tree that was rooted with eukaryotic CPSF-73 and bacterial RNase J (Figure 1A). These OGs, which we named aRNase J, aCPSF1, aCPSF1b and aCPSF2, correspond to distinct and specific subtrees. This configuration validates our classification procedure. Apart from the members of the major aCPSF1 group harboring an N-terminal extension of 110 amino acids, archaeal β-CASP candidates are commonly restricted to the β-CASP and metallo-β-lactamase core domains (composed of an average of 420 amino acids) with no additional N- or C-terminal extensions (see below). Note that the S. solfataricus aCPSF2 member (Sso 0386) with a 40 amino acids N-terminal region, is peculiar among the aCPSF2 group.
Figure 1.
The archaeal metallo-β-lactamase β-CASP protein family. (A) Phylogenetic tree of the four β-CASP OGs. The β-CASP proteins with reported structure or/and activity were added as landmarks: yeast and human CPSF 73 kD (10), M. thermautotrophicus MTH1203 (14), P. horikoshii PH1404 (16), M. mazei MM0695 (26), B. subtilis RNase J1 (48), Thermus thermophilus RNase J (21,49), P. abyssi PAB1751 (24), M. jannaschii MJ0861, MJ0162 and MJ1236 (15), S. solfataricus SSO0386 (25). The P. abyssi β-CASP members are framed. It should be mentioned that the landmarked Candidatus Korarchaeum cryptofilum β-CASP member is not considered as a CPSF1b fellow in view of its sequence. (B) Schematic representation of Pab-aRNAse J (PAB1751) and Pab-aCPSF1 (PAB1868), homologs of bacterial RNase J (Top) and eukaryal CPSF-73 (Bottom), respectively. The β-CASP and metallo-β-lactamase domains are highlighted in dark and light grey, respectively. The three β-CASP motifs (A-C), four metallo-β-lactamase motifs (1–4) and N- and C-terminus are indicated with respective features. Pab-aRNase J loop 1 and 2 insertions are indicated (24). The asterisks (*) indicate the position of conserved amino acids that have been mutated in this study, H261A and H594A.
Bacterial and aRNase J were separated from the CPSF-like OGs by a long branch (100% bootstrap support), suggesting an early evolutionary separation. Bacterial RNase J is distinguished from the archaeal homologs by a characteristic C-terminal extension (Figure 1B). The 65 aRNase J members were exclusively present in Euryarchaea as described previously (24) (Supplementary Figure S2). Recently, three members of this group have been reported to have 5′–3′ exoribonuclease activity (15,24) (Table 1) and a phylogenetic analysis showed that aRNase J has been inherited vertically, suggesting an ancient origin predating the separation of Bacteria and Archaea (24).
Table 1.
Reported archaeal β-CASP proteins with their 5′–3′ exo- and/or endoribonucleolytic activity
| β-CASP OG | Protein | Reference | Endo- | 5′–3′Exo |
|---|---|---|---|---|
| aCPSF1 | PAB1868 | This work | + | + |
| MJ1236* | Levy et al. (15) | + | − | |
| aCPSF1b | MJ0162* | Levy et al. (15) | − | + |
| aCPSF2 | Sso0386* | Hasenohrl et al. (25) | − | + |
| aRNAse J | PAB1751 | Clouet-d'Orval et al. (24) | − | + |
| TK 1409 | Clouet-d'Orval et al. (24) | − | + | |
| MJ0861 | Levy et al. (15) | − | + |
The proteins marked by asterisks were misidentified as aRNase J homologs.
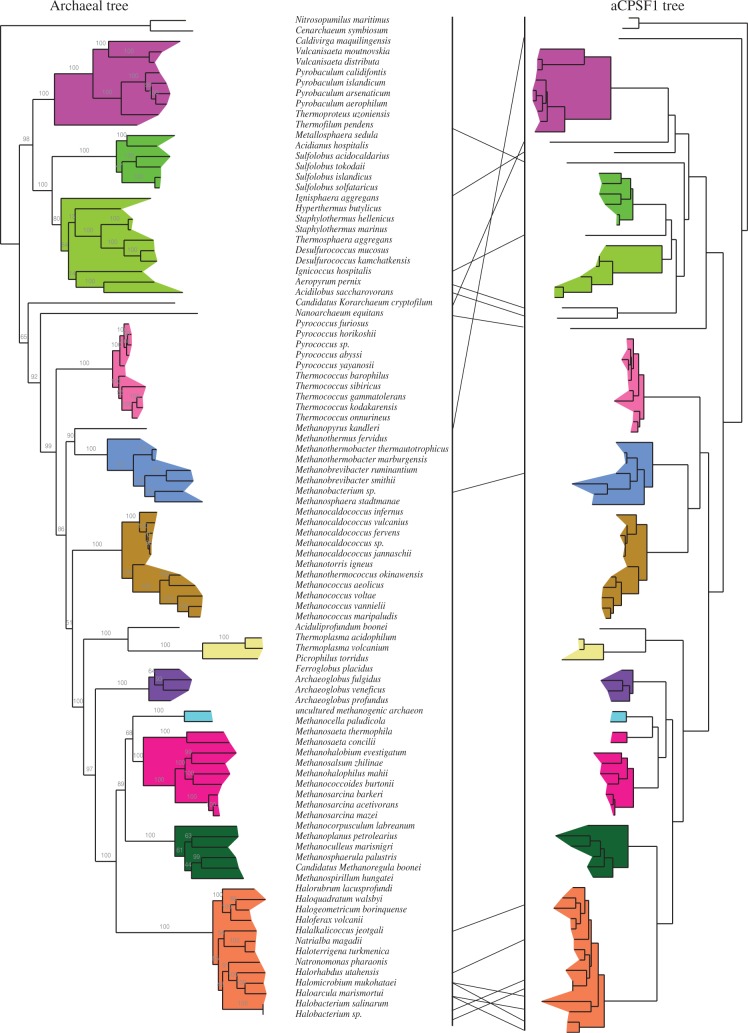
Proteins related to eukaryal CPSF-73 clustered into three OGs: aCPSF1 (112 members) and aCPSF1b (11 members) corresponding to COG1782 and aCPSF2 (80 members) corresponding to COG1236. The aCPSF2 OG is distributed among Crenarchaeota, Euryarchaeota and Thaumarcheoata (Supplementary Figure S2). One member of this subgroup, which was misidentified as RNase J orthologs, has been reported to have a 5′–3′ exonucleolytic activity (25) (Figure 1A and Table 1). The aCPSF1 OG corresponds to a highly conserved family with an N-terminal extension containing two KH RNA binding motifs specific to this group, and a C-terminal motif that is part of a protein dimer interface (Figures 1B and 2). This OG is notable because of its remarkable conservation in all Archaea with no exception to date (Supplementary Figure S2). Moreover, the congruence between the archaeal and aCPSF1 phylogenetic trees (Figure 3) shows that aCPSF1 has been inherited vertically, suggesting an ancient origin predating the emergence of Archaea. The small aCPSF1b OG branching close to the aCPSF1 OG is restricted to the Methanococcales (Figure 1A and Supplementary Figure S2). Only one member (MJ0162, misidentified as RNase J orthologs) is biochemically characterized and harbors 5′–3′ exonucleolytic activity (25) (Figure 1A and Table 1). The aCPSF1b proteins, which appear to have an undecipherable ancient origin, lack the N-terminal extension that is characteristic of the aCPSF1 family.
Figure 3.
Congruence of the archaeal and aCPSF1 phylogenetic trees. The archaeal tree was constructed from a concatenated sequence of 53 ribosomal proteins. The archaeal aCPSF1 tree was constructed from sequences that were available at the time of analysis. Individual archaeal clades are colored to facilitate the comparison between the two trees.
The crystal structure of three aCPSF1 members has recently been reported (14,16,26) as well as endoribonuclease activity for one of them (15) (Figure 1A and Table 1). However, little is known about the substrate specificity and enzymatic properties of the aCPSF1 members. To better characterize the ubiquitous aCPSF1 OG, we investigated the properties of the P. abyssi member of this group. The P. abyssi genome contains three open reading frames with β-CASP protein signatures. PAB1751, PAB1868 and PAB1035 are members of the aRNase J, the aCPSF1 and the highly divergent αβCy cluster, respectively (Figure 1A and Supplementary Figure S1A). PAB1035 was not further considered here. PAB1751, denoted as Pab-aRNase J, corresponds to the recently identified ortholog of the bacterial RNase J (Figure 1B) (24,36). This protein has been shown to have a highly processive 5′-end-dependent exonuclease activity with a 5′–3′ directionality (24). In the following sections, we analysed the mode of ribonuclease cleavage and substrate specificity of Pab-aCPSF1 as well as the function of the N-terminal KH domains and C-terminal protein dimer interface.
Pab-aCPSF1 has endo- and exoribonuclease activity
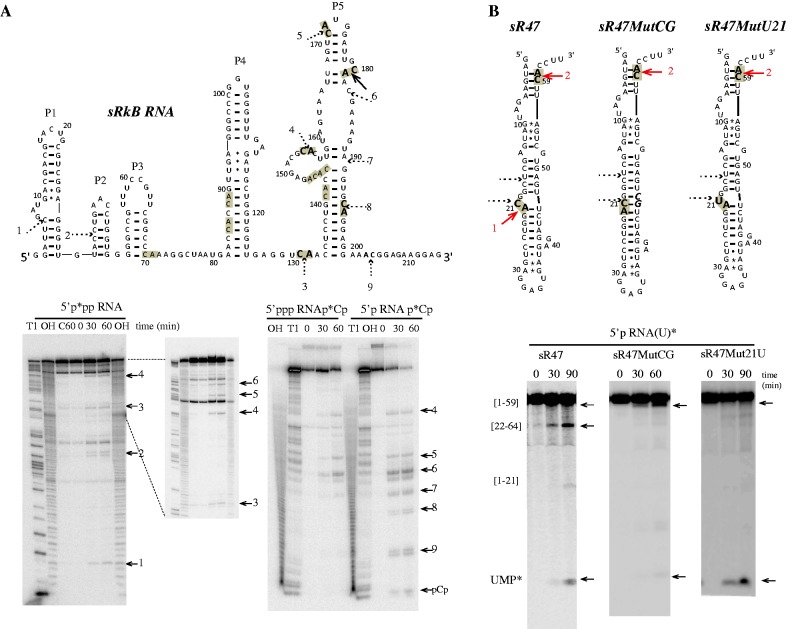
We investigated the enzymatic activity of recombinant Pab-aCPSF1 (untagged version) by performing assays in enzyme excess using the well-characterized 64-nt sR47 RNA substrate, corresponding to a P. abyssi box C/D guide RNA (24,34) (Figure 4). Incubation of Pab-aCPSF1 with 5′-end-labeled triphosphorylated RNA (5′p*pp RNA) yielded two major products of 21 and 59 nt in length (Figure 4A). These products correspond to cleavages after cytosines C21 and C59, which are located in the only two CA dinucleotides in the sR47 RNA substrate (Figure 4A). Assays with triphosphorylated 3′-end-labeled (5′ppp RNAp*Cp) and uniformly labeled (5′ppp RNA(U)*) substrates yielded two and four RNA products, respectively, corresponding to cleavages at the CA dinucleotides (Figure 4A). The addition of divalent ions to the reaction buffer did not stimulate activity, nor did the addition of EDTA inhibit the reaction. However, the activity of the enzyme was strongly inhibited by addition of 1,10-phenanthroline, a potent Zn2+ chelator (Supplementary Figure S3A). Furthermore, the substitution of conserved residues in the β-lactamase and β-CASP motifs (H261A and H594A in motifs 2 and B, respectively, Figure 1B) abolished ribonuclease activity (Supplementary Figure S3B). The recently published crystal structures of several aCFSF1 orthologs show that these histidines are involved in coordinating two zinc ions that are essential for catalysis (14,16,26). Altogether, these results reveal that Pab-aCPSF1 is a bona fide β-CASP protein and that the activity reported here is not due to a contaminating ribonuclease. We performed similar assays with sRkB, a 216 nt non-coding RNA recently identified in P. abyssi (35) (Figure 5A). sRkB was cleaved at nine positions: five corresponded to CA dinucleotides and the other four to GC, CC, AG or AC dinucleotides (Figure 5A). Two CA dinucleotides located in the highly stable P4 helix of sRkB were not cleaved suggesting a preference for single-stranded CA dinucleotides. This conclusion is supported by results with the sR47MutCG and sR47MutU21, which are not cleaved at position 21 (Figure 5B). C21 and A22 are embedded in an extended RNA helix in the sR47MutCG variant and C21 is replaced by a U in the sR47MutU21 variant (Figure 5B, higher panel). Altogether, these results show that Pab-aCPSF1 has endoribonuclease activity with a preference for cleavage at single-stranded CA dinucleotides.
Figure 4.
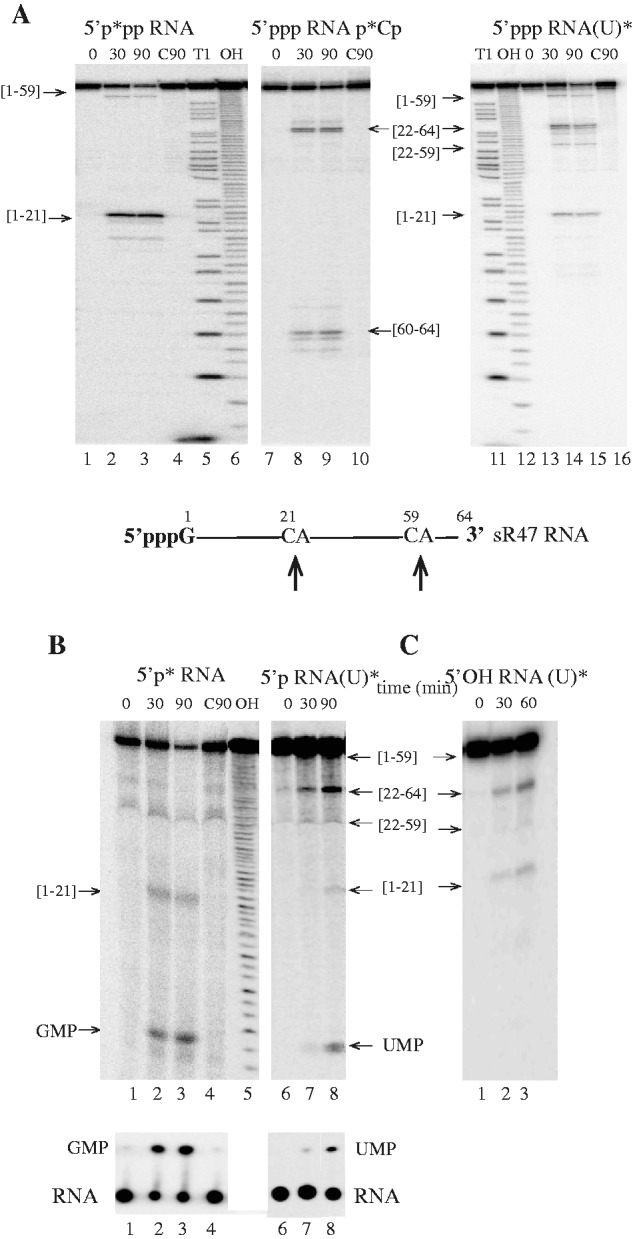
In vitro activity of Pab-aCPSF1. The products of the reaction were analysed on a 10% PAGE and in parallel on TLC (B, Bottom). (A) Kinetic analysis of RNA cleavage of 5′-end-triphosphate sR47 (<1 nM) by an excess of Pab-aCPSF1 (6 µM) at 65°C for the indicated times (0, 30 and 90 min). 5′-end-labeled sR47 RNA carrying a 5′ γ-[P32] (5′p*pp RNA), lanes 1–4; 3′-end-labeled with [P32]Cp (5′ppp RNAp*Cp), lanes 7–10; uniformly labeled with α-[P32]UTP (5ppp RNA(U)*), lanes 13 to 16. Substrates incubated 90 min without protein (C90) are shown in lanes 4, 10 and 16; RNase T1 ladder (T1) in lanes 5 and 11; hydroxyl ladders (OH) in lanes 6 and 12. The cleavage products are indicated by arrows and their position is indicated in the sR47 schematic drawing. The boundaries of the cleavage products are given in brackets. (B) Kinetic analysis of RNA cleavage of 5′-end-monophosphate sR47 by Pab-aCPSF1 (6 µM) at 65°C for the indicated time. 5′-end-labeled sR47 RNA carrying a 5′ α-[P32] (5′p* RNA) is shown in lanes 1–4; uniformly-labeled sR47 (5′p RNA(U)*) in lanes 6–8 and (C90) and (OH) in lanes 4 and 5, respectively. (C) Kinetic analysis of RNA cleavage of 5′-end hydroxyl uniformly labeled sR47 (α-[P32] UTP, 5′OH RNA(U)*) by Pab-aCPSF1 (7 µM) at 65°C at the indicated times (lanes 1–3).
Figure 5.
In vitro activity of Pab-aCPSF1 on sRkB and sR47 variant RNA substrates. (A) Cleavage sites on the sRkB map are indicated by solid and dotted arrows for strong and weak, respectively. The cleavage sites are numbered from 1 to 9 and the 11 CA dinucleotides of sRkB RNA are highlighted in grey (Top). The kinetic analysis of cleavage of the 5′p*pp RNA, 5′ppp RNAp*Cp and 5′p RNAp*Cp sRkB substrates at 65°C is shown (Bottom). (B) Cleavage sites induced by Pab-aCPSF1 on the sR47, sR47MutCG and sR47Mut21U maps (Top). The CA dinucleotides are highlighted in grey and the cleavage sites are indicated by red arrows. The sR47MutCG and the sR47Mut21U mutants possess an extra CG dinucleotide (bold italic characters, forming base pairs with G20 and C21) and a U at position 21, respectively. The CG insertion impedes cleavage at C21 and reduced the exonucleolytic degradation pathway as shown by the kinetic analyses of uniformly labeled sR47 and sR47MutCG (5′p RNA(U)*). The replacement of the U by a C impedes cleavage at position 21 but does not affect the exonucleolytic degradation of 5′p sR47Mut21U (U)* (Bottom). See Figure 2 legend for symbols.
To test whether the phosphorylation state of the 5′-end of the RNA substrate affects Pab-aCPSF1 activity, we performed assays with 5′ monophosphorylated sR47 (Figure 4B). The degradation of 5′p RNA contrasts markedly with that of 5′ppp RNA owing to the production of GMP or UMP (Figure 4B). The 5′p*RNA generated radiolabeled GMP, which corresponds to the 5′ teminal base in the sR47 substrate, whereas the uniformly labeled 5′p RNA(U)* generated radiolabeled UMP (Figure 4B). Comparable results were observed with the sRkB RNA substrate (Figure 5A). The 3′-end-labeled 5′p RNA yielded radiolabeled p*Cp, but not the 3′-end-labeled 5′ppp RNA. These results strongly suggest that Pab-aCPSF1 has a 5′ monophosphate-dependent 5′–3′ exoribonucleolytic activity. This dependence is strict because neither 5′ppp (Figure 4A and Figure 5A) nor 5′hydroxyl (Figure 4C) transcripts can be degraded exonucleolytically. However, it should be noted that the distal products of endonucleolytic cleavage of the 5′ppp and 5′hydroxyl substrates do not appear to be degraded by the exoribonucleolytic activity as evidenced by the absence of UMP production. This suggests that products of endonucleolytic cleavage are somehow protected from exoribonucleolytic digestion. Interestingly, the exoribonucleolytic activity appears to be slowed or impeded by RNA secondary structure (compare the production of UMP* with the sR47, sR47MutCG and sR47 Mut21U substrates) (Figure 5B, lower panel). Finally, like other exoribonucleases from the β-CASP family [see ref. (24)], Pab-aCPSF1 can partially degrade 5′-end-labeled DNA oligonucleotides to mononucleotides (Supplementary Figure S3C). In conclusion, Pab-aCPSF1 has a dual activity: an endoribonuclease activity that preferentially cleaves at single-stranded CA dinucleotides, and exoribonuclase activity that is restricted to 5′-monophosphorylated RNA substrates.
Pab-aCPSF1 N-terminal and C-terminal extensions are involved in RNA binding and protein dimerization
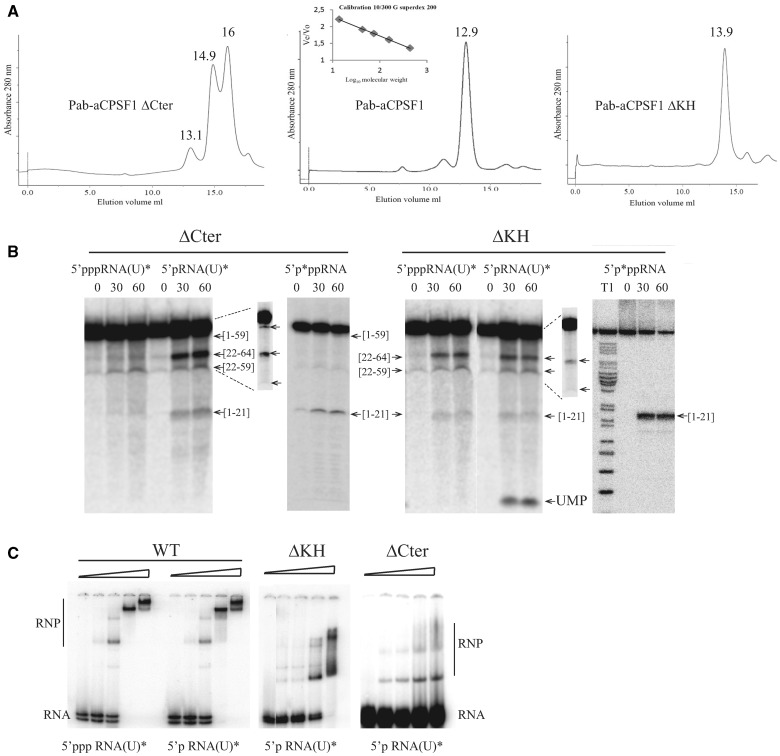
To investigate further the properties of Pab-aCPSF1, we produced a variant deleted for the last 12 residues of Pab-aCPSF1 (Pab-aCPSF1ΔCter) (Figure 2). In the crystal structures of M. mazei (Mm) and M. thermautotrophicus aCPSF1 (14,26), these residues form a network of hydrogen bonding interactions at the interface of the dimeric holoenzyme. Furthermore, the interacting residues correspond to a sequence motif that is conserved in the aCPSF1 family (Figure 2). Gel filtration shows that the ΔCter variant is mostly monomeric, whereas the wild type protein is dimeric (Figure 6A, left and middle panels), thus validating the role of the C-terminus in dimerization. Note that the Pab-ΔCter recombinant protein is highly sensitive to proteolysis in the region linking the N-terminal KH domain and the core β-CASP metallo-β-lactamase domains (Supplementary Figure S4). We assayed the activity of the ΔCter variant using the 5′p*pp RNA, 5′ppp RNA (U)* and 5′p RNA (U)* substrates. Our data show that the exonucleolytic activity of Pab-aCPSF1ΔCter is impaired as evidenced by the absence of UMP production with the 5′p RNA(U)* substrate (Figure 6B, left panel). Although endonucleolytic cleavage of 5′ppp RNA (U)* appears to be weak, cleavages at C21 and C59 are clearly detected with the 5′p*pp RNA and 5′p RNA(U)*.
Figure 2.
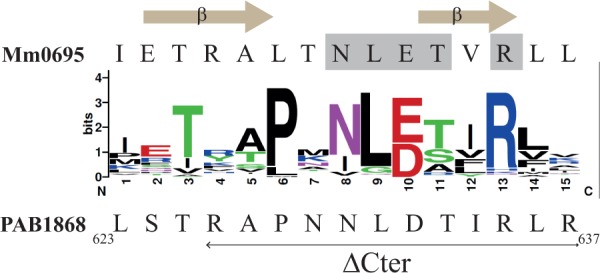
Sequence conservation of the last 16 residues of the 110 members of aCPSF1 is shown using Weblogo representation (http://weblogo.berkeley.edu). The amino acid C-terminal sequence of M. mazei Mm0695 and of P. abyssi PAB1868 (Pab-aCPSF1) are specified on the top and bottom, respectively. The residues establishing interacting hydrogen bonds in the protein–protein interface of the dimeric structure of Mm0695 (26) are highlighted in grey and the β-sheets formed by these residues are indicated. The last 12 residues that were deleted in aCPSF1ΔCter variant are indicated by a horizontal arrow under the P. abyssi sequence.
Figure 6.
Pab-aCPSF1ΔCter (ΔCter) and Pab-aCPSF1ΔKH (ΔKH) variants. (A) Size exclusion chromatography of Pab-aCPSF1 and its variants. The proteins were separated on Superdex 200 and the apparent molecular masses defined using the inset calibration curve. Gel filtration yielded a minor peak at 13.1 ml and major peaks at 14.9 and 16 ml for Pab-aCPSF1ΔCter corresponding to dimeric protein (148 kDa), monomers (55 kDa) and a proteolysis products (30 kDa) (Supplementary Figure S4), respectively. It should be noted that the monomer elutes at higher volume than expected (55 kDa instead of 72 kDa) (Supplementary Figure S4) (left). Gel filtration yielded a peak at 12.8 ml for Pab-aCPSF1 corresponding to a dimeric protein (159 kDa) (middle) Gel filtration yield a peak at 13.9 ml for Pab-aCPSF1ΔKH corresponding to dimeric protein (92 kDa) (right) (B) Kinetic analysis of RNA cleavage of 5′p*pp RNA, 5′ppp RNA(U)* and 5′p RNA(U)* substrates by protein variants (6 µM) at 65°C for the indicated times. Symbols are as in Figure 2. The products of the reaction were analysed on a 10% PAGE. Long migrations are shown in the inset (dotted lines) for 5′p RNA(U)* products. (C) Gel retardation assay with 10 fmol of 5′p RNA(U)* or of 5′ppp RNA(U)* incubated with increasing concentrations (0, 0.3, 0.6, 1.25 and 2.5 µM) of Pab-aCPSF1 (WT), ΔKH or ΔCter variants. RNA-protein complexes (RNP) are indicated.
To test the importance of the Pab-aCPSF1 N-terminus containing two KH domains (Figure 1), we produced a Pab-aCPSF1ΔKH variant missing the first 179 residues. Gel filtration shows that Pab-aCPSF1ΔKH is dimeric (Figure 6A, right panel). Thus, the N-terminal extension is not involved in dimerization. We assayed the activity of Pab-aCPSF1ΔKH using the sR47 substrate (Figure 6B). The pattern of digestion is comparable with wild type (Figure 4A) except that the 59 nt RNA corresponding to a cleavage at C59 was not produced. We conclude that the N-terminal KH domains are not necessary for catalytic activity, but are most likely involved in the recognition of certain specific sites. Because KH domains are predicted to bind nucleic acids (14,16), we examined the capacity of the ΔKH variant to bind sR47 RNA by electrophoretic mobility shift assay (EMSA) (Figure 6C). On incubation with increasing protein concentration, three major distinct RNP complexes were detected with wild type Pab-aCPSF1. sR47 was fully shifted at a protein concentration of about 1 µM. 5′-triphosphate and 5′-monophosphate RNA bound with similar affinity, suggesting that the nature of the 5′-end is not important for binding. As well, we analysed the RNP patterns of sR47MutCG and sR47Mut21U substrates, which are invalid for endonucleolytic cleavage at position 21. Preliminary data showed similar overall binding affinities and high molecular weight RNP complexes as observed in Figure 6C (data not shown). However, the intensity of each RNP band was somehow different from sR47 EMSA, which did not permit a clear conclusion. This opens the question of whether binding and endonucleolytic activity could be uncoupled in future studies. The affinity for the ΔKH variant is slightly lower and the higher molecular weight RNP complexes are less stable as evidenced by smearing in the gel (Figure 6C). We also analysed the ΔCter variant by EMSA. This variant is severely impaired in its capacity to bind RNA, suggesting that dimerization of the holoenzyme is important for RNA binding (Figure 6C). Altogether, these results show that the dimerization of Pab-aCPSF1 is important for exoribonuclease and RNA binding activity, whereas the KH domains participate in endoribonucleolytic cleavage at certain sites and are important for the stability of high molecular weight RNP complexes.
DISCUSSION
In this study, we systematically identified β-CASP proteins in Archaea and classified them according to sequence similarities to determine their phylogenetic relationships (Figure 1 and Supplementary Figure S1) and their taxonomic distribution (Supplementary Figure S2 and Figure 7). Among the seven archaeal β-CASP OGs that we identified: one is related to the bacterial RNase J (aRNase J), three are related to the eukaryal CPSF-73 (aCPSF1, aCPSF1b and aCPSF2). aRNase J, which is distributed exclusively in the Euryarchaeota, includes three members known to have 5′-end-dependent exonucleolytic activity (15,24,25) (Table 1). The aCPSF-like proteins are clearly divided into three clusters: aCPSF1 includes extremely well conserved members in all Archaea whose genome has been sequenced (this work); aCPSF2 groups more divergent members that are widespread in the Archaea and includes some that were previously misidentified as RNase J orthologs (15,25); aCPSF1b members are only present in the Methanococcales. aCPSF1b and aCPSF1 are closely related, but aCPSF1b lacks the N-terminal extension containing two KH domains. In summary, our phylogenetic analysis rectifies the misidentification of certain archaeal β-CASP proteins as aRNase J homologs (15,25) and clarifies their evolutionary origin.
Figure 7.
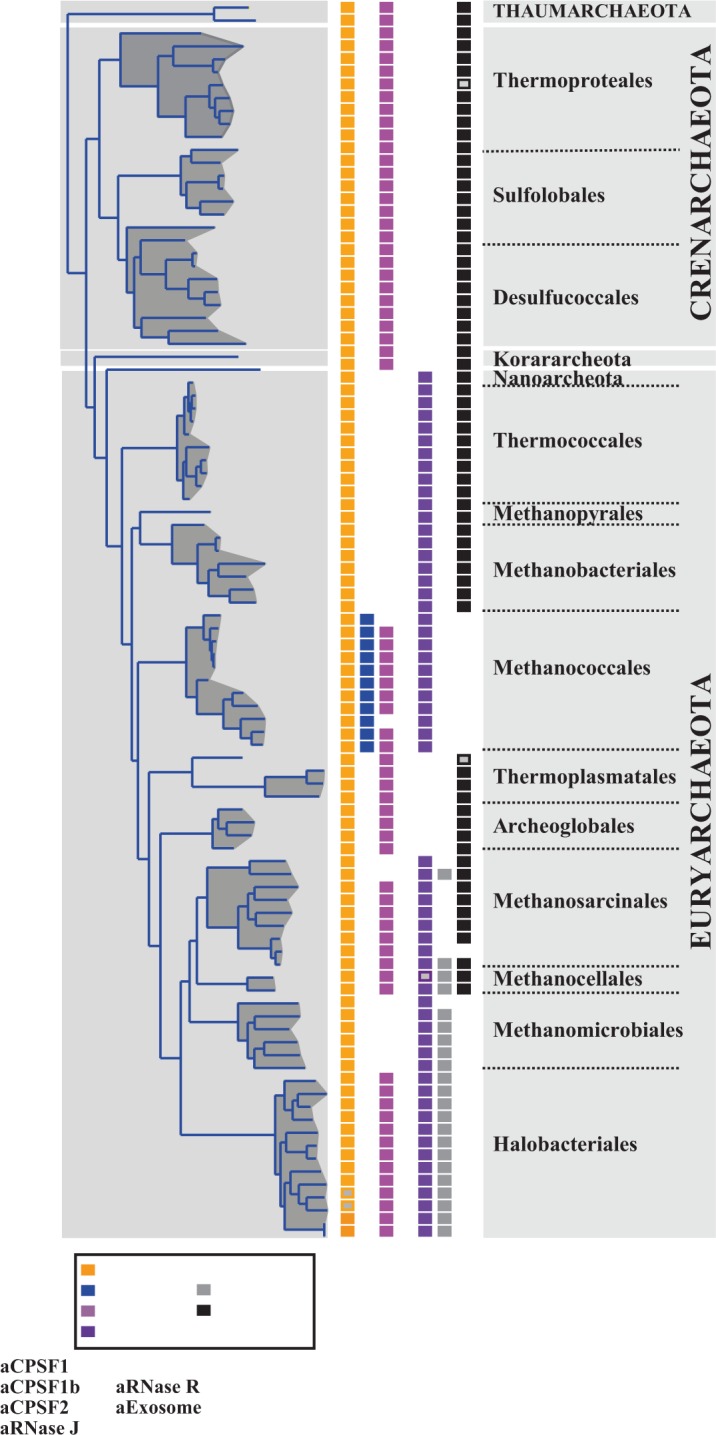
Distribution of the four archaeal β-CASP OGs, archaeal exosome and aRNase R on the archaeal lineage inferred from a set of concatenated conserved proteins (see ‘Materials and Methods’ section). Open rectangles indicate the existence of more than one member.
Here, based on biochemical studies, we report that Pab-aCPSF1 has both an endonucleolytic and 5′–3′ exonucleolytic activity. In addition, the endonucleolytic cleavage occurs in single-stranded RNA with a pronounced preference for CA dinucleotides. We reveal that the C-terminal homodimeric interface, initially identified in the crystal structures of M. mazei and M. thermautotrophicus members (14,26), is conserved amongst aCPSF1 homologs. Disruption of this interface in Pab-aCPSF1 results in monomeric enzyme that has endoribonuclease activity, but is deficient for exoribonuclease activity. In the same manner, protein interactions were shown to be required for full activity of eukaryotic CPSF-73, which forms a heterodimer with CPSF-100 (an inactive CPSF-73 paralog) (37). Deletion of the N-terminal KH domains of Pab-aCPSF1 abolishes endoribonuclease cleavage at certain specific sites and destabilizes high molecular weight RNPs, without affecting exoribonucleolytic activity and the dimeric state of the enzyme. Given the general prevalence of KH domains in proteins associated with transcriptional and translational regulation (PNPase, the exosome, NusA and ribosomal proteins) (38), it seems likely that they will have an important role in aCPSF1 specificity. Mj-aCPSF1 has been recently reported to have endonucleolytic but not exonucleolytic activity (15) similar to the activity of the Pab-aCPSF1ΔCter variant studied here. Despite this apparent inconsistency, we believe that most aCPSF1 members are likely to have endo- and exoribonucleolytic activity because this is a property of both CPSF-73 and Pab-aCPSF1 (10–12).
All archaeal β-CASP proteins characterized to date, except for Mj-aCPSF1 (15), display a 5′–3′ exoribonucleolytc activity that is dependent on the 5′ phosphorylation state of the substrate [(15,24,25), this work]. Previous biochemical work showed that the translation initiation factor a/eIF2 binds to and protects RNA with 5′-triphosphorylated ends from degradation in the Crenarcheaon S. solfataricus (25,39). This observation suggests parallels to the principal mechanisms of 5′–3′ RNA decay in Bacteria and Eukarya (1), in which Nudix hydrolases in Bacteria (40,41) and decapping enzymes in Eukarya (42) trigger mRNA degradation by producing 5′-monophosphate ends. Nevertheless, comparable enzymes remain to be discovered in the Archaea. In conclusion, the nature of the substrate 5′-end (tri- versus mono-phosphorylated) emerges as a major determinant in the activity of β-CASP ribonucleases.
To highlight the prospective archaeal RNA degradation machinery, we have summarized the distribution of the archaeal β-CASP ribonucleases, together with the archaeal exosome and aRNase R, which have 3′–5′ exonucleolytic activity (1,43,44) (Figure 7). It should be noted that the Crenarchaeaota, Thaumarcheota and Korarchaeota, which were recently described to be part of the ‘TACK’ superphylum and speculated to be at the origin of Eukarya (45), all have the same combination of RNA degrading enzymes (aCPSF1, aCPSF2 and archaeal exosome). In contrast, distribution in the Euryarcheota is heterogeneous apart from the ubiquitous aCPSF1 homolog. As previously reported (13), the exosome is missing from Methanococcales, Methanomicrobiales and Halobacteriales, illustrating the divergence in the Euryarcheaota. In the Halobacteriales, the emergence of an RNase R-like protein is believed to compensate this deficiency (46) (Figure 7). In the Methanococcales, the absence of the exosome correlates with the presence of aCPSF1b homologs, suggesting a possible functional link between the exosome and β-CASP proteins.
In conclusion, the enzymatic properties of aCPSF1 members are comparable with eukaryal CPSF-73 including 5′-end-dependent exoribonuclease activity and an endoribonuclease activity with a preference for single-stranded CA dinucleotides. The strict conservation of these orthologs throughout the Archaea suggests a fundamental role in RNA metabolism. An analogy can be made with the eukaryal CPSF-73, which is a component of the machinery required for mRNA 3′-end maturation and termination of RNA polymerase II transcription (9,47). Our results suggest that a CPSF-like β-CASP protein was present in the last common ancestor of Archaea and Eukarya. We speculate that the highly conserved aCPSF1 might be an active component of an essential RNA-processing complex involved in mRNA degradation and/or 3′-end processing and transcription termination. By analogy to CPSF-73, which is part of a multicomponent RNP, clues to the function of the archaeal homolog might come from future studies aimed at identifying archaeal complexes containing aCPSF1 homologs.
SUPPLEMENTARY DATA
Supplementary Data are available on NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Centre National de la Recherche Scientifique (CNRS) with additional funding from the Agence Nationale de la Recherche (ANR) [BLAN08-1_329396]; from the Université de Toulouse (UPS) (AO1_2011). Funding for open access charge: Agence Nationale de la Recherche (ANR) [BLAN08-1_329396].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to members of the Carpousis group for helpful discussions, G. Fichant, M. Bouvier, L. Minvielle-Sébastia and D. Flament for critical reading of the manuscript and F. Anglès and R. Simon for technical assistance. Author contributions: Y.Q., A.J.C. and B.C.O. designed research; D.K.P., D.R, P.L.G., Y.Q. and B.C.O. performed research and analysed data; Y.Q., A.J.C. and B.C.O. wrote the paper.
REFERENCES
- 1.Evguenieva-Hackenberg E, Klug G. RNA degradation in Archaea and Gram-negative bacteria different from Escherichia coli. Prog. Mol. Biol. Transl. Sci. 2009;85:275–317. doi: 10.1016/S0079-6603(08)00807-6. [DOI] [PubMed] [Google Scholar]
- 2.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 3.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′-end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominski Z. The hunt for the 3′-endonuclease. Wiley Interdiscip. Rev. RNA. 2010;1:325–340. doi: 10.1002/wrna.33. [DOI] [PubMed] [Google Scholar]
- 8.Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev. RNA. 2011;2:321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell Biol. 2009;29:31–42. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva AP, Chechik M, Byrne RT, Waterman DG, Ng CL, Dodson EJ, Koonin EV, Antson AA, Smits C. Structure and activity of a novel archaeal beta-CASP protein with N-terminal KH domains. Structure. 2011;19:622–632. doi: 10.1016/j.str.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy S, Portnoy V, Admon J, Schuster G. Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biol. 2011;8 doi: 10.4161/rna.8.6.16604. [DOI] [PubMed] [Google Scholar]
- 16.Nishida Y, Ishikawa H, Baba S, Nakagawa N, Kuramitsu S, Masui R. Crystal structure of an archaeal cleavage and polyadenylation specificity factor subunit from Pyrococcus horikoshii. Proteins. 2010;78:2395–2398. doi: 10.1002/prot.22748. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L. An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol. 1999;1:69–91. [PubMed] [Google Scholar]
- 18.Condon C, Gilet L. Nucelic Acids and Molecular Biology. Vol. 26. Heidelberg, Berlin: Springer-verlag; 2011. pp. 245–267. [Google Scholar]
- 19.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 21.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 22.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 23.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Maturation of the 5′-end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 24.Clouet-d'Orval B, Rinaldi D, Quentin Y, Carpousis AJ. Euryarchaeal beta-CASP proteins with homology to bacterial RNase J Have 5′- to 3′-exoribonuclease activity. J. Biol. Chem. 2010;285:17574–17583. doi: 10.1074/jbc.M109.095117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasenohrl D, Konrat R, Blasi U. Identification of an RNase J ortholog in Sulfolobus solfataricus: implications for 5′-to-3′ directional decay and 5′-end protection of mRNA in Crenarchaeota. RNA. 2011;17:99–107. doi: 10.1261/rna.2418211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mir-Montazeri B, Ammelburg M, Forouzan D, Lupas AN, Hartmann MD. Crystal structure of a dimeric archaeal cleavage and polyadenylation specificity factor. J. Struct. Biol. 2011;173:191–195. doi: 10.1016/j.jsb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 27.van Dongen S, Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol. Biol. 2012;804:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- 28.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 31.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 33.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolivos S, Carpousis AJ, Clouet-d'Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phok K, Moisan A, Rinaldi D, Brucato N, Carpousis AJ, Gaspin C, Clouet-d'Orval B. Identification of CRISPR and riboswitch related RNAs among novel non-coding RNAs of the euryarchaeon Pyrococcus abyssi. BMC Genomics. 2011;12:312. doi: 10.1186/1471-2164-12-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E. coli. RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell Biol. 2005;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 39.Hasenohrl D, Lombo T, Kaberdin V, Londei P, Blasi U. Translation initiation factor a/eIF2(-gamma) counteracts 5′ to 3′ mRNA decay in the archaeon Sulfolobus solfataricus. Proc. Natl. Acad. Sci. USA. 2008;105:2146–2150. doi: 10.1073/pnas.0708894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, Condon C, Belasco JG. An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol. Cell. 2011;43:940–949. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 42.Song MG, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol. Cell. 2010;40:423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 45.Guy L, Ettema TJ. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Portnoy V, Schuster G. RNA polyadenylation and degradation in different Archaea; roles of the exosome and RNase R. Nucleic Acids Res. 2006;34:5923–5931. doi: 10.1093/nar/gkl763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proudfoot N. New perspectives on connecting messenger RNA 3′-end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Newman JA, Hewitt L, Rodrigues C, Solovyova A, Harwood CR, Lewis RJ. Unusual, dual endo- and exonuclease activity in the degradosome explained by crystal structure analysis of RNase J1. Structure. 2011;19:1241–1251. doi: 10.1016/j.str.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 49.Dorleans A, Li de la Sierra-Gallay I, Piton J, Zig L, Gilet L, Putzer H, Condon C. Molecular basis for the recognition and cleavage of RNA by the bifunctional 5′-3′ exo/endoribonuclease RNase J. Structure. 2011;19:1252–1261. doi: 10.1016/j.str.2011.06.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.