Abstract
DNA concentration has been recently suggested to be the reason why different arrangements are revealed for K+-stabilized human telomere quadruplexes by experimental methods requiring DNA concentrations differing by orders of magnitude. As Raman spectroscopy can be applied to DNA samples ranging from those accessible by absorption and CD spectroscopies up to extremely concentrated solutions, gels and even crystals; it has been used here to clarify polymorphism of a core human telomeric sequence G3(TTAG3)3 in the presence of K+ and Na+ ions throughout wide range of DNA concentrations. We demonstrate that the K+-structure of G3(TTAG3)3 at low DNA concentration is close to the antiparallel fold of Na+-stabilized quadruplex. On the increase of G3(TTAG3)3 concentration, a gradual transition from antiparallel to intramolecular parallel arrangement was observed, but only for thermodynamically equilibrated K+-stabilized samples. The transition is synergically supported by increased K+ concentration. However, even for extremely high G3(TTAG3)3 and K+ concentrations, an intramolecular antiparallel quadruplex is spontaneously formed from desalted non-quadruplex single-strand after addition of K+ ions. Thermal destabilization or long dwell time are necessary to induce interquadruplex transition. On the contrary, Na+-stabilized G3(TTAG3)3 retains its antiparallel folding regardless of the extremely high DNA and/or Na+ concentrations, thermal destabilization or annealing.
INTRODUCTION
Linear eukaryotic chromosomes are protected at both ends by telomeres (1). Telomeric DNA in humans consists of the (TTAGGG)n/(CCCTAA)n repeats terminating on the 3′-end of the molecule in a single-stranded G-rich overhang (2), which folds into a G-quadruplex under suitable conditions (3). Telomeres play a key role in maintaining chromosomal integrity, control of DNA replication and protection against chromosome elongation by telomerase (4). Unregulated telomerase activity allows cancer cells to become immortal (5). As formation of G-quadruplexes at telomeres perturbs telomerase function (6), the quadruplex-stabilizing agents can suppress the proliferation of tumour cells (7,8). Searching for these agents is of great importance for cancer treatment in human medicine (9). For a better efficiency, structural properties of telomeric quadruplexes under the physiologically relevant conditions, that is, at moderate concentration of K+ ions and high local concentration of DNA, must be known properly.
Despite numerous studies, the structure adopted by human telomeric repeats under the biologically relevant conditions remains unclear. The first relevant structure solved at atomic resolution was reported by Wang and Patel in 1993 (10). Based on nuclear magnetic resonance (NMR) study and molecular dynamics simulations, the telomeric fragment AG3(TTAG3)3 was shown to adopt an antiparallel basket-type structure in the presence of Na+ (10). A year later, the same topology was evidenced by CD spectroscopy and chemical probing for quadruplexes formed by related telomeric sequences, G3(TTAG3)3 and (TTAG3)4, in both Na+ and K+ solutions (11). On the contrary, radically different intramolecular parallel quadruplex with three propeller-shaped, double-chain-reversal d(TTA)-loops was reported for the crystalline K+-form of AG3(TTAG3)3 (12). Platinum cross-linking studies revealed the formation of the basket-type antiparallel quadruplex of AG3(TTAG3)3 in both Na+ and K+ solution (13), whereas 125I-radioprobing confirmed the basket-type of AG3(TTAG3)3 in Na+ solutions, and the antiparallel chair-type in the presence of K+ (14).
Several studies also proposed that parallel and antiparallel quadruplexes could coexist in K+ solutions, for example (15). On the other hand, parallel-strand alignment derived from crystallography (12) was excluded as the major biologically relevant conformation of the human telomere quadruplex in K+ solution (16). In 2005, another type of quadruplex arrangement, so called ‘(3 + 1) structure’ containing three parallel and one antiparallel strands, was suggested for K+-stabilized G3(TTAG3)4 containing one redundant TTAG3 repeat, to explain peculiarities of its CD spectra (17). A year later, structure of the intramolecular (3 + 1) quadruplex scaffold was solved by NMR in K+ solutions for TTG3(TTAG3)3A, TAG3(TTAG3)3 (18) and A3G3(TTAG3)3AA (19); however, attempts to elucidate the K+-structures of some related sequences bearing no or few residues attached to the minimal quadruplex-forming sequence G3(TTAG3)3 failed because of their conformational heterogeneity. Another variant of the (3 + 1) topology with different order of the loop arrangements was discerned by NMR for TAG3(TTAG3)3TT stabilized by K+ ions (20). Furthermore, depending on the nature and number of the extra residues flanking the minimal four-repeat G3(TTAG3)3 unit, and the method applied, various quadruplex arrangements under the physiologically relevant K+ conditions were reported, antiparallel (17,21), mixture of hybrid (3 + 1) and chair-type antiparallel (22), mixture of basket- and chair-type antiparallel (23) and others (24–26). There was even reported CD evidence of parallel arrangement, although in aqueous solution at high K+ concentration (27). Parallel topology was later observed in the presence of organic solvents simulating crowding conditions present in cells, that is, polyethylene glycol (28,29) or ethanol (30). Recently, a new quadruplex type has been observed by NMR for G3(TTAG3)3T stabilized by K+ ions (31). This stable structure ascribed to a basket-type quadruplex contains, however, only two G-tetrad layers capped with several layers of stacked guanines and adenines on both sides of the tetrad core.
From a great deal of experimental data available to date, it follows that surprisingly high polymorphism of the human telomere quadruplex could be a rather complex resultant of the exact sequence studied, DNA concentration, stabilizing cation type and concentration, dehydrating or molecular crowding conditions, as well as different protocols of sample preparation. Notably, DNA concentration differing by orders of magnitude for various experimental methods can be responsible for contradictory structures observed under otherwise comparable conditions. For instance, recent CD study has demonstrated that K+-stabilized quadruplexes of several human telomeric fragments undergo structural transition on the increase of oligonucleotide concentration, even at the physiologically relevant K+ concentrations (30). However, more detailed CD and absorption studies at high DNA concentrations are hindered by high optical densities of the samples.
In the present work, Raman spectroscopy is reintroduced as a practical and well-established, albeit yet underappreciated, method suitable for structural studies of G-quadruplexes. Although conventional, notably non-resonant Raman spectroscopy has proved useful for investigation of nucleic acids conformations [for overview see (32,33)], relatively few articles relate specifically to G-quadruplexes (e.g. 34–41). One of advantages of Raman spectroscopy consists in its applicability to DNA samples starting from the concentrations comfortably accessible by absorption and CD spectroscopies up to highly concentrated solutions inducing molecular crowding without need of cosolutes, gels and crystals studied by X-ray diffraction, with the possibility to acquire informational rich Raman melting profiles at high quadruplex concentrations, which are still missing. Raman spectroscopy can thus bridge, in a methodologically consistent way, an information gap between experimental methods restricted to low and high DNA concentrations solely. It can contribute to reconciliation of different K+-quadruplex structures reported for aqueous solutions and clarify arrangement of human telomeric sequences under the physiologically relevant conditions.
MATERIALS AND METHODS
Samples
High-performance liquid chromatography-purified and lyophilized/highly concentrated oligonucleotides were purchased from VBC-Biotech (Vienna, Austria) and Generi Biotech (Hradec Králové, Czech Republic) in several consecutive batches as ammonium or methylammonium salts. Chemicals of analytical grade (Sigma-Aldrich) and deionized water (18 MΩ, Elga) were used for buffers. To adjust precisely type and concentration of the quadruplex-stabilizing cations in the samples differing in DNA concentration, the following procedure was used. Stock solutions of oligonucleotides (∼200–500 mM in nucleosides) were prepared by dissolving lyophilized oligomer into deionized water. To replace/remove former countercations and small molecules initially present in the lyophilisate, appropriate amounts of the stock solution were repeatedly (typically 3–5 times) washed out by a phosphate buffer (PBS, 30–150 mM according to DNA concentration, pH 6.8) containing Na+ or K+ ions at desired concentrations (100–500 mM), using centrifugal filter devices (Amicon Ultra 3K, Millipore). Final concentration of K+ or Na+ in PBS was adjusted by addition of KCl or NaCl. The same centrifugal washing but by deionized water was used for preparation of the samples without alkali cations. Non-quadruplex arrangement of the fully desalted samples and the quadruplex formation after addition of alkali cations was checked out by ultraviolet (UV) absorption (42). The DNA concentrations were adjusted by controlling the starting and final centrifuged volumes, as well as by subsequent dilution by an appropriate solvent. In addition to total and controllable solvent exchange and precise adjustment of the DNA concentration, repeated centrifugal filtration helped to remove trace contaminants causing fluorescence background in Raman spectra. The exact oligonucleotides concentration was determined by absorbance measurements of appropriately diluted samples at 95°C by using a Lambda 12 UV/VIS (Perkin-Elmer) or Unicam 5626 UV/VIS spectrophotometers and molar extinction coefficients calculated according to (43). To increase accuracy of the concentration determination of extremely concentrated DNA samples, especially important for DNA concentration series, relative concentrations were determined concurrently from Raman intensities normalized with respect to OH-stretching band of water molecules (44). If not stated explicitly otherwise, DNA concentrations are related to nucleosides throughout the present article. Quadruplex structures thermodynamically equilibrated in relevant solutions were prepared by a 15 min thermal denaturation at 95°C and subsequent slow annealing (∼5 h) to room temperature. Heated but thermodynamically non-equilibrated structures were prepared by a fast cooling (from 95 to 20°C within <1 min), using expanding stream of compressed dry air. Moreover, structural changes in the course of thermal denaturation and annealing were monitored by Raman spectroscopy as a function of actual temperature in a temperature-controlled quartz microcell.
Raman measurements
Raman spectra were excited with the 532 nm line of a continuous-wave solid-state Nd:YVO4 laser (Verdi 2, Coherent) using the radiation power at the sample ranging from 100–400 mW (according to the sample concentration). The spectra were collected in the 90° scattering geometry on a multi-channel Raman spectrograph (Jobin Yvon–Spex 270 M) equipped with a holographic notch-plus filter (Kaiser) to reject Rayleigh scattering and a liquid nitrogen-cooled CCD detector (Princeton Instruments). Raman measurements were carried out in a temperature-controlled, hermetically sealed quartz microcell (5 μl sample volume) at somewhat reduced temperature [5–10°C, in comparison with room temperature used for CD and polyacrylamide gel electrophoresis (PAGE) experiments] to eliminate potential sources of artifacts specific for Raman scattering; however, such a slight temperature difference was proven to have no apparent effect on quadruplex structures and structural conclusions. Thermal denaturation/annealing experiments were conducted in the range of 2–96°C, with an accuracy of a temperature control within ±0.5°C. If not stated otherwise, the sample was equilibrated at the desired temperature for ∼10 min before recording the spectrum. The wavenumber scales of Raman spectra were precisely calibrated using the emission spectra of a neon glow lamp taken before and after each Raman measurement. The estimated precision of the calibration procedure was better than 0.1 cm−1. The Raman contribution from the solvent was carefully subtracted, and the spectra were corrected for their non-Raman background by using advanced methods of factor analysis (44). Spectra were normalized to the peak height of the 1093 cm−1 band associated with the PO2− symmetric stretching mode, reported previously to be largely invariant to the melting of DNA duplexes (45). If appropriate, for example, for quantitative comparisons of the oligonucleotides differing in the length or in the number of specific nucleosides, normalized spectra were furthermore renormalized according to the parameter in question.
CD measurements
CD measurements were done in a Jobin-Yvon CD6 dichrograph (Longjumeau, France) in 1–0.001 cm path-length quartz cells (Hellma Analytics, Germany) placed in a temperature-controlled holder. The scan rate was 0.5 nm/s. CD signals are expressed as the difference in the molar absorption Δε of the right- and left-handed circularly polarized light. The molarities are related to nucleosides.
PAGE
PAGE was performed in a temperature-controlled electrophoretic apparatus (SE-600; Hoefer Scientific, USA). Gel concentration was 16% (29:1 monomer to bis ratio, Applichem, Germany). About 2 μg of DNA was loaded into each lane of a 14 × 16 × 0.1 cm gel. Samples were electrophoresed at 20°C for 19 h at 30 V (∼2 Vcm−1). Gels were stained with Stains All (Sigma-Aldrich) after electrophoresis and scanned using a Personal Densitometer SI, model 375-A (Molecular Dynamics, USA).
RESULTS AND DISCUSSION
Equilibrated transition of annealed K+-stabilized G3(TTAG3)3 depends on DNA concentration
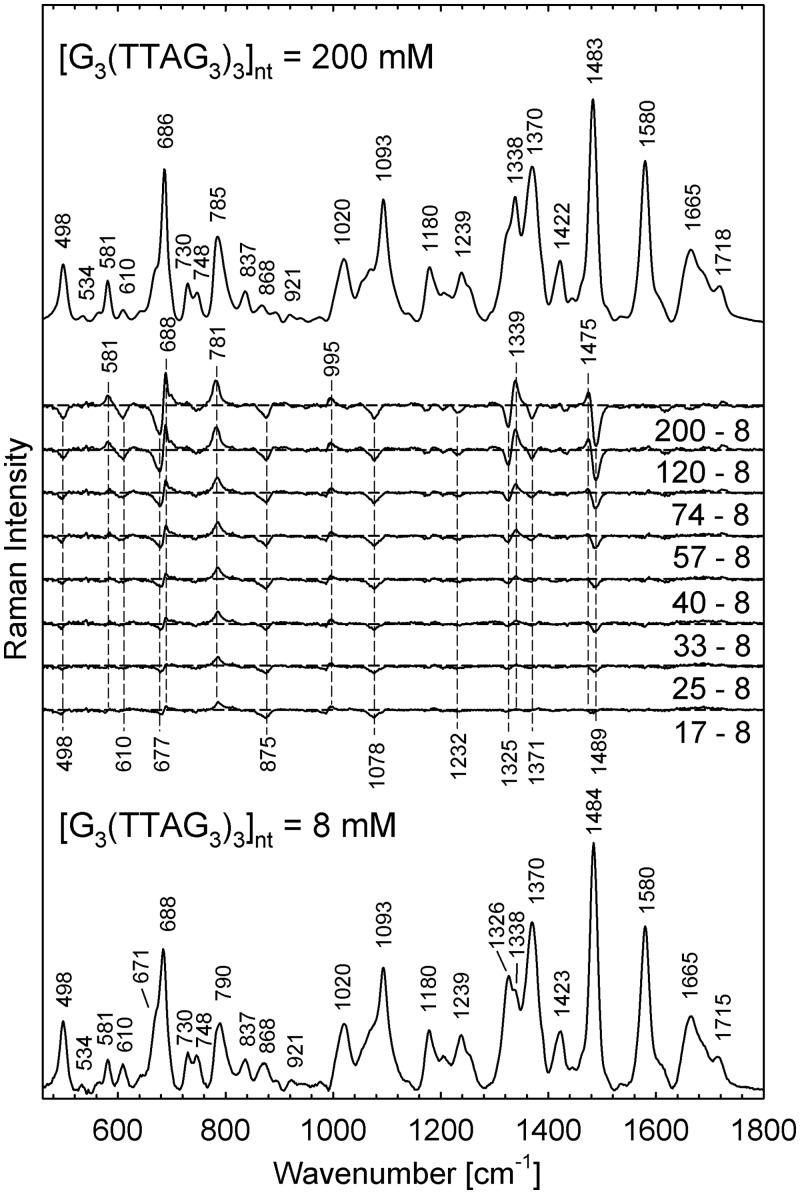
In the recent CD study (30), it was suggested that the antiparallel-to-parallel rearrangement of human telomeric sequences can be accomplished at physiologically relevant K+ concentrations simply by increasing the concentration of DNA, without addition of organic cosolutes to simulate molecular crowding conditions. To provide methodologically independent evidence of the self-crowding effect throughout a wider range of DNA concentration, Raman spectra of thermodynamically equilibrated (slowly annealed) solutions of G3(TTAG3)3 at DNA concentrations ranging from 8 to 240 mM and stabilized by 200–250 mM K+ were acquired and compared with those prepared in a similar way in Na+ buffers. As evident from differential spectra shown in Figure 1, initial quadruplex arrangement adopted by 8 mM G3(TTAG3)3 at moderate concentration of 200 mM K+ undergoes gradual structural changes on increase of DNA concentration. No similar spectral changes were observed for G3(TTAG3)3 in Na+ solutions (Supplementary Figure S3, top).
Figure 1.
Raman spectra of G3(TTAG3)3 in 200 mM K+ (30 mM of PBS, pH 6.8, t = 5°C) at the nucleoside concentrations of 8 mM (bottom trace) and 200 mM (top trace). Intermediate traces show the differences between the spectra at indicated concentration and that of the lowest one.
Differential Raman features of K+-G3(TTAG3)3 depicted in Figure 1 are mostly consistent with those reported on structural transition of Oxytricha telomeric sequence (T4G4)4 induced by increased concentration of Na+ and K+, and shown to be indicative for interquadruplex switching from intramolecular antiparallel fold to four-stranded parallel structure (35). As, to the best of our knowledge, human telomeric sequences have not been studied yet by conventional non-resonant Raman spectroscopy, assignment of Raman bands of G3(TTAG3)3 and structural interpretation applied hereinafter is based on Raman studies of interquadruplex switching of Oxytricha telomeric sequences controlled by the type, concentration and molar ratio of Na+ and K+ cations (34–39), on relevant Raman studies of various non-quadruplex DNAs (32,33,45–51), as well as on Raman spectra of model oligonucleotides (dG)15, (dT)15 and (dA)15 (Supplementary Figures S1 and S2). Raman markers used in the present work are listed and thoroughly discussed in the (Supplementary Table S1).
According to Raman markers indicative for C2-endo/syn (671 and 1326 cm−1) and C2-endo/anti (686 and 1338 cm−1) conformers of dG (32,34–36,39), structural transition of K+-G3(TTAG3)3 comprises gradual increase in the population of C2-endo/anti-dG at the expense of C2-endo/syn-dG (for detailed discussion see Supplementary Data), as can be documented by relative intensity increase of the Raman bands at 686 and 1338 cm−1, accompanied by intensity decrease of the shoulder at 671 cm−1 and the 1326 cm−1 band (Figure 1 and Supplementary Figure S3, bottom). Furthermore, intensity decrease of relatively weak but important Raman marker of C2-endo/syn-dT at 611 ± 2 cm−1 (32,36,39), sensitive to geometry of d(TTA)-loops (for detailed discussion see Supplementary Data), is consistent with quadruplex rearrangement from the fold incorporating side/diagonal loops to the quadruplex structure having loops organized in other geometry.
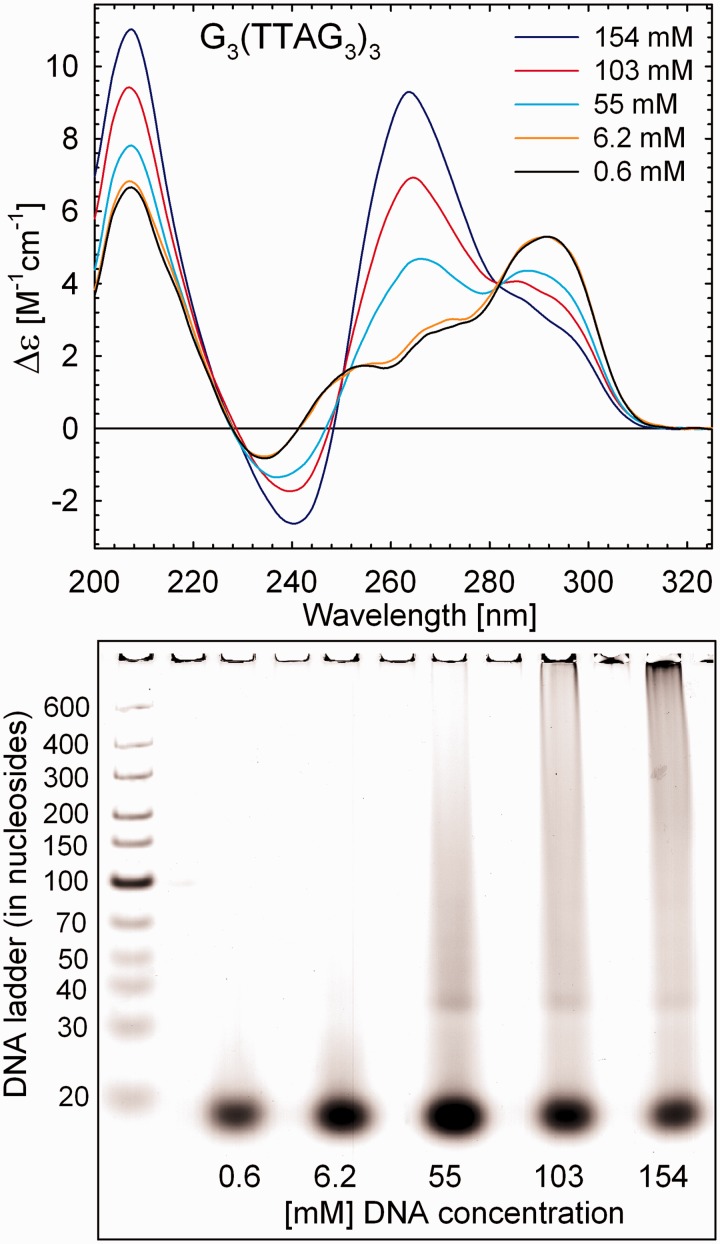
Characteristic changes in other Raman bands sensitive to syn/anti-dG ratio (∼581 ± 1 cm−1), structurally informative markers of hydrogen bonding within G-tetrads (∼1483 ± 2 cm−1), arrangement markers of d(TTA)-loops (∼499 ± 2 and ∼1371 ± 1 cm−1) and markers sensitive to sugar-phosphate backbone (∼788 ± 2 cm−1) constitute differential pattern that can serve as Raman fingerprint of interquadruplex transition. Conformational switching concluded from Raman spectra corroborates structural transition inferred from the corresponding CD spectra indicating gradual changes from predominantly antiparallel (6.2 mM) to largely hybrid (3 + 1) (55 mM) and finally to predominantly parallel folding (30) at DNA concentration of 154 mM still accessible to CD measurements (Figure 2).
Figure 2.
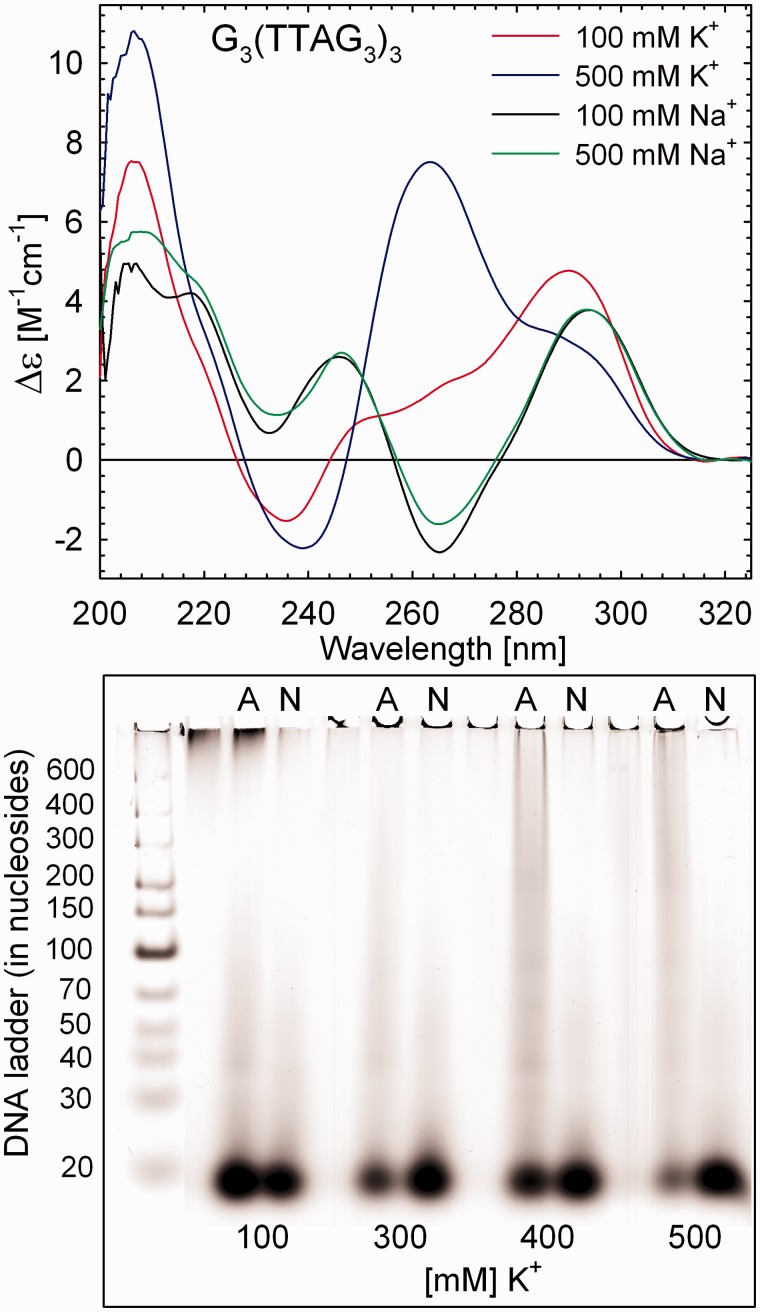
Dependence of CD spectra of G3(TTAG3)3 on DNA concentration at 23°C. Each sample was slowly annealed after dilution to the final DNA concentration. Concentration of K+ in all samples was 300 mM. Bottom: native PAGE of K+-stabilized G3(TTAG3)3 at indicated DNA concentrations. The PAGE was performed at 20°C in 30 mM of PBS containing 240 mM of K+, pH 6.7.
Linear character of interquadruplex transition indicates conformational heterogeneity
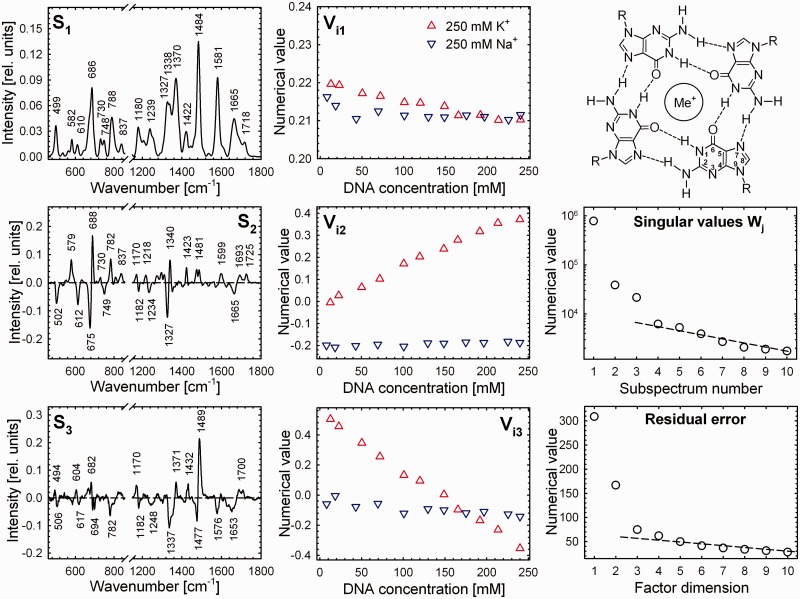
To better understand progression of the changes induced by DNA concentration and to depict different impact of the K+ and Na+ ions, Raman spectra of DNA concentration series carefully prepared according to identical protocols from the same stock solution have been analysed together by a factor analysis (44). Using a singular value decomposition (SVD) approach (44), spectral variability within the normalized Raman dataset under consideration (11 and 11 Raman spectra of G3(TTAG3)3 at the nucleotide concentrations 8–240 mM, stabilized by slightly higher concentration of 250 mM Na+ and K+ to favour parallel form, respectively) was decomposed into a set of orthonormal abstract factors (subspectra Sj), their statistical weights (singular values Wj) and coefficients Vij representing relative contributions of Sj to the i-th experimental spectrum from the dataset (Figure 3). Despite somewhat abstract character of the subspectra and less common mode of presentation, practically constant values of the coefficients Vi1–Vi3 for Na+-samples throughout full DNA concentration range tested here evidently means that their normalized Raman spectra are virtually identical. On the other hand, surprisingly linear dependency of all three coefficients Vi1–Vi3 on DNA concentration for K+-samples suggests gradual structural transition (instead of sigmoidal one expected for conformational transition between two or three distinct forms), probably not completed even at the highest DNA concentration of 240 mM studied in the series. Linear progression of the spectral changes in the range of DNA concentrations usual for NMR studies is consistent with conformational heterogeneity and coexistence of multiple quadruplex structures (18,19), the reason why attempts to elucidate K+-structure of G3(TTAG3)3 [or some related sequences bearing just few flanking nucleotides, e.g. AG3(TTAG3)3] by NMR spectroscopy failed.
Figure 3.
Dependence of normalized Raman spectra of G3(TTAG3)3 on DNA concentration in the presence of 250 mM of K+ or Na+ (150 mM of PBS, pH 6.8, 5°C). Spectral differences between K+- and Na+-stabilized quadruplex structures and changes induced by increasing DNA concentration are expressed as abstract orthonormal factors obtained by SVD analysis (44). For better insight to real spectral differences and their extent, refer to Figure 4 and Supplementary Figure S3. Right top corner: scheme of a G-tetrad.
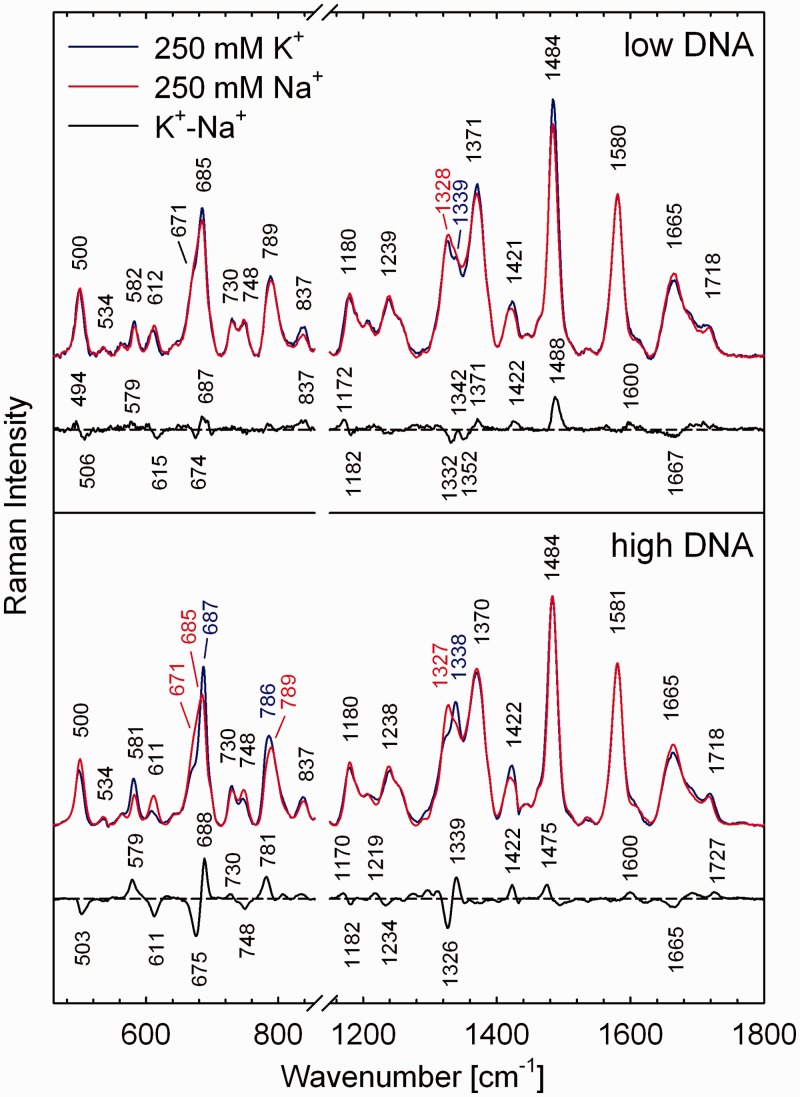
Furthermore, disclosure of three subspectra (according to singular values and residual errors shown in Figure 3) necessary for an adequate description of the spectral variability within full Raman dataset, corroborates persistence of the spectral differences between the Na+- and K+-structures beyond to the lowest DNA concentrations used in this experiment (∼8 mM). Corresponding orthogonal differences extracted by the SVD from Raman dataset are expressed predominantly by a subspectrum S3, as can be documented by substantial match with differential curve shown in Figure 4 (up), where the low-DNA spectra in both alkali cations are compared directly for better illustration. On the other hand, subspectrum S2 can be related primarily to spectral differences between the high- and low-DNA structures of K+-stabilized G3(TTAG3)3, as evident from apparent similarity with high-DNA differential curve between the K+ and Na+ structures shown in Figure 4 (bottom).
Figure 4.
Detail comparison of the spectral differences between K+- and Na+-stabilized G3(TTAG3)3 quadruplexes at low (8 mM, top) and high (240 mM, bottom) DNA concentration. Normalized Raman spectra correspond to lower and upper concentration limits of the series analysed by SVD in Figure 3.
Although the Raman spectra of Na+- and K+-stabilized G3(TTAG3)3 at the lowest DNA concentration exhibit some spectral differences (Figure 4, top), these do not concern the crucial Raman markers of the syn/anti-dG conformers discussed earlier in the text. Only the minute flip of relative intensities between 674 and 687 cm−1 bands of dG might be interpreted as a sign of slightly higher population of C2-endo/anti-dG in the K+ form. However, the changes of another syn/anti-dG markers in the 1326/1338 cm−1 region (i.e. appearance of 1332/1352 cm−1 differential features) and opposite sign of difference at 1371 cm−1 (Figure 4) are more complex and do not sustain changes in the syn/anti dG distribution or rearrangement of dT-loops expected for substantially different geometries. Furthermore, structurally sensitive band of symmetrical phosphodiester stretching mode at 789 cm−1 exhibits similar intensity and shape in both Na+- and K+-forms at low DNA concentration (Figure 4, top), in contrast to substantial difference accompanying antiparallel-to-parallel transition at high-DNA content (Figure 4, bottom). Finally, a positive differential feature at 1488 cm−1, the most distinct difference between Raman spectra of K+- and Na+-stabilized low-DNA folds of G3(TTAG3)3 is inconsistent with antiparallel-to-parallel transition, for which a negative feature at the same position is expected (Figure 1). It can be concluded that our Raman study provides methodologically independent argument in favour of prevailing antiparallel topology of the K+-G3(TTAG3)3 at low DNA concentration, even though corresponding CD spectrum is dissimilar to that of Na+-stabilized antiparallel structure (Figure 6). The CD spectrum of low-DNA K+-G3(TTAG3)3 has been previously interpreted as a sign of hybrid (3 + 1) form of K+-G3(TTAG3)3 (22,52), identifying its structure with the mixed (3 + 1) structures of TTG3(TTAG3)3A, TAG3(TTAG3)3 (18), A3G3(TTAG3)3AA (19) and TAG3(TTAG3)3TT (20) in K+ solutions resolved by NMR; however, at DNA concentrations by one or two orders higher that those studied by CD.
Figure 6.
Top: CD spectra of 50 mM G3(TTAG3)3 in the presence of 100 and 500 mM of Na+ or K+. Positive bands at ∼245 and ∼293 nm and negative band at ∼265 nm affirm antiparallel foldback of the Na+-stabilized G3(TTAG3)3, regardless of the Na+ concentration. Even though without negative band at ∼265 nm, the CD spectrum of 100 mM K+-stabilized G3(TTAG3)3 was recently proven to be indicative for essentially the same antiparallel topology as adopted by Na+-stabilized quadruplex (30), contradicting the previous interpretation attributing CD spectra of this shape to hybrid (3 + 1) form (18,19,22). Transition to the parallel structure via hybrid (3 + 1) forms is accompanied by appearance of a strong positive band at ∼263 nm, decrease of the positive band at ∼290 nm and the presence of negative band at ∼240 nm (30), the CD features visible in the presence of 500 mM K+. Bottom: native PAGE of 50 mM G3(TTAG3)3 in 100–500 mM of K+ before heating (N) and after slow annealing (A). The PAGE was performed at 20°C in 30 mM PBS containing 300 mM K+ in total, pH 6.7.
Interquadruplex transition of K+-G3(TTAG3)3 is more effective at higher concentrations of K+
Similar structural effect as induced by DNA concentration at a fixed concentration of K+ can be seen on the increase of K+ concentration at a fixed concentration of G3(TTAG3)3. Representative Raman spectra of thermodynamically equilibrated (slowly annealed) solutions of 50 mM G3(TTAG3)3, the concentration of the order common for NMR studies, in 100 and 500 mM of K+ or Na+ are shown in Figure 5. Corresponding CD spectra and native PAGE of the samples are presented in Figure 6.
Figure 5.
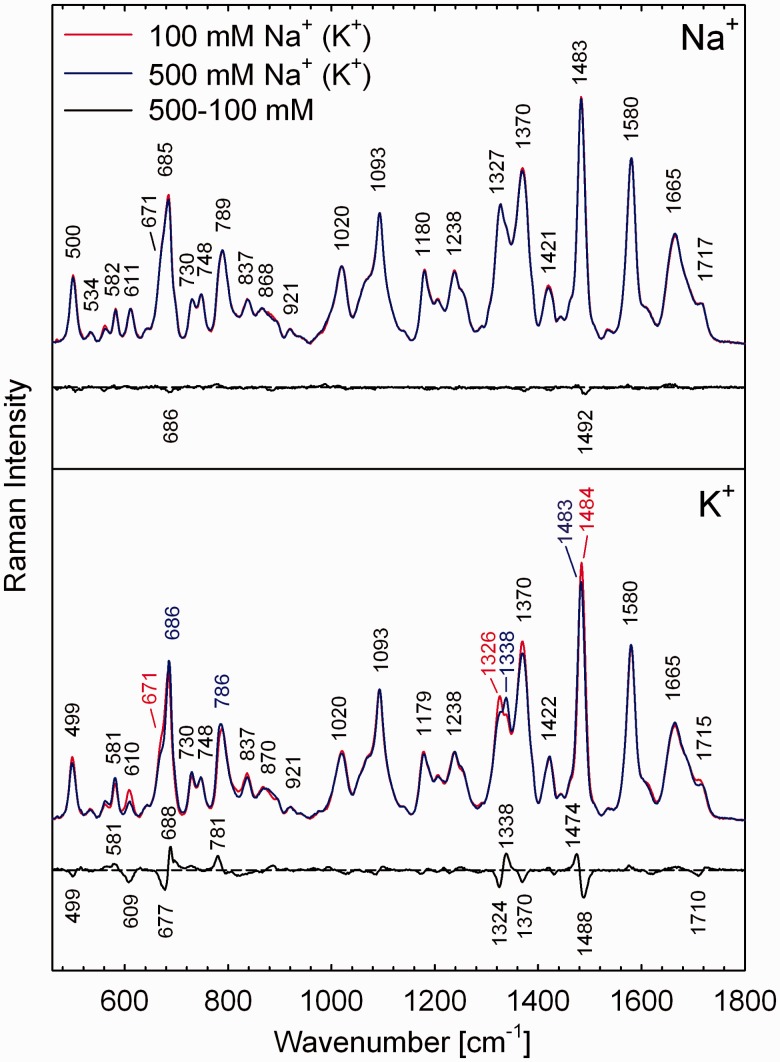
Raman spectra of 50 mM G3(TTAG3)3 in the presence of 100 and 500 mM of Na+ (top) or K+ (bottom) at 10°C. Difference spectra between the high- and low-salt structures are shown to highlight cation-specific spectral variance. Differential Raman features are labelled according to their apparent maxima/minima that can differ from the maxima/minima of the respective Raman bands.
According to CD, low-salt quadruplex structure of G3(TTAG3)3 at moderate DNA concentration (Figure 6) is identical to that of low-DNA G3(TTAG3)3 at moderate concentration of K+ (Figure 2). Corresponding Raman spectrum (Figure 5, bottom) corroborates structural similarity, exhibiting Raman features indicative for antiparallel structure in the same extent as for low-DNA K+-G3(TTAG3)3 in 250 mM K+ (Figure 1). Increase in the K+ concentration to 500 mM is followed by the same spectral changes signalling structural transition towards parallel form, as described above on interquadruplex transition induced by DNA concentration. Difference between high- and low-salt spectra of 50 mM G3(TTAG3)3 (Figure 5, bottom) is virtually identical to Raman fingerprint pattern observed on DNA increase (Supplementary Figure S3, bottom). On the other hand, no remarkable structural changes occur for 50 mM G3(TTAG3)3 on raising the Na+ concentration (Figure 5, top). The effect of K+ concentration is more profound for higher DNA concentrations, as will be shown later.
Rearrangement of G3(TTAG3)3 from antiparallel to (3 + 1) structure is accompanied by a transition of one of the three side/diagonal d(TTA)-loops to double-chain-reversal conformation. In a fully parallel quadruplex, all three d(TTA)-loops have to be double-chain-reversal. However, on the interquadruplex transition induced by high DNA (Figure 1) or K+ concentration (Figure 5), intensity of the dT-loop marker at 611 ± 2 cm−1 is reduced approximately to half of its initial value, whereas intensity of the C2-endo/anti-dG marker at 581 ± 1 cm−1 increases for ∼20%. Consequently, a fraction of G3(TTAG3)3 quadruplexes seems to preserve their loops in side/diagonal arrangement. The finding seems to be in qualitative agreement with CD spectra (Figures 2 and 6, top) that affirm partial preservation of (3 + 1) and/or even antiparallel form, as can be concluded from still intense positive CD shoulder at 293 nm. However, up to 20% of the 611 cm−1 band intensity persists even under the conditions strongly favouring parallel quadruplex. As will be shown later, the band is still visible even at the highest concentrations of DNA and K+ used in the present study, well beyond DNA concentrations for which CD spectra exhibit just CD features of parallel arrangement. For instance, in the Raman spectrum of 315 mM G3(TTAG3)3 slowly annealed in the presence of 500 mM K+ (Figure 8, bottom), normalized intensity of the 611 cm−1 band still preserves ∼20% of the intensity observed for 8 mM G3(TTAG3)3 in 100 mM K+, the later sample being considered as the least prone to interquadruplex transition. Beyond persisting conformational heterogeneity of the quadruplexes, the apparent quantitative disagreement between intensity of the 611 cm−1 band, its structural assignment and real arrangement of the loops, may stem from diverse conformations of particular dT residues constituting d(TTA)- or dT4-loops.
Figure 8.
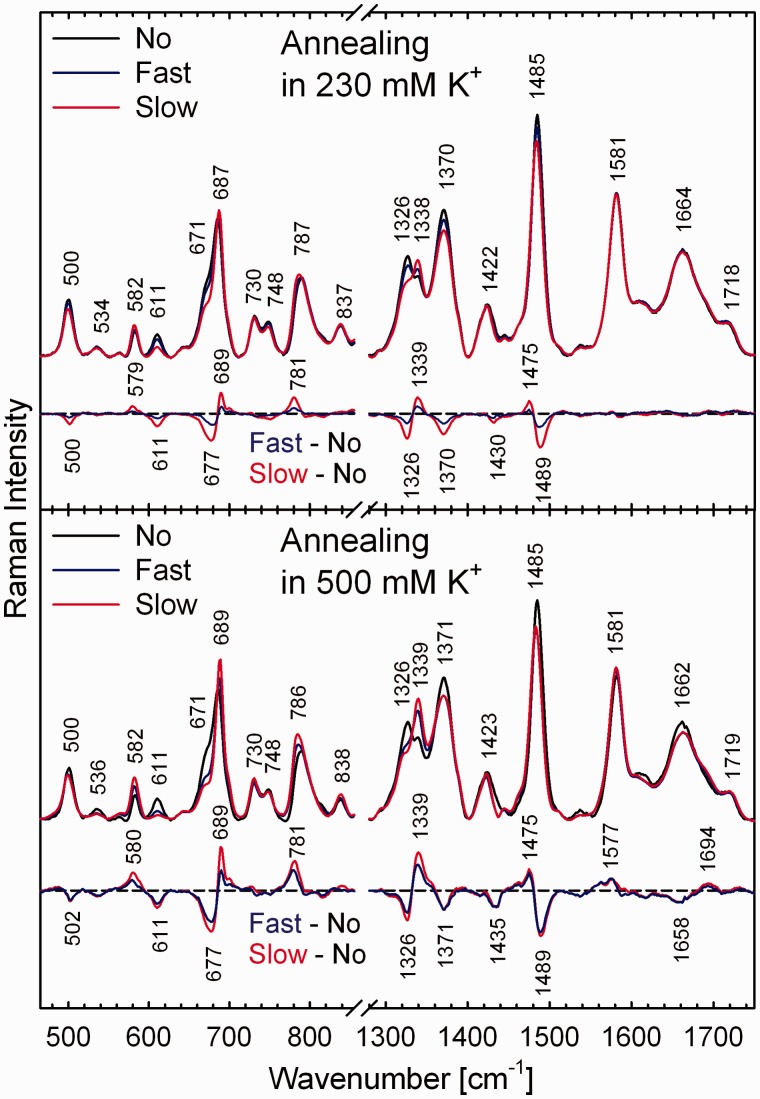
Effect of the annealing mode and K+ concentration on G3(TTAG3)3 quadruplex structure at extremely high DNA concentration. Raman spectra of 330 and 315 mM G3(TTAG3)3 in the presence 230 and 500 mM of K+, respectively, before thermal denaturation (No) and after fast (Fast) and slow (Slow) annealing. Differential spectra between thermally destabilized, thermodynamically equilibrated and initially adopted K+-stabilized G3(TTAG3)3 structures are shown to depict differences.
Effect of thermal destabilization and annealing on interquadruplex transition
As demonstrated above, antiparallel-to-parallel transition of G3(TTAG3)3 can be induced by increasing K+ and/or DNA concentrations, both factors acting in synergy. However, thermal destabilization of the antiparallel folding spontaneously adopted by G3(TTAG3)3 in the course of quadruplex formation under the high-DNA and/or high-K+ conditions is a third prerequisite (30). Incidentally, protocols of sample preparation used in some NMR studies (18,19) do not mention heating or annealing. Effect of heating and annealing at high DNA concentrations, a task for which Raman spectroscopy comes exceptionally useful, was thus studied in detail.
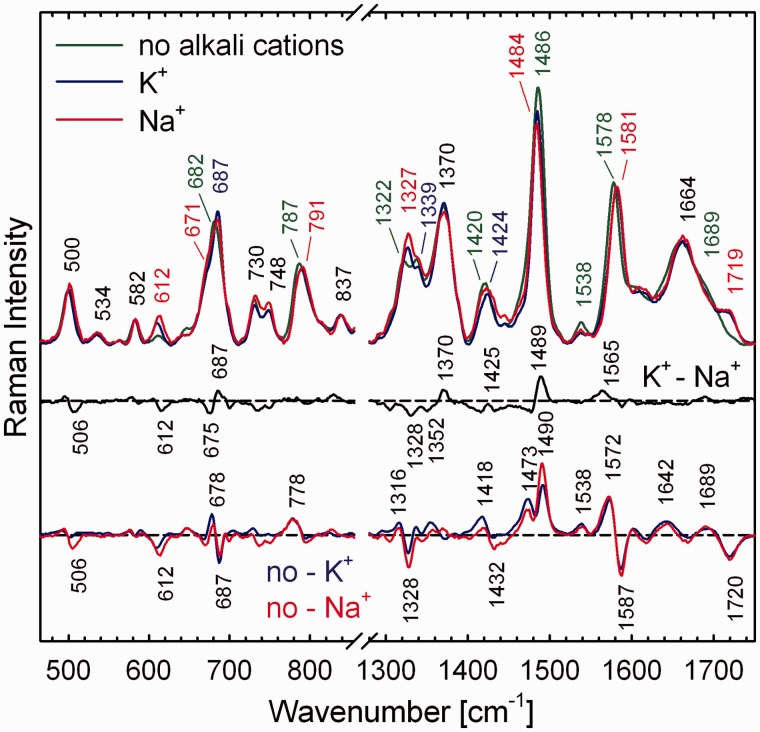
As shown in Figure 7, desalted sample of G3(TTAG3)3 prepared by repeated centrifugal washing with deionized water adopts non-quadruplex structure even at DNA concentration as high as 520 mM. Common quadruplex markers signalling tight base-pairing within G-tetrads (when located at 1484, 1581 and 1719 cm−1) are shifted to the wavenumbers typical for dG hydrogen bonding with H2O (1486, 1578 and 1689 cm−1). However, even in the absence of stabilizing cations, G3(TTAG3)3 is probably ordered, as its spectrum contains conformational marker at 837 cm−1, which is present with regular B-DNA-family backbone (32), and, except for the band at 1486 cm−1, no Raman hypochromism expected for substantially destacked bases (32) is observed. After addition of K+ ions (230 mM) but omitting annealing, the high-DNA G3(TTAG3)3 spontaneously adopts antiparallel quadruplex structure, as can be documented by appearance of syn-dG markers (671 and 1327 cm−1) and dT marker of lateral/diagonal d(TTA)-loops (612 cm−1). The K+-structure is close to the antiparallel Na+-folding formed under the same DNA and Na+ concentrations, as seen from the match of their Raman spectra and respective differential curves (Figure 7, no-K+ and no-Na+ curves). Although Raman difference spectrum between ‘unheated’ K+- and Na+-stabilized quadruplexes at high-DNA (330 mM) reveals some spectral differences (Figure 7, K+-Na+ curve), they are virtually identical with those depicted in K+–Na+ difference spectrum of G3(TTAG3)3 slowly annealed at low-DNA conditions of 8 mM (Figure 4, top). It can be suggested that, according to Raman markers, structure of K+-stabilized G3(TTAG3)3 quadruplex formed at extremely high DNA concentration before heating/annealing is virtually identical with that of the thermodynamically equilibrated K+-structure formed at low-DNA conditions by a standard annealing. Raman spectra of freshly prepared unheated K+-samples were found to be independent of G3(TTAG3)3 concentration. However, if the unheated high-DNA solutions remain standing for several days at room temperature, Raman signs of antiparallel-to-parallel transition begin to appear slowly.
Figure 7.
Spectral differences between non-quadruplex G3(TTAG3)3 structure at extremely high DNA concentration (520 mM; no alkali cations present) and initial quadruplex structures adopted by highly concentrated G3(TTAG3)3 (330 mM) after adjustment of K+ or Na+ concentrations to 230 mM, but before annealing.
The spontaneously adopted antiparallel structure of K+-stabilized G3(TTAG3)3 is evidently not a thermodynamically most stable quadruplex arrangement, especially at extremely high DNA concentrations. For example, when heated to 95°C and rapidly (in <1 min) cooled down to room temperature, partial (∼50% according to differential Raman features) switching to the parallel form is achieved, as evident from the comparison with a slowly annealed sample (Figure 8, top). The effect is synergically amplified by increasing concentration of K+, as the difference between rapidly cooled and slowly annealed differential Raman spectra becomes smaller in 500 mM K+ (Figure 8, bottom). By decreasing the G3(TTAG3)3 concentration, the differences between unheated, rapidly cooled and slowly annealed Raman spectra decrease rapidly, becoming virtually indiscernible <100 mM DNA in 230 mM K+, although yet still well apparent for 30 mM G3(TTAG3)3 in 500 mM K+. For Na+-stabilized G3(TTAG3)3, no such dependence on heating/annealing was observed.
To better understand thermal destabilization of initial antiparallel K+-form of G3(TTAG3)3 and interquadruplex switching under DNA concentrations inaccessible to CD or absorption measurements, annealing process was monitored by Raman scattering on the temperature increase and decrease. The 330 mM of G3(TTAG3)3 in 230 mM of K+ was heated continuously from 10°C to 95°C and then cooled down at the rate of ∼1.5°C/min. The SVD analysis of the entire Raman dataset sorted out spectral changes common for an overall denaturation/renaturation process of both quadruplex structures (S2, Vi2), and separated them from the features reflecting structural differences between respective quadruplexes (S3, Vi3; Supplementary Figure S4). As evidenced by a close match of temperature dependencies of the Vi2 coefficients on the temperature increase and decrease, overall renaturation curve (giving rise to the parallel quadruplex) follows well denaturation curve of the antiparallel quadruplex (Supplementary Figure S4). On the contrary, orthogonal subspectrum S3 and coefficients Vi3 clearly discriminate between antiparallel and parallel folds, except for the temperatures >77°C when Raman spectra measured on temperature increase and decrease become identical.
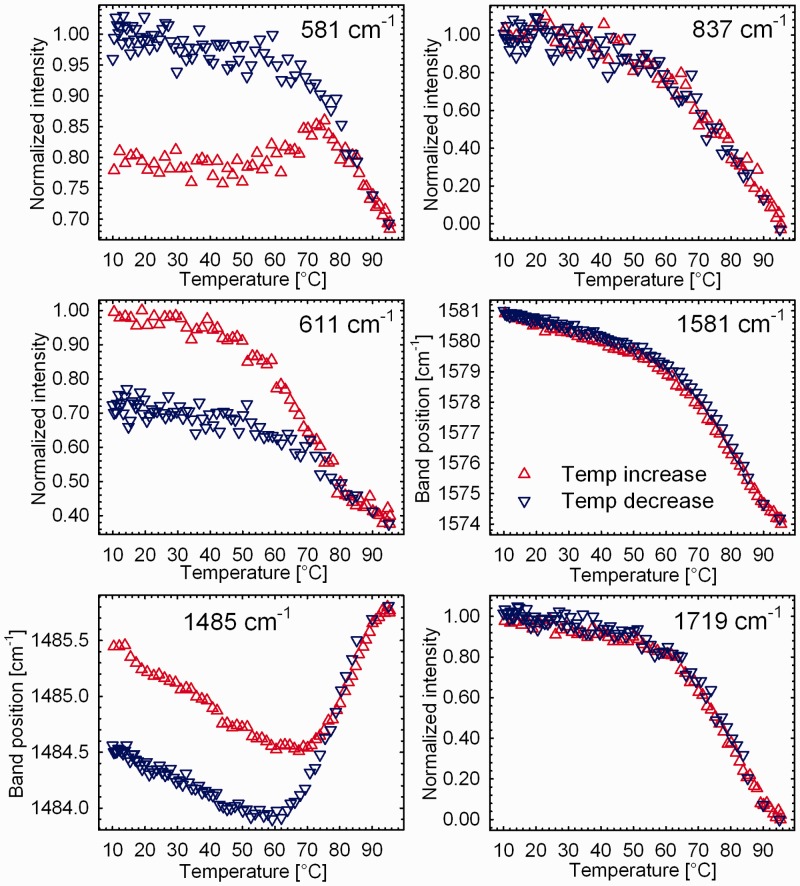
To better depict different temperature dependencies of the Raman features sensitive and insensitive to quadruplex arrangement, spectral parameters of selected bands are shown in Figure 9. Raman marker of regular B-DNA-family backbone (837 cm−1) and general markers of interbase hydrogen bonding within G-tetrads (position and intensity of the ∼1581 and ∼1719 cm−1 bands, respectively) affirm reversible melting of the sugar-phosphate backbone and similar strengths of the N2H and O6 hydrogen bonds within G-tetrads in K+-stabilized antiparallel and parallel forms under the present DNA and K+ concentrations. On the contrary, newly established C2-endo/anti-dG marker at 581 cm−1, the 611 cm−1 marker of the lateral/diagonal arrangement of d(TTA)-loops and an indicator of strong Hoogsteen hydrogen bonding at N7-dG (1485 cm−1) exhibit more complicated temperature dependences (Figure 9). Convergence of the 581, 611 and 1485 cm−1 melting curves of unheated antiparallel G3(TTAG3)3 towards renaturation curves leading to formation of the parallel quadruplex at low temperatures (indicated by the 837, 1581 and 1719 cm−1 bands; Figure 9) implies thermally induced structural switching of the antiparallel folding to a parallel-like arrangement that precedes complete quadruplex unfolding and complete disintegration of G-tetrads at higher temperatures. Denaturation/renaturation profiles exhibiting similar convergence were reported recently by CD spectroscopy for K+-stabilized G4(T2G4)3 Tetrahymena quadruplex (53), and interpreted as a sign of thermally induced conversion of intramolecular antiparallel conformers to more stable parallel forms. Poor reversibility observed during the first cooling of the melted sample was ascribed to the slow kinetics of the formation of antiparallel conformers (53). A similar preference for parallel structure after thermal destabilization of spontaneously preformed antiparallel quadruplex can be seen here for K+-stabilized G3(TTAG3)3. Once slowly annealed, K+-stabilized parallel form of G3(TTAG3)3 melts reversibly without any hysteresis, as can be evidenced by a close match of denaturation and renaturation profiles visualized by a factor analysis (Supplementary Figure S6).
Figure 9.
Heating and cooling Raman profiles of selected Raman bands sensitive (581, 611 and 1485 cm−1) and insensitive (837, 1581 and 1719 cm−1) to antiparallel-to-parallel switching of the K+-stabilized G3(TTAG3)3 quadruplex. Initially, antiparallel K+-quadruplex (330 mM), spontaneously formed on addition of 230 mM K+, was heated from 10°C to 95°C and then cooled down at the same rate of ∼1.5°C/min.
It is worthwhile to note that under the extreme DNA and K+ concentrations, unheated samples constitute clear and relatively non-viscous solution, which turns irreversibly into a solid gel after annealing. Increased viscosity or gelation after annealing was observed also for extremely high-DNA samples at moderate K+ concentrations (200–300 mM). As the interquadruplex transition induced by DNA concentration assumes some physical contacts between G3(TTAG3)3 molecules, a question about molecularity of the parallel form arises. According to electrophoretic mobilities, the samples identified by CD and Raman spectra as K+-stabilized parallel quadruplexes migrate preferentially as monomers, although slowly migrating continuous smear indicates the presence of multi-molecular structures, especially in the samples annealed at the highest K+ and DNA concentrations (Figures 2 and 6). Except for a weak band at the position corresponding to a bimolecular form visible in Figure 2, the smear lacks any structure. We suppose that the presence of the monomer band and slowly migrating, unstructured smear corresponds better to a wide distribution of the self-stacked associates consisting of intramolecular parallel quadruplexes (30) than to four-molecular, four-stranded parallel structures or multi-molecular, locally four-stranded parallel structures with out-of-register alignment of adjacent strands. As four- and multi-molecular parallel quadruplexes are more thermodynamically and kinetically stable than the higher-order self-associates of monomeric quadruplexes, their formation as well as disintegration should be less probable and slower than in the case of self-associates. The latter can be thus, under the diluting conditions of electrophoresis, disintegrated into the monomers much easier and faster than the four- and multi-molecular parallel quadruplexes, and it could better explain appearance of the monomer band in PAGE. In the case of molecular crowding induced by DNA itself, the self-association by stacking is a factor stabilizing parallel arrangement of each individual G3(TTAG3)3 quadruplex. On the other hand, such a self-association requires quadruplexes with parallel or at least (3 + 1) intramolecular arrangement (30).
CONCLUSIONS
It is generally accepted that quadruplex structures determined for crystalline states cannot be uncritically transferred to solutions. Less accepted is a fact that even quadruplex structures determined by NMR in solutions at relatively high DNA concentrations can differ from those existing in more dilute solutions investigated by absorption and CD spectroscopies.
By means of Raman spectroscopy that can be applied over wide range of DNA concentrations, we demonstrate here that quadruplex structure of K+-stabilized G3(TTAG3)3, the shortest four-repeat model of human telomeric quadruplex, depends on DNA concentration, as well as on the mode of sample preparation. Complex interplay of DNA and K+ concentrations with other factors affecting quadruplex structure, for example, presence or absence of thermal destabilization and annealing in protocols, storage conditions and time elapsed between the sample preparation and measurement, could be the reason why distinct K+-stabilized quadruplex structures of human telomeric sequences were reported by various experimental methods using DNA concentrations differing by orders of magnitude. Raman spectroscopy could be of great benefit in the current effort to better understand quadruplex structural properties under the crowding conditions existing in cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–6.
FUNDING
Grant Agency of the Charles University in Prague [3222/2008]; Grant Agency of the Czech Republic [P202/09/0193, P205/12/0466]; Ministry of Education of the Czech Republic [MSM0021620835]; ‘CEITEC—Central European Institute of Technology’ [CZ.1.05/1.1.00/02.0068] European Regional Development Fund. Funding for open access charge: Grant Agency of the Czech Republic [P202/09/0193].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Londono-Vallejo JA, DerSarkissian H, Cases L, Thomas G. Differences in telomere length between homologous chromosomes in human. Nucleic Acids Res. 2001;29:3164–3171. doi: 10.1093/nar/29.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murchie AIH, Lilley DMJ. Tetraplex folding of telomere sequences and the inclusion of adenine bases. EMBO J. 1994;13:993–1001. doi: 10.1002/j.1460-2075.1994.tb06344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Telomeres shorten during aging of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann S, Martens UM. Telomeres and telomerase as targets for cancer therapy. Cell Mol. Life Sci. 2007;64:906–921. doi: 10.1007/s00018-007-6481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 11.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 12.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 13.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Platinum cross-linking of adenines and guanines on the quadruplex structures of the AG3(T2AG3)3 and (T2AG3)4 human telomere sequences in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He YJ, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying LM, Green JJ, Li HT, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrus A, Chen D, Dai JX, Bialis T, Jones RA, Yang DZ. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi JY, Shafer RH. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorgan. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Chang CC, Chien CW, Lin YH, Kang CC, Chang TC. Investigation of spectral conversion of d(TTAGGG)4 and d(TTAGGG)13 upon potassium titration by a G-quadruplex recognizer BMVC molecule. Nucleic Acids Res. 2007;35:2846–2860. doi: 10.1093/nar/gkm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 25.Antonacci C, Chaires JB, Sheardy RD. Biophysical characterization of the human telomeric (TTAGGG)4 repeat in a potassium solution. Biochemistry. 2007;46:4654–4660. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- 26.Dai JX, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyoshi D, Karimata H, Sugimoto N. Drastic effect of a single base difference between human and Tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem. Int. Ed. Engl. 2005;44:3740–3744. doi: 10.1002/anie.200462667. [DOI] [PubMed] [Google Scholar]
- 29.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 30.Renciuk D, Kejnovska I, Skolakova P, Bednarova K, Motlova J, Vorlickova M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009;37:6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim KW, Amrane S, Bouaziz S, Xu WX, Mu YG, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benevides JM, Overman SA, Thomas GJ., Jr Raman, polarized Raman and ultraviolet resonance Raman spectroscopy of nucleic acids and their complexes. J. Raman Spectrosc. 2005;36:279–299. [Google Scholar]
- 33.Thomas GJ., Jr Raman spectroscopy of protein and nucleic acid assemblies. Annu. Rev. Biophys. Biomol. Struct. 1999;28:1–27. doi: 10.1146/annurev.biophys.28.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Miura T, Thomas GJ., Jr Structural polymorphism of telomere DNA: interquadruplex and duplex-quadruplex conversions probed by Raman spectroscopy. Biochemistry. 1994;33:7848–7856. doi: 10.1021/bi00191a012. [DOI] [PubMed] [Google Scholar]
- 35.Miura T, Benevides JM, Thomas GJ., Jr A phase diagram for sodium and potassium-ion control of polymorphism in telomeric DNA. J. Mol. Biol. 1995;248:233–238. doi: 10.1016/s0022-2836(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 36.Miura T, Thomas GJ., Jr Structure and dynamics of interstrand guanine association in quadruplex telomeric DNA. Biochemistry. 1995;34:9645–9654. doi: 10.1021/bi00029a042. [DOI] [PubMed] [Google Scholar]
- 37.Laporte L, Thomas GJ., Jr Raman spectral studies of nucleic acids part LXVI—structural basis of DNA recognition and mechanism of quadruplex formation by the β subunit of the Oxytricha telomere binding. Biochemistry. 1998;37:1327–1335. doi: 10.1021/bi972400d. [DOI] [PubMed] [Google Scholar]
- 38.Laporte L, Thomas GJ., Jr A hairpin conformation for the 3′ overhang of Oxytricha nova telomeric DNA. J. Mol. Biol. 1998;281:261–270. doi: 10.1006/jmbi.1998.1938. [DOI] [PubMed] [Google Scholar]
- 39.Krafft C, Benevides JM, Thomas GJ., Jr Secondary structure polymorphism in Oxytricha nova telomeric DNA. Nucleic Acids Res. 2002;30:3981–3991. doi: 10.1093/nar/gkf517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abu-Ghazalah RM, Irizar J, Helmy AS, Macgregor RB. A study of the interactions that stabilize DNA frayed wires. Biophys. Chem. 2010;147:123–129. doi: 10.1016/j.bpc.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Pagba CV, Lane SM, Wachsmann-Hogiu S. Conformational changes in quadruplex oligonucleotide structures probed by Raman spectroscopy. J. Biomed. Opt. 2011;2:207–217. doi: 10.1364/BOE.2.000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 43.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 44.Palacký J, Mojzeš P, Bok J. SVD-based method for intensity normalization, background correction and solvent subtraction in Raman spectroscopy exploiting the properties of water stretching vibrations. J. Raman Spectrosc. 2011;42:1528–1539. [Google Scholar]
- 45.Movileanu L, Benevides JM, Thomas GJ., Jr Determination of base and backbone contributions to the thermodynamics of premelting and melting transitions in B DNA. Nucleic Acids Res. 2002;30:3767–3777. doi: 10.1093/nar/gkf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisz K, Leitner D, Krafft C, Welfle H. Structural heterogeneity in intramolecular DNA triple helices. Biol. Chem. 2000;381:275–283. doi: 10.1515/BC.2000.036. [DOI] [PubMed] [Google Scholar]
- 47.Thomas GJ, Jr, Benevides JM, Overman SA, Ueda T, Ushizawa K, Saitoh M, Tsuboi M. Polarized Raman spectra of oriented fibers of A-DNA and B-DNA: anisotropic and isotropic local Raman tensors of base and backbone vibrations. Biophys. J. 1995;68:1073–1088. doi: 10.1016/S0006-3495(95)80282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Movileanu L, Benevides JM, Thomas GJ., Jr Temperature dependence of the Raman spectrum of DNA. Part I–Raman signatures of premelting and melting transitions of Poly(dA–dT)·Poly(dA–dT) J. Raman Spectrosc. 1999;30:637–649. doi: 10.1002/bip.10022. [DOI] [PubMed] [Google Scholar]
- 49.Peticolas WL, Kubasek WL, Thomas GA, Tsuboi M. Biological applications of Raman spectroscopy. In: Spiro TG, editor. Raman Spectra and the Conformations of Biological Macromolecules. New York: John Wiley & Sons; 1987. pp. 81–133. [Google Scholar]
- 50.Benevides JM, Thomas GJ., Jr A solution structure for Poly(rA).Poly(dT) with different furanose pucker and backbone geometry in rA and dT strands and intrastrand hydrogen bonding of adenine 8CH. Biochemistry. 1988;27:3868–3873. doi: 10.1021/bi00410a051. [DOI] [PubMed] [Google Scholar]
- 51.Serban D, Benevides JM, Thomas GJ., Jr HU protein employs similar mechanisms of minor-groove recognition in binding to different B-DNA sites: demonstration by Raman spectroscopy. Biochemistry. 2003;42:7390–7399. doi: 10.1021/bi030050r. [DOI] [PubMed] [Google Scholar]
- 52.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Víglaský V, Bauer L, Tlučková K. Structural features of intra- and intermolecular G-quadruplexes derived from telomeric repeats. Biochemistry. 2010;49:2110–2120. doi: 10.1021/bi902099u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.