Abstract
The ENCODE Project has generated a wealth of experimental information mapping diverse chromatin properties in several human cell lines. Although each such data track is independently informative toward the annotation of regulatory elements, their interrelations contain much richer information for the systematic annotation of regulatory elements. To uncover these interrelations and to generate an interpretable summary of the massive datasets of the ENCODE Project, we apply unsupervised learning methodologies, converting dozens of chromatin datasets into discrete annotation maps of regulatory regions and other chromatin elements across the human genome. These methods rediscover and summarize diverse aspects of chromatin architecture, elucidate the interplay between chromatin activity and RNA transcription, and reveal that a large proportion of the genome lies in a quiescent state, even across multiple cell types. The resulting annotation of non-coding regulatory elements correlate strongly with mammalian evolutionary constraint, and provide an unbiased approach for evaluating metrics of evolutionary constraint in human. Lastly, we use the regulatory annotations to revisit previously uncharacterized disease-associated loci, resulting in focused, testable hypotheses through the lens of the chromatin landscape.
INTRODUCTION
The sequencing of the human genome produced the complete recipe for a human being encoded in digital form, and much of the past decade of molecular biology has been devoted to deciphering the meaning of this code. On this premise, the ENCODE Project Consortium sought to discover a complete catalog of all functional elements in the human genome (1), analogous to delineating sentences and words that comprise the human genome, and understanding the type of function each element plays. Such a catalog will undoubtedly never be complete, given the diversity of cell types where elements may activate, the diversity of experimental assays needed to probe them, and the specific conditions and stimuli to which they may respond. The scale-up phase of the ENCODE project, however, has made substantial advances toward the goal of a comprehensive catalog. It has carried out a daunting 1640 total experiments in 147 cell types, using multiple distinct biochemical assays, including ChIP-seq, DNase-seq, FAIRE-seq and RNA-seq. Interpreting the resulting information is arguably more complex than interpreting the primary sequence of the human genome. The four nucleotides of the sequence have been replaced by a large vector of numerical values, each representing the result of a different biochemical assay in a given condition and a given cell type, at each position of each chromosome. The challenge at hand is thus to turn these vectors of numerical values into an interpretable annotation, namely, the list of functional elements that the ENCODE project set out to annotate.
To address this challenge, we and others have developed a variety of computational techniques that seek to identify functional elements from high-throughput genomic datasets. These techniques fall into two groups: supervised learning methods that attempt to find instances of one or more pre-determined classes of elements, and unsupervised learning methods that seek to simultaneously discover functional classes and annotate their instances de novo. Supervised learning methods have been widely used for automatic gene finding methods that can recognize protein-coding transcripts using sequence features, cDNA sequence and evolutionary conservation of known examples (2,3). Supervised models have also been successfully used to recognize promoters (4), enhancers (5) and microRNAs (6), based on known examples. As supervised learning methods require a training set of known examples, they are incapable of discovering novel types of functional elements. Unsupervised methods, in contrast, identify candidate functional elements without the need for previously defined classes or known examples, thereby avoiding biases toward well-understood phenomena. Despite their generality, peak-finding algorithms for ChIP-seq analysis can be seen as supervised learning methods, seeking to recognize ‘peak-like’ behavior. Moreover, peak-finding methods have difficulty generalizing to the joint analysis of dozens of tracks of functional genomics data, where the diversity of possibly interesting patterns is very high. Such integrative analyses are central to the mission of ENCODE, which aims not only to produce such data but also to make sense of the resulting collection of datasets.
In this work, we apply unsupervised chromatin state annotation methods that simultaneously discover the locations of functional elements in the human genome and assign to each element one of a small number of labels, which can be interpreted as functional annotations. As input, our methods receive a collection of functional genomics datasets and a user-specified parameter for the number of distinct labels that the method should discover. The input datasets consist of ChIP-seq assays for multiple histone modifications, general transcription factors and chromatin accessibility assays. We restrained ourselves to chromatin-level information in the initial annotation stage, and did not use RNA-seq information as an input to our models, instead reserving it for later validation. Our computational analysis provides as output an annotation of the human genome. This annotation consists of a segmentation into non-overlapping segments, and a labeling of each segment using one of a small set of labels, which we refer to as chromatin states. The goal of the chromatin state annotation is to capture the similarities of segments that show the same patterns across many experiments by assigning them the same label, thus summarizing a very large collection of data into a more meaningful form. The resulting segment labels typically correspond to an intuitive, human interpretable biological function, which we use to summarize them, even though we recognize that the underlying biology is usually more complex. Other times, the segment labels may remain uninterpreted until we learn more about additional functions that may be distinguishable by their specific combinations of chromatin marks, but whose biological roles may not yet be understood until additional biological processes become elucidated, or until additional datasets become available. The unsupervised nature of these chromatin state annotations may thus identify novel instances of known classes of functional elements, suggest novel subdivisions of classes into subclasses, or hypothesize the existence of entirely new types of functional elements.
During the pilot phase of ENCODE, Thurman et al. combined a hidden Markov model (HMM) with wavelet smoothing to produce a two-label segmentation of the ENCODE pilot regions into ‘active’ and ‘repressed’ regions (7,8). A variety of segmentation models have been described subsequently, employing HMMs with flat (9,10) or hierarchical (11) structures, or generalizing the HMM to a hierarchical change-point model (12).
For the second phase of ENCODE, two research groups within the consortium independently developed chromatin state annotation algorithms, ChromHMM (13,14) and Segway (15). Although the methods were designed and initially implemented independently of one another, they share many key features. Most significantly, the methods employ closely related probabilistic models. ChromHMM is implemented as an HMM, in which the ‘time’ axis is the chromosomal coordinate and the various ENCODE datasets are the observed variables. Similarly, Segway employs a dynamic Bayesian network (DBN) approach, which is a generalization of the HMM framework. The HMM/DBN approach offers multiple important advantages, including efficient algorithms for carrying out inference and a modeling paradigm in which the model’s internal variables have well-defined semantics.
Key differences between the two chromatin state annotation methods are summarized in Table 1. Broadly speaking, ChromHMM aims to take more of a birds-eye view of the data, opting to compress each data track to a single Boolean value for each 200-bp segment of the genome. This approach makes ChromHMM computationally efficient, enabling training on the entire genome, and reduces the chances that artifacts related to scaling of the data or local patterns of missing data due to mapping problems will mislead ChromHMM. The 200-bp resolution also ensures that ChromHMM produces reasonably large nucleosome-sized segments without having to implement complex constraints on segment length distributions. Segway, in contrast, operates on the full data matrix at 1-bp resolution. Segway handles missing data in a principled fashion by marking each missing data point as a hidden variable and marginalizing over all possible values. To ensure that Segway produces segments of a reasonable size, we employ both hard constraints (for example, enforcing a minimum length of 100 bp per segment) and soft constraints (length priors). For efficiency, we trained the Segway models described here only on 1% of the human genome, but training on the entire genome is possible. Finally, ChromHMM and Segway differ in the choice of algorithm used to assign the final labeling: ChromHMM assigns to each segment the label with maximum posterior probability, whereas Segway selects the series of labels that jointly achieves the highest probability over the entire segmentation path.
Table 1.
Major differences between ChromHMM and Segway as applied to the ENCODE data
| ChromHMM | Segway | |
|---|---|---|
| Modeling framework | Hidden Markov model | Dynamic Bayesian network |
| Genomic resolution | 200 bp | 1 bp |
| Data resolution | Boolean | Real value |
| Handling missing data | Interpolation | Marginalization |
| Emission modeling | Bernoulli distribution | Gaussian distribution |
| Length modeling | Geometric distribution | Geometric plus hard and soft constraints |
| Training set | Entire genome | ENCODE regions (1%) |
| Decoding algorithm | Posterior decoding | Viterbi |
| Learning across six cell types | Single model for all cell types | One model per cell type |
The work presented here constitutes the first systematic integration of chromatin elements across the entire ENCODE project. The two methods used reveal functional chromatin elements at different levels of resolution, making it possible to study both the transitions between different types of chromatin states at single-nucleotide resolution, and to obtain a robust annotation that can tolerate small variations in large chromatin domains. These two annotations form the basis of the integrative analysis for the ENCODE project, and they provide a systematic view of the chromatin landscape. This integrated viewpoint will be of great value to epigenomics research. In addition, this work describes a manually curated chromatin annotation that synthesizes the two complementary methodologies.
The resulting annotations capture the remarkable diversity of genomic functions encoded by distinct chromatin states, are robust across different cell types, and are reliably recovered by the two methods used here. We also created a combined segmentation that contains features of both. Our systematic annotation of chromatin elements has important implications for the study of the human genome.
The annotation successfully and automatically recovers much of what is already known about genome organization, including transcript-associated chromatin states and diverse classes of regulatory elements, based solely on an unsupervised analysis of chromatin data.
The annotation reveals the important relationship between biochemical activity for chromatin functions and RNA transcription, and shows important differences between the two.
The annotation points to the surprising finding that a large portion of the human genome exists in a quiescent state, which holds across multiple cell types.
The annotation provides an unbiased view of functional non-coding regulatory elements, enabling us to evaluate different metrics and methods for measuring human evolutionary constraint. In particular, we report the first genome-wide experimental demonstration of the functional relevance of evolution-based inference of constraint for pairs of nucleotides, rather than individual nucleotides at each position (16).
The annotation enables us to revisit disease-associated regions, identified via genome-wide association studies (GWAS), that previously lacked any functional annotations, providing focused, testable hypotheses revealed through the lens of the chromatin landscape.
MATERIALS AND METHODS
Track selection
For the coordinated segmentations, we selected all the available combined ENCODE histone modification, Pol2 and CTCF ChIP-seq (plus control) and open chromatin (DNase-seq and FAIRE-seq) tracks that were available in the January 2011 data freeze for each of the six Tier 1–2 cell types. A full list of tracks used is in Supplementary Table S1.
Signal track generation
We used a uniform signal-processing pipeline to generate genome-wide normalized signal coverage tracks for different types of ENCODE datasets (Supplementary Figures S1–S3, Supplementary Table S2). We used different subsets of these tracks as input to the segmentation algorithms. We combined signal from multiple replicates of each experiment. In addition, for a select subset of experiments that had equivalent datasets from multiple labs, we combined signal across all datasets. We downloaded read alignment files from the ENCODE portal (http://genome.ucsc.edu/ENCODE/downloads.html) and filtered them to remove multi-mapping reads. For each of the replicates (datasets) that we combined to produce a unified signal track, we estimated a characteristic read-shift parameter from the data and shifted reads by this estimated value in the 5′–3′ direction. We computed the effective fragment coverage for each position in the genome by first computing the read-count coverage followed by a smooth aggregation (using kernel smoothing) in a pre-specified window around each position. We then normalized the fragment coverage for the total number of mapped reads over all replicates, effective read-extension and smoothing lengths, local mappability around each location and the overall mappable size of the genome. We expressed the final signal values at each position in terms of a fold-change over the expected signal from an equivalent uniform distribution of reads over all mappable locations in the genome. We explicitly represented as missing values those signal values at unreliable locations consisting of genomic positions surrounded by a large number of unmappable locations and those in assembly gaps. Supplementary Methods contain a complete description of this procedure.
Segmentation input preprocessing
Before applying ChromHMM, we converted the normalized signal tracks into binarized data at a 200-bp resolution. We used the maximum signal for a mark in each 200-bp interval to represent the mark in that interval. The threshold for each mark was the maximum of 4.0 and the value corresponding to a Poisson tail distribution probability of 0.0001. Requiring a fold threshold, in addition to the tail distribution threshold, enabled more meaningful binarization of some of the most deeply sequenced datasets. We excluded regions that associated with repetitive elements such as α- and β-satellite repeats, ribosomal and mitochondrial DNA (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/wgEncodeDukeMapabilityRegionsExcludable.bed.gz).
For Segway, we excluded the ENCODE Data Analysis Consortium Blacklisted Regions (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/wgEncodeDacMapabilityConsensusExcludable.bed.gz), comprising a comprehensive set of regions in the human genome that exhibit anomalous or unstructured read-counts in next gen sequencing experiments, independent of cell line and type of experiment. To identify these regions, we used 80 open chromatin tracks (DNase and FAIRE datasets) and 20 ChIP-seq input/control tracks spanning ∼60 human tissue types/cell lines in total. The regions tend to have a very high ratio of multi-mapping to unique mapping reads and high variance in mappability. Some of these regions overlap pathological repeat elements such as satellite, centromeric and telomeric repeats. However, simple filters based on mappability do not account for most of these regions.
Training
We trained ChromHMM in concatenated mode on the six cell types using a two-stage nested parameter initialization approach, considering models of up to 30 states, using Euclidean distance for the state pruning distance, and setting the emission and transition smoothing parameters to 0.01 and 0.5, respectively, for the second stage parameter initialization (13,17). For each pass, we completed 200 training iterations. ChromHMM mnemonics and brief state descriptions are in Supplementary Table S3.
We trained Segway on the 1% of the genome selected as the ENCODE pilot regions (7). We performed expectation maximization training until either (i) the log likelihood of the model minus the log likelihood from the previous iteration divided by the log likelihood was <10−5, or (ii) until 100 training iterations. We assigned mnemonic labels according to Supplementary Table S3.
The ChromHMM and Segway chromatin state annotations can be accessed by loading the ENCODE Analysis Hub at http://encodeproject.org/cgi-bin/hgHubConnect.
RESULTS
We applied both chromatin state annotation methods to a set of chromatin signal tracks from the ENCODE Tier 1 (GM12878, H1 hESC, K562) and Tier 2 (HeLa-S3, HepG2, HUVEC) cell types. For this analysis, we included data from the kinds of experiments that have the potential to reveal the most about chromatin state—the open chromatin assays DNase-seq and FAIRE, and ChIP-seq on histone modifications, RNA polymerase 2 (Pol2) and CTCF. Generally, we used only the signal tracks that were available for all six Tier 1–2 cell types, combining data from multiple laboratories when appropriate. Figure 1 displays the full list of signal tracks used, with data sources in Supplementary Table S1.
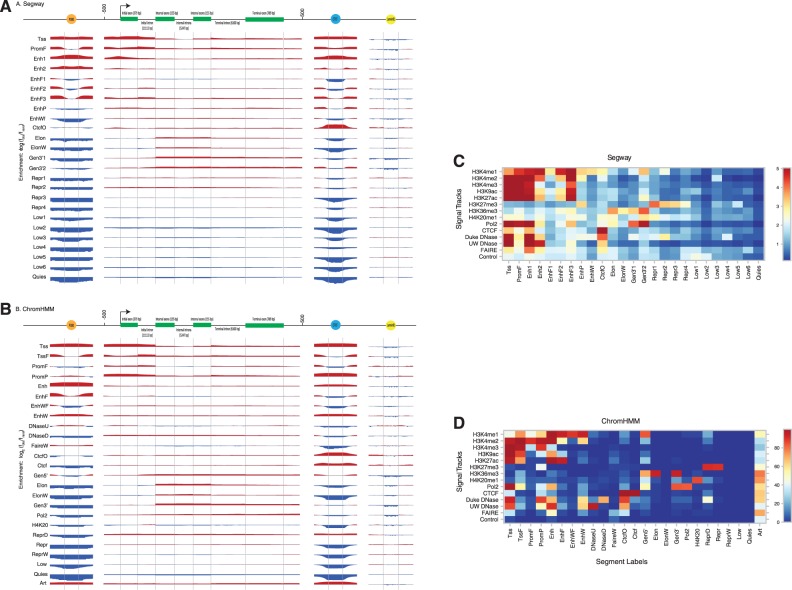
Figure 1.
Enrichment of various segment labels (vertically, labeled by green panels) from (A) Segway and (B) ChromHMM K562 segmentations over positions on an idealized p300 binding site, gene, CTCF binding site, and LaminB1 binding site. We calculated enrichment as the base-2 logarithm of the observed frequency of a label at a particular position along an annotation divided by the expected frequency of the label from its prevalence in the genome overall. Enriched positions are shown in red, and depleted positions are shown in blue. The labels for idealized gene components at the top include the mean length of that component in parentheses. (C) Heat map of parameters from Segway training for 14 GM12878 signal tracks against 25 segment labels. Color indicates the mean of a Gaussian according to the color bar on the right. (D) Heat map of parameters from ChromHMM concatenated training on 84 signal tracks from 6 ENCODE Tier 1–2 cell types. Color indicates the probability of a present mark, as a percentage, according to the color bar on the right.
We trained Segway on each of the six Tier 1–2 cell types independently giving a separate Segway model for each cell type, and ChromHMM jointly using a virtual concatenation of the six cell types giving a single ChromHMM model applicable to all six cell types. We specified that the methods should find 25 chromatin states. We picked this number because it is large enough to describe many interesting functional elements while still being small enough for a biologist to interpret easily, given our previous experience with Segway (15) and ChromHMM (13,17).
The chromatin state procedure is semi-automated in that the two algorithms assign to each segment an integer label, but one must ascertain the functional semantics of these labels in a post hoc analysis step. Based on a variety of types of evidence, including investigation of known genomic loci and examination of the model parameters, we assigned names such as ‘TSS’ or ‘enhancer’ to each of the integer labels. These names are summarized in Figure 1, and the basis for these naming assignments is shown in Figure 1, Supplementary Figures S4–S7 and described more fully below. The proportion of the genome covered by each label is shown in Supplementary Figure S8, and the distribution of segment lengths is shown in Supplementary Figure S9. Figure 2 shows a sample locus on chromosome 13 with the two chromatin state annotations displayed along the top. For ease of visualization, we have colored all the chromatin states using a reduced palette of 10 colors derived from Ernst et al. (2011).
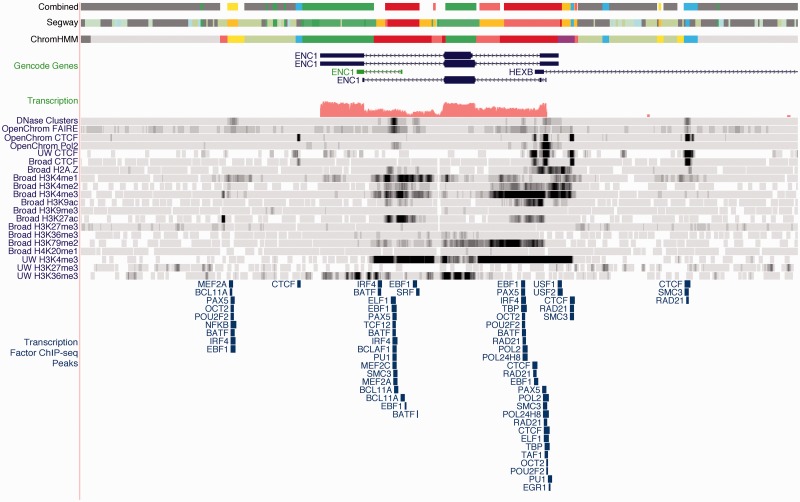
Figure 2.
View of the ENC1 locus on the minus strand using the ENCODE GM12878 segmentations. The unusual state pattern in middle of the gene in all three segmentations reveals a potential intronic regulatory element, which is confirmed by H3K4me1, H3K27ac, DNaseI hypersensitivity and transcription factor binding, and overlaps a putative GENCODE processed transcript.
In addition to the separate chromatin state annotation produced by Segway and ChromHMM, we considered it desirable to produce a simpler summary-level classification to provide more immediate interpretability and data display for a more general audience. Toward this end, we identified closely related states both within and across methods, and then defined a rule-based metric (Supplementary Methods, Supplementary Results, Supplementary Tables S4 and S5, Supplementary Figure S10) to classify them into seven classes, emphasizing biologically meaningful differences: Transcription Start Site (TSS), Promoter Flanking (PF), Enhancer (E), Weak Enhancer (WE), CTCF binding (CTCF), Transcribed Region (T) and Repressed or Inactive Region (R). Based on these rules, we produced a combined annotation of each of the Tier 1 and Tier 2 cell lines that achieved high coverage of the assayable genome (94.4–96.5%, see Supplementary Table S6 and Supplementary Figures S11 and S12) in each cell type. For the rest of the article we primarily consider the detailed primary chromatin state segmentations, both in aggregate and at individual loci such as ENC1 (Figure 2), NOD2 (Figure 3) and HBB (Supplementary Results and Supplementary Figure S13).
Figure 3.
(A) Enrichment or depletion of GWAS SNPs (and several comparison SNP sets) in function-associated segments. The bars extend to the level of enrichment or depletion of each SNP set in the 25 segmentation classes from Segway (top) and ChromHMM (bottom) in GM12878. The results for 1000 random samplings of the SNPs matched to the phenotype-associated SNPs are displayed as a box plot, with the box extending from the 25th to the 75th percentiles, the whiskers extending to 1.5 times the interquartile range, and any outliers beyond shown as circles. If the enrichment for the phenotype associated, GWAS lead SNPs exceeded the 95th percentile of the results from the matched SNPs, then the bar is colored red (orange if otherwise). (B) An example of Crohn’s disease SNPs in non-coding sequences that could serve to regulate expression of NOD2. The figures show gene models from the GENCODE group (version 12), locations of SNPs associated with Crohn’s disease by GWAS, results of ChromHMM and Segway segmentations, selected histone modifications measured in GM12878 and HUVEC cells, locations of DNase hypersensitive sites in several cell types, and sites of occupancy by selected transcription factors. Regions discussed in the text are outlined by blue rectangles.
Chromatin states recover genes and regulatory elements
Genes
Perhaps the best-understood type of functional element in the human genome is the protein-coding gene. Given that these genes account for a large proportion of the evolutionary constraint observed in multi-species alignments, it is reassuring that both ChromHMM and Segway devote a considerable proportion of their model parameters to identifying and characterizing protein-coding genes. Both types of model contain approximately eight labels that strongly correlate with various genic components. Figure 1 illustrates how these gene-related labels aggregate around protein-coding genes.
Promoters
Segway and ChromHMM both learn labels associated with promoters generally, and more specifically with regions of varying proximity to the TSS (Supplementary Figure S14). Both methods show excellent recall of TSSs, in a manner dependent on the cell type and data (Table 2). The high-resolution Segway Tss labels have higher log2 fold enrichment of TSSs (Segway: 7.2–8.0; ChromHMM: 6.9–7.3), and the high-continuity ChromHMM Tss label has better base-level recall (Segway: 53–91%; ChromHMM: 79–94%). In particular, Segway has the ability to find high-resolution patterns at TSSs localized to the level of stable nucleosome-free promoter regions, whereas ChromHMM finds larger TSS-associated segments (Supplementary Figure S15).
Table 2.
Cumulative precision (top value in each cell) and recall (bottom value) for prediction of TSSs with CAGE support in a given cell type using the TSS-proximal labels of ChromHMM and Segway
| Cell type | Segway |
ChromHMM |
Combined TSS | ||||
|---|---|---|---|---|---|---|---|
| Tss | +TssF | +PromF | Tss | +TssF | +PromF | ||
| K562 | 15.65 | 6.46 | 10.9 | 5.62 | 4.44 | 8.37 | |
| 60.6 | 65.9 | 92.6 | 93.5 | 93.6 | 97.0 | ||
| GM12878 | 18.0 | 6.96 | 3.61 | 7.73 | 3.89 | 2.63 | 5.62 |
| 90.6 | 93.7 | 94.9 | 93.6 | 93.9 | 94.2 | 96.9 | |
| H1 hESC | 15.27 | 7.90 | 2.87 | 12.84 | 8.71 | 5.22 | 9.08 |
| 82.9 | 94.0 | 94.2 | 78.9 | 79.3 | 79.4 | 95.7 | |
| HeLa-S3 | 11.78 | 5.82 | 8.63 | 5.44 | 3.88 | 11.06 | |
| 64.3 | 68.3 | 84.6 | 84.9 | 85.2 | 87.8 | ||
| HepG2 | 20.30 | 6.01 | 8.10 | 4.23 | 2.95 | 9.18 | |
| 52.7 | 53.8 | 81.5 | 81.8 | 82.0 | 88.6 | ||
| HUVEC | 12.72 | 6.96 | 11.35 | 7.29 | 5.61 | 11.22 | |
| 83.4 | 89.2 | 91.3 | 91.9 | 92.1 | 94.2 | ||
Additionally, both methods identify other labels likely to occur upstream and downstream of the TSS. These TSS-flanking patterns occur to a certain extent around other transcriptionally active regions such as enhancers. Often, the Segway TSS-flanking TssF label can be tightly associated with well-positioned nucleosomes, flanking a narrow Tss label of a consistent nucleosome-free region. The promoter-associated segments can lead to new discoveries. For example, the ENC1 gene in Figure 2 is annotated with promoter-associated segments not only at the canonical TSS, but also in intron 2. The GENCODE group has identified this as the start site for an internal, non-coding RNA (green gene model).
Some Segway models (H1 hESC and HepG2) and the ChromHMM model contain a ‘poised promoter’ PromP label associated with both the activating H3K4me3 and the repressing H3K27me3 modifications. This matches a previously identified bivalent chromatin structure that silences genes but keeps them ready for activation (18).
Candidate transcriptional terminators
The chromatin state annotations present a more sophisticated and nuanced view of the chromatin landscape than focusing on observations from a single assay. For example, the Gen3′ label discovered by both ChromHMM and Segway has a high frequency for H3K36me3, H4K20me1 and Pol2, with low frequency of other marks (Figure 1). Consistent with previous findings (13), we observe that these states show enrichment around 3′ ends of protein-coding genes (Supplementary Figure S16). Interestingly, ChromHMM also discovers a Pol2 label (associated with high frequency of RNA polymerase II and low frequency of all other signals) that displays a peak in enrichment 1–2 kbp after the 3′ end of the genes. The accumulation of Pol2 could be explained by transcriptional pausing in this region, part of the mechanism of transcription termination (19–21).
Enhancers
Both chromatin state annotations contain labels associated with enhancer type sequences: ChromHMM states Enh and EnhW, and Segway state Enh. These have emission parameters associated with chromatin features particularly suggestive of enhancers (Figure 1). This includes the presence of DNase hypersensitivity and a relatively higher H3K4me1 signal compared to H3K4me3 (7,22). The Enh states show a strong enrichment for transcription factor binding (Supplementary Table S7), while having minimal enrichment with regions proximal to annotated starts of genes (Supplementary Figure S17). The EnhF and EnhWF states also have histone modifications similar to Enh and EnhW states, respectively, and frequently transition to them, but had substantially lower DNase hypersensitivity as well as lower enrichment for transcription factor binding peaks. Such an arrangement is consistent with a central region with no or highly remodeled nucleosomes, flanked by nucleosomes with highly modified histone tails (23,24). The ChromHMM and Segway Enh states show the strongest enrichment for the transcriptional coactivator p300, often found at enhancers (25) (Supplementary Figure S18).
Repressed regions
The histone modification H3K27me3, generated by the Polycomb repressor complex 2, covers some repressed genes. Hence, the chromatin states labeled with the prefix Repr, enriched in H3K27me3 signal (Figure 1), are strong indicators that the genes within them have been silenced. ChromHMM state ReprD has a relatively high frequency of H3K27me3 and DNase sensitivity (Duke DNase), and Segway state Repr2 (in GM12878) has some similar features. ChromHMM state Repr as well as Segway states Repr3 and Repr4 (in GM12878) have strong Polycomb repression signal but lack the DNase signal. While ChromHMM state ReprW has low emission probabilities, it frequently transitions to state Repr, suggesting that it is part of broader repressed regions. In contrast, ChromHMM state Low is also associated with low signal but more frequently transitions to active elements, therefore representing low activity domains of the genome near active elements.
Insulators
Insulators are cis-regulatory modules that restrict the effect of long-range regulatory modules, such as enhancers, so that they act on the appropriate promoter target (26,27). One way to do this is via an enhancer-blocking activity, which requires the protein CTCF (28). Both methods produce one or two states enriched for CTCF occupancy, one of which (CtcfO) is also enriched for DNase-sensitive, open chromatin (Figure 1, Supplementary Figure S19).
Chromatin states are predictive of RNA transcription
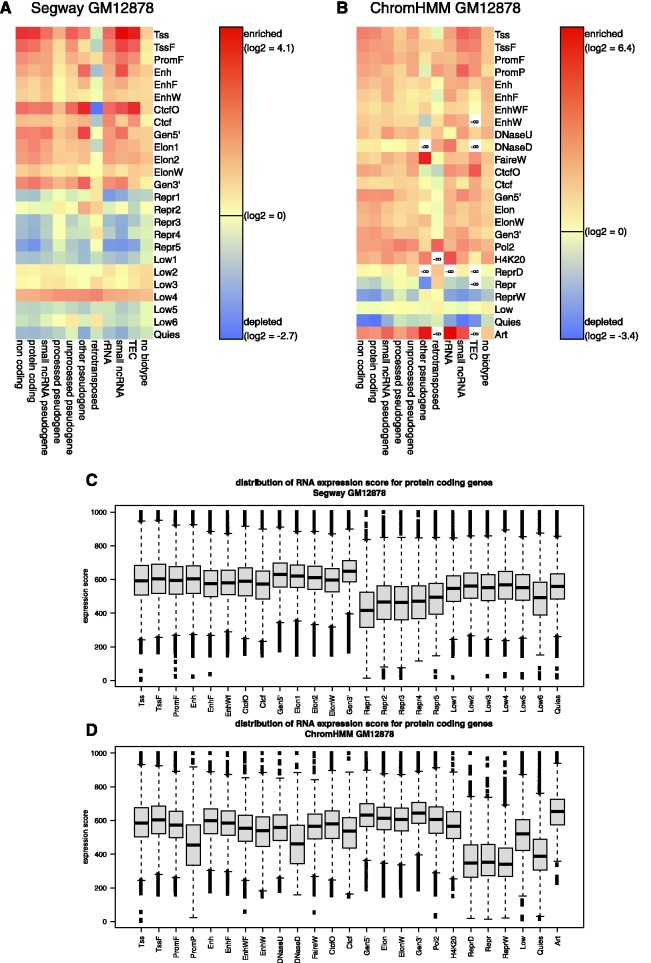
The extensive transcriptome mapping, largely by RNA-seq, from the ENCODE consortium was not used as input in our chromatin state annotations. Thus, we can evaluate the transcriptional activity of the DNA segments in each state, both for interpretation of the chromatin states and for discovery of novel relationships. As expected, the chromatin states associated with genes are highly enriched for transcripts in GM12878 cells (Figure 4). This is the case for almost all the biotypes, i.e. RNAs from different categories of genes; the only exception is for RNA from retrotransposed elements in the Segway states in GM12878. When protein-coding genes in each chromatin state are examined, we find that almost all (80–90%) of the genes overlapping the classes with labels for promoters and gene bodies are transcribed (Supplementary Figure S20), and the expression levels within these genes are high (Figure 4). Also as expected, the DNA segments in states associated with repression are depleted for transcripts (Figure 4), only a minority of the protein-coding genes overlapping these states are transcribed (Supplementary Figure S20), and the expression levels (29) for these genes are low (Figure 4).
Figure 4.
Distribution of various classes of transcripts in the segmentations. Enrichment (red) or depletion (blue) of RNA-seq transcript categories (‘biotypes’) in each state for two 25-state segmentations: (A) Segway GM12878 and (B) ChromHMM GM12878. White cells indicate an absence of an RNA biotype in the corresponding state. Distribution of expression levels in segmentation states. The level of expression of each protein-coding RNA-seq contig intersecting a protein-coding gene in each state for (C) Segway GM12878 and (D) ChromHMM GM12878 was extracted from the data in Djebali et al. (29). The distribution of those values for all RNA contigs in the DNA segments for each state is shown as a box plot.
Notably, the DNA segments in states associated with enhancers are also transcribed, indicating that the transcription of enhancers (30,31) is widespread. Interestingly, states with a high frequency of CTCF are also enriched for transcripts; further work could evaluate how often CTCF-bound regions of different categories (e.g. insulator versus non-insulator) are transcribed.
An unexpected result is that many of the chromatin states with low frequency of most of the epigenetic marks (Low classes) are also enriched for transcripts. Most (75–80%) of the protein-coding genes within the ChromHMM Low class are transcribed, i.e., captured by RNA-seq contigs (Supplementary Figure S20), but they tend to be transcribed at a lower level than the protein-coding genes overlapping the promoter and gene body-associated classes (Figure 4). Thus, the chromatin-based annotations reveal a subset of a genome with a low frequency of histone modifications, but that nonetheless supports transcription. This observation contrasts starkly with the quiescent states described in the next section. Future work should examine the classes of genes and other elements in these regions with transcriptional activity but infrequent appearance of the histone modifications and chromatin features examined here.
A large proportion of the genome resides in a quiescent state consistently across cell types
Both ChromHMM and Segway collect a large portion of the genome into states with very little signal for any of the features used as inputs, which we term quiescent. We label the quiescent states as Quies in both ChromHMM and Segway. Furthermore, the Low states have emission parameters that are nearly as low as the quiescent states, which have near-zero emission parameters for most signals (Figure 1). Together, the Quies and Low states comprise a majority of the genome for most cell types in both annotations (Supplementary Figure S8). The Quies states showed a consistent enrichment for the nuclear lamina domains mapped previously in human lung fibroblasts (32) (Supplementary Figure S21).
The low signal for epigenetic marks in these quiescent regions is not a result of a failure to map sequencing reads to them. The mappability and repeat content of the quiescent states is not dramatically different from that in other states (Supplementary Figure S22). In addition, we find that the nucleosome content is not significantly lower from that of other segmentation classes. Thus, we interpret these quiescent segments as being in a chromatin structure that is largely devoid of the histone modifications included in the segmentation. The quiescent regions are also strikingly depleted for all annotated transcript categories examined here, including coding and non-coding transcripts (Figure 4). Restricting the analysis to protein-coding genes, we find that slightly more than half of these genes in the Quies states overlap with RNA-seq contigs, much fewer than for genes overlapping promoter and gene body-associated labels (Supplementary Figure S20). Moreover, transcription factors bind much less frequently in the quiescent states than in more active states, showing a level of depletion of TF occupancy comparable to or more depleted than that seen in most of the states associated with repression (Supplementary Table S7). These results indicate that the quiescent segments are inactive for dynamic histone modifications and are transcribed infrequently. Thus, the DNA in these regions is largely inactive.
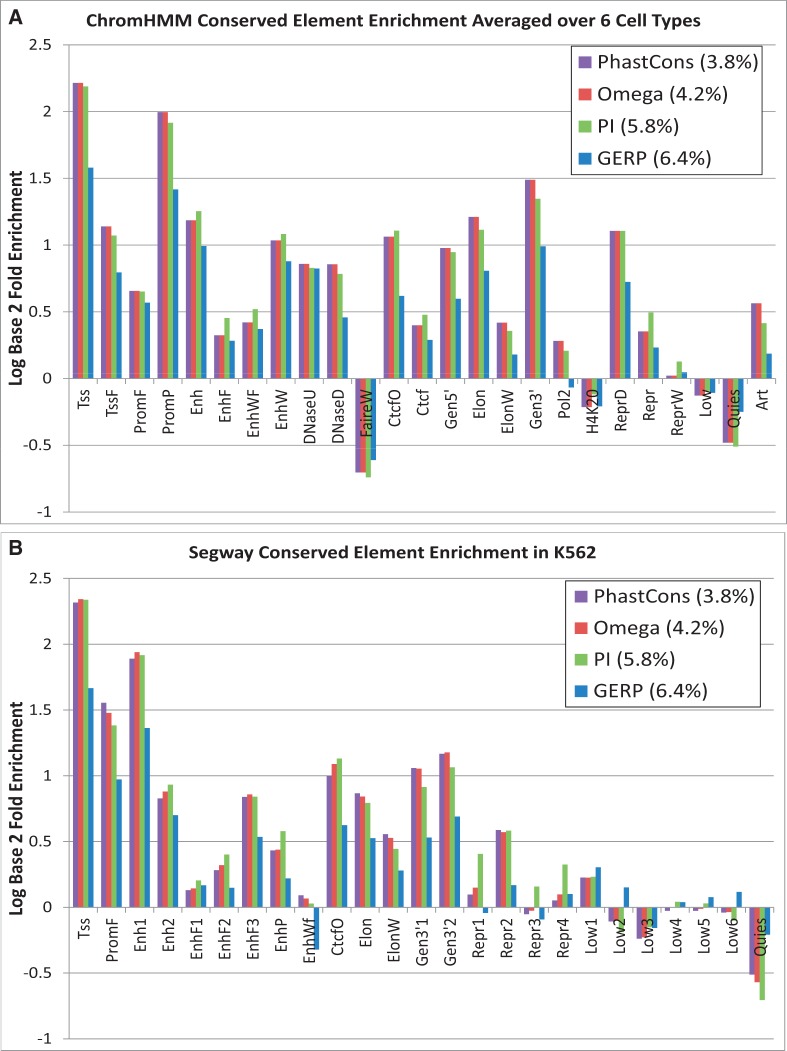
The apparent reduced functionality of these regions, as indicated by the limited transcriptional activity and absence of histone modifications, suggests that most of these sequences are evolving like neutral DNA, and indeed, we find that the DNA in quiescent states is depleted of constrained sequences (Figure 5). It is likely that the DNA in this low activity, quiescent state is similar to the ‘black’ state previously described in Drosophila melanogaster (9).
Figure 5.
(A) Average log2 enrichment or depletion of four different conserved element sets—PhastCons (33), SiPhy-ω, SiPhy-π (16,35), and GERP (34)—for the 25 ChromHMM states averaged across all 6 cell types. (B) The same comparison for Segway states, but restricted to the K562 segmentation.
One would expect that some DNA regions are quiescent only in specific cell types or conditions, being activated in other cell types in response to appropriate signals. Other regions could become dormant and not used again, such as for terminally differentiated cells. Still other DNA could be quiescent in all cell types. Indeed, we find that 635 Mbp of genomic DNA (21% of the genome) is in the ChromHMM quiescent state in all 6 cell types examined, including the embryonic stem cell line H1, which may be expected to have a large fraction of its genome active. Of course, some of this DNA may be activated in the many cell types that have not yet been interrogated. It will be informative to observe how small a fraction of genomic DNA remains classified in an apparently constitutively inactive state as the histone modification and transcription profiles are thoroughly examined in a broad set of cell types.
Chromatin state annotations provide support for larger fraction of genome under constraint
The annotations provide an unbiased view of candidate functional non-coding regulatory elements, enabling us to evaluate different methods for measuring human evolutionary constraint. Accordingly, we overlapped the 25-state ChromHMM annotation defined across 6 cell types with 4 different conserved element sets: PhastCons based on 46 mammals (16,33), GERP based on 33 mammals (16,34), SiPhy-ω and SiPhy-π elements defined based on 29 mammals (16,35). GERP identifies conserved elements by identifying locations of highly rejected substitutions. PhastCons and SiPhy-ω detect elements based on the overall substitution rate. SiPhy-π uses a novel strategy to detect conserved elements based on substitution patterns that deviate from the neutral pattern found in bases evolving without selective pressure. SiPhy-π enables detection of constraint for pairs of nucleotides, rather than individual nucleotides at each position. The PhastCons and SiPhy-ω sets are the smallest of the four, covering 3.8% and 4.2% of the bases considered in the ChromHMM annotation, whereas SiPhy-π and GERP are larger, covering 5.8% and 6.4%, respectively.
It was previously noted that many additional bases detected by SiPhy-π were found in non-coding regions (16) without additional supporting evidence for the biological function of these bases. We observe here that PhastCons, SiPhy-ω and SiPhy-π enrich in most of the ChromHMM states that are suggestive of function with no appreciable decline of enrichments in the larger SiPhy-π set (Figure 5, Supplementary Figure S23). This observation demonstrates at a genome-wide scale the functional relevance of the additional constrained elements detected by SiPhy-π (16). In contrast, for the larger GERP element set, we see substantially lower enrichment in most states on average across cell types, suggesting the additional bases in this set may be less functionally relevant (Figure 5). These phenomena were also observed based on a similar analysis using Segway (Figure 5, Supplementary Figure S23).
Chromatin state annotations help interpret disease association signals from GWAS
The majority of genetic variants associated with phenotypes by GWASs are not in protein-coding regions (36). The large-scale annotation of the human genome, including the non-coding portion, by the ENCODE project showed promise as a guide to interpretation of phenotype-associated SNPs (1), and indeed the regions of the genome associated with function by ENCODE data are enriched in phenotype-associated SNPs (17,37,38). Here, we show that the 25-state annotations from both ChromHMM and Segway provide a higher resolution view of the types of function-associated regions that are enriched for genetic variants with potential phenotypic consequences.
The SNPs on a genotyping array that exhibit the most significant associations with a phenotype of interest are called the lead SNPs in a GWAS. These lead SNPs are not expected necessarily to be the functional SNPs, but they should be in linkage disequilibrium with the functional SNP (39). However, the SNPs on the genotyping arrays are enriched for function-associated regions, and many examples are being found of lead SNPs either being a functional SNP or at least very close to it. For example, the ENCODE Project found such an example in the 8q24 locus (1). We have therefore examined the enrichment of the GWAS lead SNPs in the various chromatin states and compared them to a range of null models, i.e., other SNP sets either not expected to be in functional regions or not currently implicated in function. We use the chromatin state annotations based on epigenetic features in six Tier 1–2 human cell types, regardless of the cell types most likely to be involved in disease susceptibility. While it is preferable to use epigenetic data in cell types of close physiological relevance when available, the studies described here and elsewhere (17,37,38) show that phenotype-associated SNPs are enriched in DNA segments associated with function in the Tier 1–2 cell types, perhaps reflecting regions functional in multiple cell types (39). It is possible that an even stronger enrichment would be found in more relevant cell types, and the analysis presented here may reflect a lower bound estimate.
The phenotype-associated lead SNPs were compiled from the GWAS Catalog in the summer of 2011 (36). This collection was filtered to produce a non-redundant set of 4492 SNPs associated with 362 phenotypes (4860 SNP-phenotype associations, some SNPs are associated with more than one phenotype). The distribution of these SNPs in different chromatin states was compared to the distribution of SNPs not expected to be functional, i.e., two largely unbiased genome-wide collections of SNPs generated by whole-genome sequencing of multiple humans (40) and a compilation of SNPs in 24 published individual human genomes (41). The distribution of SNPs on the 1M Illumina genotyping array was also analysed, along with SNPs from the genotyping array matched to the GWAS SNPs for allele frequency in CEU, distance from a transcription start site and location with respect to gene structure (components such as intronic, exonic and intergenic) (37). This latter set of GWAS-matched SNPs is a particularly rigorous comparison, and we determined the enrichment statistics for 1000 random samplings from the GWAS-matched SNP set. These SNPs are not currently associated with phenotype by GWAS, but they could be in or close to functional regions, e.g., those close to transcription start sites.
In an analysis carried out in the GM12878 cell line, we find that the SNPs in the GWAS Catalog are depleted in the quiescent segments compared to the population and individual SNPs (1000 Genomes and 24 personal genomes) (Figure 3). In contrast, the phenotype-associated SNPs are enriched in many function-associated chromatin states compared to the population and individual SNPs. The genotyping SNPs show a similar pattern to that of the GWAS SNPs, likely reflecting a bias in the latter set for functional regions. We therefore focused on comparing the GWAS-matched SNPs to the GWAS SNPs as a stringent test for significance of the enrichment or depletion. We sampled these matched SNPs from genic regions and the matched SNPs show an even greater enrichment in function-associated chromatin states than do the SNPs on the genotyping arrays (Figure 3). Because 1000 random samplings of the GWAS-matched SNPs were evaluated, we determined whether the observed enrichment for the GWAS SNPs exceeded that found for the lower 950 samplings of the matched SNPs (or conversely, was less than that found for the upper 950 samplings in the case of depletion), establishing an empirical P-value threshold of 0.05. The Segway chromatin states passing this threshold (denoted by red bars in the figure) for significant enrichment for GWAS SNPs are Enh, EnhF, EnhWf, Repr2 and Repr3. The categories showing significant depletion are Quies, ElonW, Low1 and Low6. For ChromHMM, states Enh, EnhF, EnhW, EnhWF, TssF and ReprW showed significant enrichment while states Quies and ElonW showed significant depletion.
The NOD2 locus provides an illustrative example of the use of chromatin states for interpretation of phenotype-associated SNPs. This gene has been associated with Crohn’s disease (42,43), but several of the SNPs map into non-coding regions (Figure 3). Three of these SNPs fall into chromatin states associated with enhancers in GM12878 (leftmost), enhancers in HUVEC (middle) and a transcribed region in GM12878 (rightmost outlined in blue). The three highlighted GWAS SNP clusters are close to DNase hypersensitive sites in a lymphoblastoid cell line (GM12878), T-cells (Th2 and Treg cells) and HUVECs, among others. Specific transcription factors binding to these sites are NF-YA, NF-YB, PU.1, SRF, GATA2 and c-Fos, with distinct factors binding to different sites in specific cell lines. All of these observations can be used to formulate testable hypotheses about the genetic variants associated with Crohn’s disease. For example, a SNP can affect the affinity for one of these transcription factors in an allele-specific manner, leading to alterations in the level of expression of NOD2 and affecting the inflammatory response associated with Crohn’s disease. We note that the DNase sensitivity data include T-cells, which could be implicated in an autoimmune disease such as Crohn’s disease. The GATA2 binding data from HUVEC could be pointing to potential sites of occupancy by GATA3, which regulates expression of many genes in T-cells. This is an example of a hypothesis derived from data from the currently studied ENCODE lines that can be readily tested by direct experiments in cell types more relevant to the phenotype.
DISCUSSION
The chromatin state annotations provided here provide a foundation for interpreting the non-coding portion of the human genome that has so far been difficult to comprehend. For example, while promoter and enhancer regulatory elements contain within them regulatory sequence motifs, their purely sequence-driven identification has remained an unsolved challenge in genomics, while our segmentations provide their systematic annotation independent of the motifs they contain. Similarly, while genomic regions showing extreme sequence conservation across related species frequently show enhancer activity, such extreme conservation is not a general property of enhancer regions, while our systematic annotation contains both conserved and non-conserved elements. Chromatin signatures provide a general approach for discovering active regulatory elements based on their biochemical properties, and beyond regulatory elements, an unbiased genome-wide view of the likely functional roles of every region of the genome. By capturing both high-continuity segments and high-resolution transitions between them, we provide a summary of dozens of genome-wide datasets into a directly interpretable and information-rich resource.
A reader might wonder which chromatin state annotation to use. One significant difference, evident in Supplementary Figure S9, is the relative sizes of the segments, with Segway producing smaller segments on average compared to ChromHMM. Beyond this distinction—high resolution for Segway and high continuity for ChromHMM—the chromatin state annotations exhibit many subtle differences, and each has distinct advantages in different applications. We therefore recommend that the user examine both annotations in regions of interest, because each might capture a different aspect of the underlying biology. As for the joint segmentation, it is not meant to replace the two primary annotations, because their differences can be important and should not be overlooked. Instead, the joint segmentation is meant to help introduce a new user to our chromatin states in a simple and approachable way. Once the user is familiar with our annotations, however, we recommend browsing the joint segmentation in parallel with the two primary chromatin state annotations, to exploit the full richness of the chromatin landscape.
It is important to note that the number of chromatin states presented here was chosen as a compromise between capturing all of the potential complexity of chromatin mark combinations (which requires very large numbers of states) and generating models that are easily interpretable and maximally useful for interpreting genomic features (which requires maintaining a small number of states). In our experience, model selection methods such as the use of the Bayesian information criterion or Akaike information criterion invariably suggest models with higher number of states beyond the point of feasibility for human interpretation. The penalties imposed by these criteria prove insufficient to overcome the increase in likelihood due to increased numbers of states and the consequent increased numbers of parameters. As such, we focused on a relatively small number of 25 states, that is well suited to the number of chromatin marks and other input data tracks that were available, and that allows us to annotate the likely functional roles of each chromatin state. However, as the number of chromatin properties that can be elucidated on a genome-wide scale increases, we expect that additional chromatin states will be discovered. These states will likely provide further functional subdivisions of the states presented here or reveal new types of chromatin elements that we may not even suspect yet.
In addition to the combinations of chromatin marks summarized in our models’ emission parameters, the topology of the graph representing allowed transitions between labels contains important information about their genomic relationships. The transition probability matrices (Supplementary Figure S7) capture the structure of these connections. For example, transitions between Tss and TssF or Prom labels occur more frequently, revealing their genomic proximity. In ChromHMM, Low and Quies states both have similar emission profiles with low probabilities of histone modifications, but show distinct transition probabilities and also distinct biological enrichments. In contrast, the transition probabilities have a relatively small impact on the final segmentation for Segway, relative to the emission distributions. This is primarily because the minimum segment length is 100 bp, and thus the Segway Viterbi path probability includes >100 emission probabilities for every transition probability. Indeed, we have observed that various manipulations of a model's transition probability distribution have relatively small impact on the resulting segmentation (data not shown).
Continued methodological improvements will likely be needed to capture additional types of chromatin information, such as three-dimensional interactions between distal regions, looping of chromatin domains or larger-scale region behavior. Thus, the chromatin states presented here may constitute the words of much more complex chromatin sentences whose grammar remains to be elucidated, as new technologies enable deeper and more complex epigenomic maps.
We have tried to illustrate in this article, and in related companion papers, the many applications of the chromatin state annotations, ranging from revealing new genes and functional elements, to interpreting disease datasets, to measuring allele-specific activity or human selection. As genome data have become personalized, we expect that the epigenomes of the future will be genotype and individual specific, and not just cell-type specific. These types of data can have profound implications for understanding the epigenomic consequences of disease, not just its genomic predisposition, and chromatin states will form a necessary component of personal epigenomes. Beyond the applications that we have explicitly listed, however, there are many others that we have not even begun to explore, and we expect these applications to be as rich and diverse as the countless uses of an encyclopedia of genomic knowledge, or a map for navigating our genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7, Supplementary Figures 1–23, Supplementary Methods, Supplementary Results and Supplementary References [44–62].
FUNDING
National Institutes of Health [HG004695 to E.B., HG006259 to M.M.H., HG005334 and HG004570 to M.K., DK065806 and HG005573 to R.C.H.]; National Science Foundation [0905968 to J.E.]. Funding for open access charge: National Human Genome Research Institute.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweikert G, Zien A, Zeller G, Behr J, Dieterich C, Ong CS, Philips P, De Bona F, Hartmann L, Bohlen A, et al. mGene: accurate SVM-based gene finding with an application to nematode genomes. Genome Res. 2009;19:2133–2143. doi: 10.1101/gr.090597.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abeel T, Van de Peer Y, Saeys Y. Toward a gold standard for promoter prediction evaluation. Bioinformatics. 2009;25:i313–i320. doi: 10.1093/bioinformatics/btp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S, Li Q, Liu J, Zhou XJ. A novel computational framework for simultaneous integration of multiple types of genomic data to identify microRNA-gene regulatory modules. Bioinformatics. 2011;27:i401–i409. doi: 10.1093/bioinformatics/btr206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007;17:917–927. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaschek R, Tanay A. Spatial clustering of multivariate genomic and epigenomic information. RECOMB. 2009;5541:170–183. [Google Scholar]
- 12.Lian H, Thompson WA, Thurman R, Stamatoyannopoulos JA, Noble WS, Lawrence CE. Automated mapping of large-scale chromatin structure in ENCODE. Bioinformatics. 2008;24:1911–1916. doi: 10.1093/bioinformatics/btn335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 19.Vandenbergh DJ, James-Pederson M, Hardison RC. An apparent pause site in the transcription unit of the rabbit α-globin gene. J. Mol. Biol. 1991;220:255–270. doi: 10.1016/0022-2836(91)90011-t. [DOI] [PubMed] [Google Scholar]
- 20.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 23.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 27.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 29.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 32.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 33.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardison RC. Genome-wide epigenetic data facilitate understanding of disease susceptibility association studies. J. Biol. Chem. 2012;287:30932–30940. doi: 10.1074/jbc.R112.352427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 43.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 44.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kundaje A, Jung YL, Kharchenko PV, Wold BJ, Sidow A, Batzoglou S, Park PJ. Adaptive calibrated measures for rapid automated quality control of massive collections of ChIP-seq experiments. 2012 In preparation. [Google Scholar]
- 47.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 48.Kundaje A, Li Q, Rozowsky J, Brown JB, Harmanci A, Wilder S, Gerstein M, Batzoglou S, Sidow A, Birney E, et al. Reproducibility measures for adaptive thresholding and quality control of ChIP-seq experiments. 2012 In preparation. [Google Scholar]
- 49.Smit AFA, Hubley R, Green P. 1996 RepeatMasker Open-3.0. http://www.repeatmasker.org/ [Google Scholar]
- 50.Buske OJ, Hoffman MM, Ponts N, Le Roch KG, Noble WS. Exploratory analysis of genomic segmentations with Segtools. BMC Bioinform. 2011;12:415. doi: 10.1186/1471-2105-12-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7:S4. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuan D, Feingold E, Newman M, Weissman SM, Forget BG. Different 3′ end points of deletions causing δβ-thalassemia and hereditary persistence of fetal hemoglobin: implications for the control of γ-globin gene expression in man. Proc. Natl. Acad. Sci. USA. 1983;80:6937–6941. doi: 10.1073/pnas.80.22.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feingold EA, Forget BG. The breakpoint of a large deletion causing hereditary persistence of fetal hemoglobin occurs within an erythroid DNA domain remote from the beta-globin gene cluster. Blood. 1989;74:2178–2186. [PubMed] [Google Scholar]
- 57.Anagnou NP, Perez-Stable C, Gelinas R, Costantini F, Liapaki K, Constantopoulou M, Kosteas T, Moschonas NK, Stamatoyannopoulos G. Sequences located 3′ to the breakpoint of the hereditary persistence of fetal hemoglobin-3 deletion exhibit enhancer activity and can modify the developmental expression of the human fetal Aγ-globin gene in transgenic mice. J. Biol. Chem. 1995;270:10256–10263. doi: 10.1074/jbc.270.17.10256. [DOI] [PubMed] [Google Scholar]
- 58.Feingold EA, Penny LA, Nienhuis AW, Forget BG. An olfactory receptor gene is located in the extended human β-globin gene cluster and is expressed in erythroid cells. Genomics. 1999;61:15–23. doi: 10.1006/geno.1999.5935. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hintze JL, Nelson RD. Violin plots: a box plot-density trace synergism. Am. Stat. 1998;52:181–184. [Google Scholar]
- 61.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.