Abstract
The degradation of most eukaryotic mRNAs is initiated by removal of the poly(A) tail, and the major deadenylase activity is associated with the CCR4/CAF1/NOT complex (NOT complex). We here study the role of CNOT10, a protein that is found in human and trypanosome, but not in yeast, NOT complexes. Trypanosome (Tb) CNOT10 is essential for growth. TbCNOT10 interacted with the deadenylase TbCAF1 and the scaffold protein TbNOT1; TbCAF1 also interacted with TbNOT1 in a yeast two-hybrid assay. In both trypanosomes and human embryonic kidney cells, approximately half of CAF1 was associated with the NOT complex. Depletion of CNOT10 from human cells did not affect this association. In contrast, depletion of TbCNOT10 in trypanosomes caused a decrease in the level of TbNOT1, detachment of TbCAF1 from the complex and pronounced stabilization of most trypanosome mRNAs. Artificial tethering of TbCAF1 to a reporter mRNA in vivo resulted in mRNA degradation, and this was not affected by TbCNOT10 depletion. We conclude that in trypanosomes, TbCNOT10 may stabilize the interaction between TbCAF1 and the NOT complex. The results further suggest that TbCAF1 is only able to deadenylate mRNA in vivo if it is recruited to the mRNA through other NOT complex components.
INTRODUCTION
The degradation of mRNAs is important for control of gene expression. In eukaryotes, the degradation of most mRNAs is initiated by removal of the poly(A) tail. Decay then proceeds either from the 3′ end, by the exosome, or from the 5′ end by decapping and subsequent digestion by the exoribonuclease XRN1 (1). Three cytosolic deadenylases have been described in eukaryotes: the poly(A) ribonucleases; the poly(A) nuclease Pan2 with its co-factor Pan3; and CCR4 and CAF1, both of which are part of the CCR4–CAF1–NOT complex (hereafter simply called ‘the NOT complex’) (2). The NOT complex has been found to be the most important effector of mRNA deadenylation (3–5). In addition, it has been implicated in the regulation of transcription, the DNA damage response, nuclear surveillance, mRNA export and translation repression (6).
In all eukaryotes examined so far, the NOT complex consists of at least seven subunits. In yeast, the universally conserved subunits are called Caf1p, Caf40p and Not1p, Not2p and Not5p (5,7–11). Not1p (human CNOT1, Drosophila and trypanosome NOT1) acts as a platform for the remaining core subunits. In human cells, it is also recruited by GW182/TNRC6 for miRNA-induced mRNA decay (12) and by Tis11 proteins, which mediate degradation of mRNAs with AU-rich elements (6,13). The central role of NOT1 as a scaffold is shown by the fact that in human cells and yeast, its depletion disrupts the complex and abrogates mRNA deadenylation (14,15). The major deadenylase activity in the human, Trypanosoma and Drosophila complexes is CAF1; there are two paralogues in humans, called Caf1a/b or CNOT7 and CNOT8. Animal cells and yeast have an additional deadenylase called CCR4 or Ccr4p; in yeast, Ccr4p is more active than Caf1p (4–6). Recent crystal structures for relevant fragments of the yeast and human Not1 and Caf1 proteins have shown that they are held together mainly through hydrophobic interactions (16,17).
The functions of the remaining core subunits are not clear. Yeast has two proteins equivalent to NOT5: Not3p and Not5p. Although these are 44% identical, the corresponding mutant strains showed specific gene expression changes, suggesting that the two subunits have distinct roles (15). NOT2 (human CNOT2) is important for complex integrity, as it seems to link NOT3/5 to NOT1 (15,18,19). The role of Caf40p (human Rcd-1) is unknown, although Rcd-1 can dimerize and bind single-stranded DNA (20). In yeast, Not4p is associated with the NOT complex (21); it is an E3 ligase involved in degrading aberrant polypeptides that lack a stop codon (22) and may act independently of the other subunits. The homologues in human cells (CNOT4) and Drosophila are not stably associated with the complex (9,10) and there is no homologue in trypanosomes (5).
The remaining subunits are less widely distributed in evolution. Caf130p, which is found exclusively in fungi, interacts with the N- and C-termini of Not1p, but is not important for the stability of the complex (15,23). Two more subunits have been identified in human cells: TAB182 and C2ORF29 (9). This article concentrates mainly on CNOT10, which has been described in human cells but is absent from fungi. Tandem affinity purification (TAP) of human CNOT10 retrieved all core subunits of the complex, but not the deadenylases (9); apart from that nothing is known about CNOT10 function.
The parasite Trypanosoma brucei, and related kinetoplastid protists, have an exceptional genome organization. Protein-coding genes are arranged in large polycistronic units transcribed by RNA polymerase II (24). Cleavage and maturation of the precursor mRNA are initiated by trans-splicing, which places identical leader sequences at the 5′-end of every mRNA; this is coupled to polyadenylation of the mRNA immediately upstream (25,26). As a consequence of this gene arrangement, the parasite lacks transcriptional control of individual protein-coding genes. Transcript levels are determined mainly by gene copy number and mRNA degradation rates (27). We have previously shown that the degradation of most trypanosome mRNAs is initiated by deadenylation (27). TAP of T. brucei CAF1 identified all core subunits of the complex, including a distant relative of Homo sapiens (Hs) CNOT10 (5). In this report, we analyse the role of TbCNOT10 in trypanosomes and compare some results with those seen for its counterpart in human cells.
MATERIALS AND METHODS
Cell culture
The experiments were done with bloodstream-form T. brucei that stably express the tetracycline repressor, with or without T7 polymerase expression. Culturing, transfections and RNAi experiments were conducted as described earlier (28). mRNA decay was analysed after 24 h of RNAi targeting TbCNOT10. Cells were treated with Sinefungin (final concentration 2 µg/ml) for 5 min, and then actinomycin D was added to a final concentration of 10 µg/ml.
Human embryonic kidney (HEK) 293 cells were cultured in Dulbecco’s-modified Eagle medium (DMEM) containing 10% fetal bovine serum (PAA Laboratories), 2 mM l-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin (all PAN Biotech) at 37°C/5% CO2. Cells were transfected with DNA using polyethyleneimine (PEI) (Polysciences Europe; 1 mg/ml, pH 7.0) at a ratio of 1:2 (DNA:PEI) in serum-free DMEM without antibiotics. For transfection of siRNAs, Lipofectamine RNAimax (Invitrogen) and Optimem (Gibco) were used according to the manufacturer’s protocol. siRNAs were transfected at a concentration of 100 nM with Lipofectamine RNAimax twice over a time period of 4 days. Medium was changed to regular DMEM 4 h after transfection of DNA.
Lists of plasmids, primers and siRNAs used in this article are in Supplementary Table S1.
RNA extraction and northern blot analysis
Total RNA was extracted using peqGold Trifast (peqLab, Germany). RNA was run on 1.5% formaldehyde gels or 4.5% polyacrylamide-urea gels and blotted onto Nytran membranes (GE Healthcare). RNase H digestion with oligo d(T) was adapted from the method described previously (5). Ten micrograms of RNA was mixed with 170 pmol oligo d(T) and RNAse H buffer (New England Biolabs) and filled up with water to 50 µl. The mixture was denatured at 85°C for 5 min, incubated at 42°C for 10 min, then further cooled over 15 min at 32°C. One microlitre of RNasin® RNase Inhibitor (Promega) and 5 U RNaseH (New England Biolabs) were added and the reaction incubated at 37°C for 1 h.
Northern blots were hybridized with radioactively labelled DNA (Prime-IT RmT Random Primer Labelling Kit, Stratagene) or RNA (MAXIscript, Ambion) probes. Signals were measured using a phosphorimager. For trypanosome RNA, either the signal of the signal recognition particle (7SL) or a large subunit rRNA (LSU1) was used for normalization. The half-lives were estimated using only those segments of the time course that gave exponential decay curves (fitted with a linear correlation coefficient generally exceeding 0.9).
Co-immunoprecipitation
To make trypanosome extracts, 5 × 107 trypanosomes were re-suspended in immunoprecipitation (IP) lysis buffer (10 mM NaCl, 10 mM Tris–HCl, pH 7.5, 0.3% IGEPAL and protease inhibitors [Protease Inhibitor Mixture {ethylenediaminetetraacetic acid (EDTA)-free}; Roche Applied Science]) and passed five times through a 27-gauge syringe. The resulting extracts were centrifuged for 20 min at 4°C, at 16 000 × g. The supernatant was transferred into a new tube and the salt concentration was adjusted to 180 mM NaCl. To test the RNA dependence of interactions, RNaseA was added to the lysis and wash buffers at a concentration of 200 µg/ml, with or without RNasin® Ribonuclease inhibitor (Promega) at a concentration of 48 µg/ml.
For IP, 30 µl of anti-V5-antibody- or anti-c-myc-antibody-coupled agarose (both from Bethyl Laboratories) was washed four times with 1× phosphate buffered saline (PBS) and once with IP lysis buffer adjusted to 180 mM NaCl at 4°C, with centrifugation at 0.9 × g for 2 min. A sample equivalent to 5 × 105 cells was taken from the input lysate and 2× Laemmli buffer was added. The remainder of the lysate was then added to the beads, and the mixture incubated for 1–1.5 h at 4°C with rotation. After centrifugation, the supernatant was transferred to another tube and a sample equivalent to 5 × 105 cells was again taken and 2× Laemmli buffer was added. The remaining beads were washed four times for 5 min at 4°C with salt-adjusted lysis buffer, with intermediate centrifugation, then boiled in 2× Laemmli buffer.
For human cells, 24 h after transient transfection, HEK293 cells from a confluent 10 cm dish were collected and lysed in 400 μl ice-cold RNA-IP lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1 mM MgCl2, 2% NP-40 and 10% glycerol with freshly added protease inhibitors [Complete; Roche]). Nuclei were removed by centrifugation at 500 × g for 5 min at 4°C. A total of 30 μl of streptavidin sepharose beads (GE Healthcare) was added for an additional 1.5–2 h and washed six times in NET2 buffer (50 mM Tris [pH 7.5], 150 mM NaCl and 0.5% Triton X-100). Protein complexes were eluted with 50 μl sodium dodecyl sulphate (SDS) sample buffer with 100 mM dithiothreitol (DTT). Proteins were resolved on 5–20% gradient polyacrylamide gels and transferred onto a 0.2-μm pore size nitrocellulose membrane (Peqlab) for western blotting. Horseradish peroxidase-coupled secondary antibodies (Jackson Immunoresearch) in combination with Western Lightning-enhanced chemiluminescence substrate (Perkin Elmer) were used for detection. Streptavidin sepharose beads (GE Healthcare) were used to purify myc-Strep-tagged (myc-SG) proteins.
Two-hybrid analysis
Complete trypanosome open reading frames (ORFs) were cloned into pGADT7 (GAL4 activation domain vector) and pGBKT7 (GAL4 DNA-binding domain vector) (Matchmaker 3 System, Clontech). The two-hybrid yeast strain AH109 was then transformed with these plasmids in all possible combinations. The expression of the fusion protein was confirmed by western blot analysis using antibody to the HA epitope of the GAL4 activation domain fusion proteins, and antibody to the c-myc epitope of the GAL4 DNA-binding domain fusions. Transformants were selected on four drop-out (SD/-Trp/-Leu/-His/-Ade) culture plates after 4 days of incubation at 30°C, and the resulting positive clones were assayed for β-galactosidase activity using a colony-lift filter assay. Alternatively, reporter activation was tested by replica plating on triple drop-out plates: SD/-Trp/-Leu/-His (medium stringency). As negative controls for self-activation, we used combinations of the TbNOT-complex subunits with CLONTECH vectors pGAD-T-antigen and pGBKT7-p53. Further, p53 (pGBKT7-p53) and SV40 large T-antigen (pGADT7-T) cotransformants were used as positive controls, whereas lamin C (pGBKT7-Lamin) and pGADT7-T cotransformants were used as negative controls.
Glycerol gradients
Trypanosomes (3 × 107), or HEK293 cells from a confluent 10 cm dish, were washed with 1 × PBS, then frozen in liquid nitrogen and stored at −80°C until use. The cell pellets were thawed on ice and re-suspended in ice-cold buffer containing 10 mM Tris–HCl, pH 7.6, 10 mM NaCl, 10 mM MgCl and protease inhibitors (Protease Inhibitor Mixture [EDTA-free]; Roche Applied Science), then forced five times through a 27-g needle. IGEPAL CA-630 (Sigma) was added to a final concentration of 0.1% and the cells were again forced through the needle five times. The lysate was centrifuged first for 20 min at 4°C, 10 000 × g, then the supernatant was spun for 1 h at 100 000× at 4°C. The final supernatant was layered onto a 12-ml 10–30% glycerol gradient (200 mM KAc, 20 mM Tris–HCl, pH 8.0, 5 mM MgAc, 1 mM DTT and protease inhibitors [Protease Inhibitor Mixture {EDTA-free}; Roche Applied Science]), which was then spun at 229 900 × g for 17 h at 4°C. Twenty-seven fractions were taken from the top at 23-s intervals. The fractions were incubated on ice overnight with 15% trichloroacetic acid, 0.1% DOC and 10 µg bovine serum albumin (BSA). Precipitates were washed with acetone and re-suspended in Laemmli buffer.
Tandem affinity purification
Trypanosomes 2.5 × 1010 were harvested and used for TAP as previously described (29). The eluate was run 1 cm into a 10% SDS–polyacrylamide gel electrophoresis (PAGE) resolving gel and stained with colloidal Coomassie. The protein-containing gel area was cut into five slices, which were analysed by mass spectrometry.
Protein detection
Trypanosomes or HEK cells were washed once with PBS then dissolved in 2× Laemmli buffer. Protein samples were run on a SDS–PAGE, then blotted onto an Optitran BA-S 85 reinforced NC 0.45 µm membrane (Whatman, Dassl, Germany). The blots were probed with appropriate antibodies and then developed with Western Lightning®-ECL solution (Perkin Elmer).
Immunofluorescence (IF) microscopy was conducted as described earlier in (5). The following antibodies were used in western blotting: monoclonal mouse Anti-V5 (Santa Cruz), monoclonal mouse anti c-myc (Santa Cruz), monoclonal mouse anti-GFP (Santa Cruz), ECL™ Anti-Rabbit IgG (GE Healthcare), ECL™ Anti-Mouse IgG (GE Healthcare), polyclonal rabbit antibodies anti-myc (A-14, sc-789, Santa Cruz), anti-HsCNOT10 (15938-1-AP, Protein Tech) and anti-HsCNOT1 (14276-1-AP, Protein Tech); polyclonal anti-trypanosome aldolase (30). Polyclonal rabbit anti-Caf1a was kindly provided by Ann-Bin Shyu (University of Texas, Houston, TX, USA) and monoclonal antibody to trypanosome tubulin was from Keith Gull (31).
We expressed recombinant His-tagged TbCNOT10 (full length) in Escherichia coli but it was very poorly soluble and we were unable to elute it specifically from the nickel column. A preparation of adequate quality for immunization was not obtained.
Database searches
To find potential homologues of Tb927.10.8720, (old number Tb10.6k15.1820), the protein sequence was analysed by BLASTp, psi-BLAST and tBLAST (32). We used standard parameters and excluded kinetoplastids (taxid:5653) from the search. Domains were identified by a SUPERFAMILY search (33). To find conserved regions in the protein sequence, multiple sequence alignments were done with ClustalW.
RESULTS
Phylogenetic sequence analysis of TbCNOT10
The protein encoded by trypanosome locus Tb927.10.8720 was previously identified as a potential component of the NOT complex through TAP of TbCAF1 (5). It is 555 amino acids long, with tetratricopeptide repeat (TPR)-like domains in positions 199–313 and 471–550. BLASTp and psi-BLAST searches on the whole Genbank database (excluding kinetoplastids) identified, as matches, >40 orthologues of CNOT10 in plants and animals, with E-values ranging from 3 × 10−4 to 1 × 10−7. The regions that matched covered 20–60% of the proteins, with identities of 20–30%. Reciprocal BLASTp of human CNOT10 (HsCNOT10) against the T. brucei genome yielded Tb927.10.8720 as the best match (E-value 3 × 10−4). Most matches with other CNOT10s were located in the beginning of the first TbCNOT10 TPR domain (Figure 1A). We also did directed BLASTp searches using human or trypanosome CNOT10 to probe different eukaryotic phyla and kingdoms: we were unable to find equivalents to CNOT10 in Apicomplexa, Diplomonadida, Alveolates and fungi, but it is present in plants. We concluded that Tb927.10.8720 might be homologue of human CNOT10, so we provisionally named it TbCNOT10.
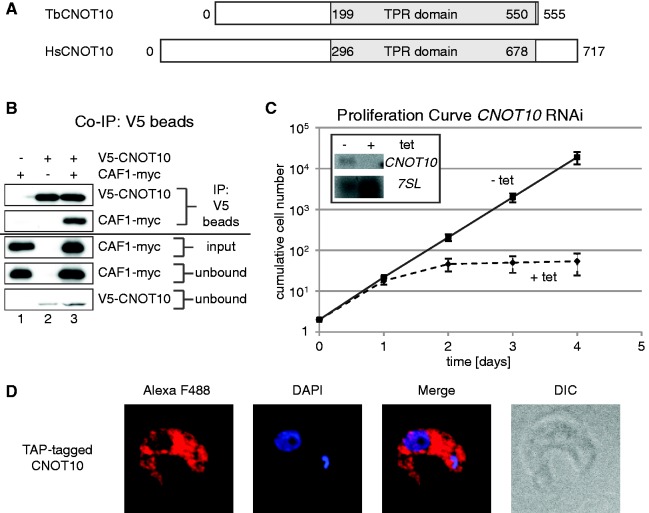
Figure 1.
TbCNOT10 is an essential and cytoplasmic protein in T. brucei. (A) Cartoon comparison of T. brucei and human CNOT10s. The conserved TPR domains are in grey. (B) Using anti-V5 beads, V5-TbCNOT10 was pulled down from extracts of trypanosomes expressing TbCAF1-myc (lane 1) V5-TbCNOT10 (lane 2) or both tagged proteins (lane 3). A western blot was probed to detect the myc and V5 tags. The top two panels show the precipitated V5-TbCNOT10 or TbCAF1-myc from the three different cell lines. The next panel below is 1% of the input extract, and the two bottom panels are the unbound fractions. (C) Proliferation was measured in trypanosomes with tetracycline-inducible RNAi targeting TbCNOT10 mRNA. Diamonds and dashed line represent TbCNOT10 RNAi and black boxes and solid line represent uninduced cells. The northern blot (inset) shows the efficiency of RNAi after 24 h, with 7SL RNA as control. (D) TAP-tagged TbCNOT10 was detected by IF using rabbit polyclonal anti-protein A at 1:50 000. DAPI indicates the positions of the nucleus and kinetoplast (mitochondrial DNA). Differential interference contrast (DIC) microscopy shows the whole parasite.
TbCNOT10 is a component of the NOT complex
To confirm TbCNOT10 as a member of the NOT complex, we conducted IPs. We used trypanosomes expressing tagged versions. The TbCNOT10 gene was tagged in situ, by homologous recombination with a sequence encoding an N-terminal V5-tag. TbCAF1 with a C-terminal myc tag (CAF1-myc) was expressed from a tetracycline-inducible promoter. A pull-down of V5-tagged TbCNOT10 (V5-TbCNOT10) resulted in coprecipitation of TbCAF1-myc (Figure 1B) and vice versa (Supplementary Figure S1). RNaseA treatment did not disrupt the interaction (not shown).
In order to identify further proteins that are stably associated with TbCNOT10, we generated trypanosomes in which a TAP-tag-coding sequence was integrated directly upstream of the TbCNOT10 ORF. In IF microscopy, TAP-tagged TbCNOT10 was detected in a rather granular pattern in the cytoplasm (Figure 1D). After TAP of TbCNOT10, all associated proteins were analysed by mass spectrometry (Supplementary Table S2). We found all previously known members of the trypanosome NOT complex except TbCAF1; they included the uncharacterized Tb927.8.1960 protein, which had been seen in the previously published purification (5). These results confirmed that TbCNOT10 is a stable component of the trypanosome NOT complex. Previous results from human cells were similar: after purification of the TAP-tagged HsCNOT10, neither HsCNOT6/6L (CCR4) nor HsCNOT7 (CAF1a) was found (9). TbCAF40 (Tb927.4.410), which had not been found in our published purification using TAP-tagged CAF1 (5), was detected in the CNOT10 preparation. We also found TbDHH1 (as seen previously with TbCAF1).
There was also a number of novel potential NOT-associated proteins. These included three ATP-dependent DEAD/H RNA helicases (Tb927.3.2600, Tb927.8.1510 and Tb09.211.3510), two of which are potential homologues of yeast Ded1p and Dbp2p (34). We have, however, found the putative Ded1p homologue (Tb09.211.3510) in several other purifications, so the specificity of the interaction is unclear. PUF2 was pulled down, but subsequent co-IPs failed to show a specific interaction (not shown). The only other interaction of note was with a potential homologue of the yeast karyopherins Pse1p and Kap123p, encoded by Tb11.01.7010; this may have parallels in human cells since some components of the human NOT complex interact with the nuclear pore (9). Additional possible interaction partners included kinetoplastid-specific proteins of unknown function (see Supplementary Table S2).
Most NOT complex components are essential for the survival of T. brucei
To see whether TbCNOT10 was essential in trypanosomes, we made permanent cell lines with tetracycline-inducible RNAi targeting TbCNOT10 mRNA. Using the parasite form that lives in the mammal (bloodstream forms), we observed a severe proliferation defect within 36 h after RNAi induction (Figure 1C). In the slower growing procyclic form, a parasite stage that multiplies in Tsetse flies, it took ∼60 h to observe a decrease in cell numbers. For comparison, we knocked down TbNOT2 (Tb927.6.850), TbNOT3/5 (Tb927.3.1920) and TbCAF40 (Tb927.4.410) RNAs. Proliferation defects were seen for TbNOT3/5 and TbCAF40, but not for TbNOT2 (Supplementary Figure S2). In contrast, in the published high-throughput RNAi screen, TbNOT2 RNAi also gave a growth defect (35). Our negative result for TbNOT2 therefore could have been due to inadequate RNAi. The high-throughput screen (35) revealed growth defects for Tb927.8.1960 only during differentiation, and we too saw no defect after bloodstream-form RNAi (not shown).
Knowing that CNOT10 was essential, we could confirm the functionality of TAP-tagged CNOT10. In the line expressing the tagged version, we knocked out the remaining wild-type gene and found that the cells grew normally.
Depletion of TbCNOT10 inhibits mRNA turnover and deadenylation
We next investigated the effect of TbCNOT10 depletion on mRNA degradation, by inhibiting mRNA synthesis and processing, then measuring mRNA levels by northern blotting. Since all trypanosome mRNAs have the 39mer-spliced leader at the 5′-end, the turnover of bulk mRNA can be assessed by hybridization of northern blots with a spliced leader probe (36). Using this method with wild-type trypanosomes, half of the mRNA remained after 30 min. After depletion of CNOT10, however, virtually no mRNA degradation was seen (Figure 2A). This indicated that TbCNOT10 was important for overall mRNA degradation. Intriguingly, the spliced leader precursor (arrow) also appeared to have been stabilized.
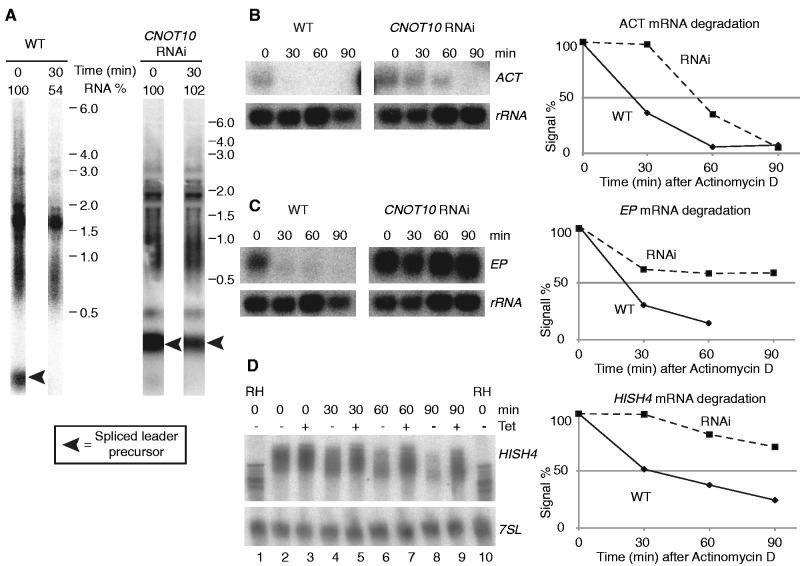
Figure 2.
Effect of TbCNOT10 depletion on mRNA degradation in trypanosomes. (A) Bulk mRNA turnover of wild-type cells and TbCNOT10-depleted cells was analysed by hybridization of northern blots with a spliced leader probe. Transcription was inhibited and the signal was measured for two time points (T = 0 and 30 min). In each case, the signal of rRNA was used as a loading control. The northern blots from T = 0 and 30 min originate from the same gel, but were cut due to the loading of other samples in between. (B–D) TbCNOT10 was depleted for 36 h by RNAi in Tb, then transcription was inhibited. RNA was prepared 0, 30, 60 and 90 min after the addition of actinomycin D. Wild-type cells were used as control. RNA levels were measured from northern blots probed for ACT (B), EP (C) and HISH4 (D) mRNAs. In each case, the signal of rRNA was used as a loading control. The mean signal for ACT normalized to rRNA (n = 6) was plotted. The dashed line and square box represent RNAi and, diamonds and line represent wild type. The same was done for HISH4 (n = 4) and for EP (n = 6). Sample agarose gels used for the half-life measurement are shown in (B) and (C). For HISH4 (D, labelled as H4), we instead show a blot from a 4.5% denaturing polyacrylamide gel; mRNAs from cells with and without RNAi are alternately loaded in order to enable mRNA length comparisons. For lanes 1 and 10, RNA was treated with RNaseH and oligo d(T) in order to remove the poly(A) tails.
Next, we studied three individual mRNAs, encoding actin (ACT), EP procyclin (EP) and histone H4 (HISH4). Degradation of each was inhibited (Figure 2B–D). In TbCNOT10-depleted trypanosomes, both degradation of ACT mRNA was delayed. Moreover, the steady-state level of ACT increased ∼6-fold after TbCNOT10 depletion (Figure 2B). These effects were even more marked for EP mRNA, with the steady-state level increasing 3–4-fold (Figure 2C). We previously showed that EP mRNA degradation is biphasic, with an initial rapid degradation mediated by the 5′–3′ pathway, followed by a deadenylation-dependent phase which was inhibited by CAF1 RNAi (37). Exactly the same was seen now for CNOT10 RNAi (Figure 2C). The HISH4 mRNA was also more stable after the RNAi, but there was no evident change in the steady-state level (Figure 2D, lanes 2 and 3). To visualize deadenylation, we ran a sample on a polyacrylamide gel, flanked by marker lanes of fully deadenylated mRNA (treated with oligo d(T) and RNase H). Several polyadenylation sites are used (Figure 2D, lanes 1 and 10), and full-length polyadenylated mRNA forms a smear between 500 and 700 nt (lanes 2 and 3) (5). After transcription inhibition, the smear becomes progressively shorter (lanes 6 and 8), and this process was clearly delayed in cells with CNOT10 RNAi (lanes 7 and 9). These results indicate that TbCNOT10 is important for deadenylation and degradation of mRNAs in T. brucei.
TbCNOT10 is important for association of TbCAF1 with the NOT complex
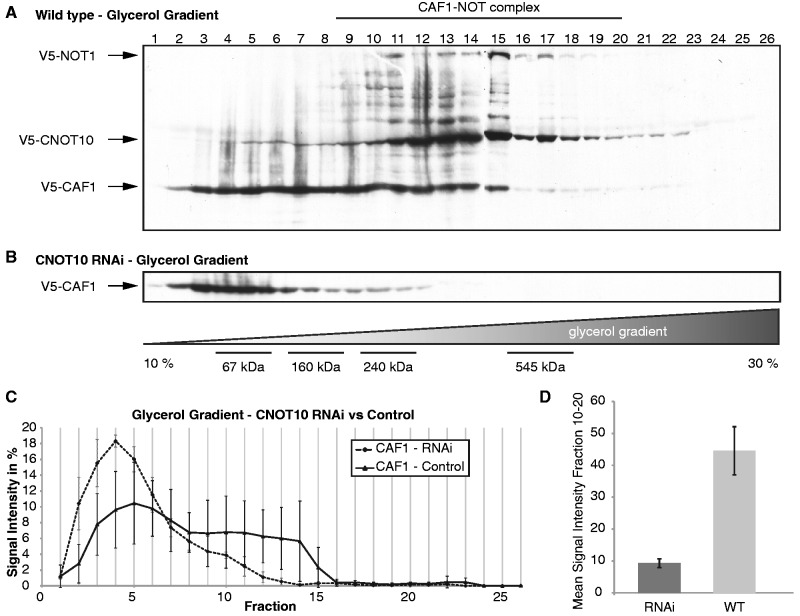
To assess the effect of TbCNOT10 depletion on the NOT complex, we analysed migration of different complex components by glycerol gradient centrifugation. For these experiments, we used equal mixtures of three cell lysates. Two lysates were always from cells expressing in situ V5-tagged versions of TbNOT1 or TbCNOT10. These were used to follow the position of the complex. Both TbNOT1 and TbCNOT10 migrated between fractions 9 and 20 (Figure 3A). The third lysate came from trypanosomes with in situ V5-tagged CAF1, with or without CNOT10 RNAi. In cells without RNAi, about half of the CAF1 was present in fractions 9–20 of the gradients, while the remainder was in lighter fractions (Figure 3A, C and D). We concluded that the NOT complex was concentrated in fractions 13–18, but that half of the CAF1 was not associated with it. In contrast, when we knocked down TbCNOT10 by RNAi, TbCAF1 no longer co-migrated with the complex at all (Figure 3B–D). This result suggested that TbCNOT10 is essential for the interaction of TbCAF1 with the rest of the NOT complex.
Figure 3.
Depletion of TbCNOT10 results in loss of TbCAF1 from the trypanosome complex. (A) Extracts of three different trypanosome lines (in situ V5-tagged TbCAF1, TbCNOT10 and TbNOT1) were mixed and run on a 10–30% glycerol gradient. The gradient was fractionated into 26 fractions. The additional bands between TbNOT1 and TbCNOT10 were degradation products of TbNOT1. Size indications are shown in figure (B) at the bottom. (B) Glycerol gradient of TbCNOT10-depleted cells with in situ V5-tagged TbCAF1. (C, D) Quantification of the signals from three independent gradients. (C) For each fraction, the level of signal is expressed as the percentage of the total signal on the blot. V5-TbCAF1 in TbCNOT10-depleted cells (dashed line; n = 3) and control (black line; n = 3). A comparison of the V5-TbCAF1 western blots signal from fractions 10–20 is shown in (D).
TbCNOT10 depletion reduces the level of TbNOT1
To define the interactions within the trypanosome NOT complex in more detail, we analysed them using the yeast two-hybrid system. The experiments showed that TbCNOT10 interacted with TbCAF1, TbNOT5 and the N- and C-terminal parts of TbNOT1 (Figure 4 and Supplementary Figure S3). The interaction between TbCNOT10 and TbNOT5 was seen only with TbCNOT10 as the prey and TbNOT5 as the bait. TbCAF1 interacted with TbCNOT10 and the N-terminal part of TbNOT1, but not with TbNOT5. Interaction of TbCAF1 with the C-terminal part of TbNOT1 was only seen in one direction, under low stringency conditions (Supplementary Figure S3).
Figure 4.
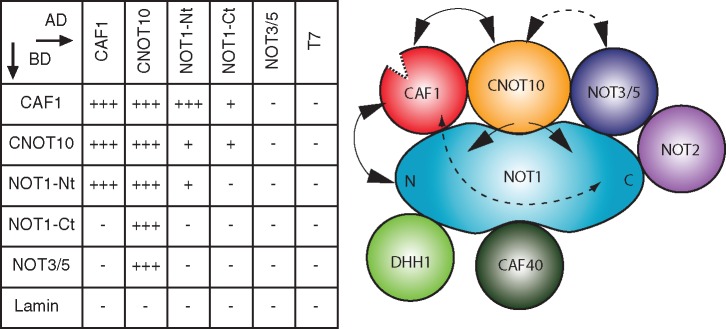
Yeast two-hybrid interactions in the trypanosome NOT complex. Left: minus and plus signs indicate the absence or presence of interaction detected by the activation of the His3 and LacZ reporters; multiple pluses indicate the activation of the Ade2, His3 and LacZ reporters. Lamin and T antigen (7) are negative controls. Right: illustration of the trypanosome NOT complex. Lines indicate interaction between different subunits, when the line is dashed, the interaction was shown only in one direction.
To find out whether TbCNOT10 depletion directly affects the level of TbCAF1 or TbNOT1 in trypanosomes, we knocked down TbCNOT10 in cells expressing in situ-tagged V5-TbCAF1 or V5-TbNOT1, and measured the proteins by western blot. The amount of V5-TbCAF1 was not influenced by depletion of TbCNOT10 (Figure 5A, lanes 3 and 4), but TbNOT1 was decreased (Figure 5B, lanes 3 and 4).
Figure 5.
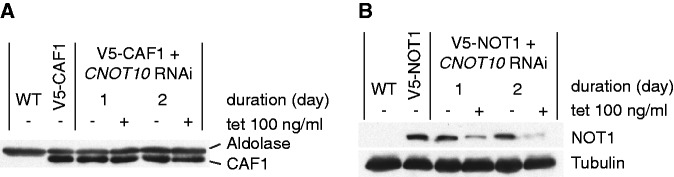
TbCNOT10 knock-down leads to decrease of TbNOT1 in trypanosomes. (A) The level of in situ V5-tagged TbCAF1 was measured after TbCNOT10 depletion. Extracts were made from wild-type cells (WT), cells with in situ V5-tagged TbCAF1, and the same cells with inducible RNAi targeting TbCNOT10, with RNAi induced for either 1 or 2 days. The level of V5-TbCAF1 was then measured by western blotting, with the aldolase signal as a control. (B) The effect of TbCNOT10 RNAi on the level of in situ V5-tagged TbNOT1 was analysed. The experiment is the same as in (A) except that in this case, the tagged protein was TbNOT1 and tubulin was used as a control.
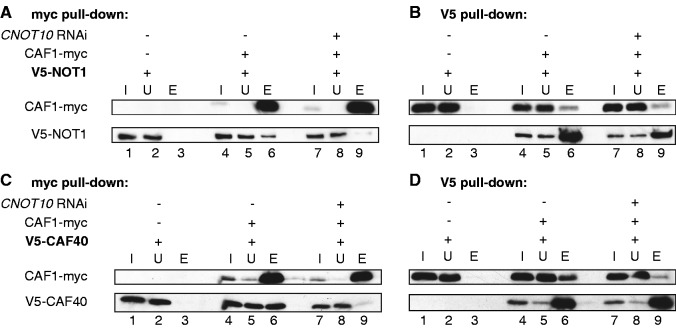
The effect of TbCNOT10 depletion on TbCAF1 association was next investigated by co-IP. We first used trypanosomes with V5-tagged TbNOT1 and inducible TbCAF1-myc; note that the V5-NOT1 is likely to be at roughly endogenous levels whereas the CAF1-myc is over-expressed. Pull-down of TbCAF1-myc yielded V5-TbNOT1 as expected (Figure 6A, lane 6), though most of the V5-TbNOT1 appeared not to be associated with TbCAF1 (Figure 6A, lane 6). If all TbNOT1 is associated with TbCAF1, this could mean that there is much more untagged TbCAF1 than TbCAF1-myc in the cells; alternatively, perhaps some TbNOT1 is not associated with TbCAF1. When TbCNOT10-depleted trypanosomes were used, the amount of co-precipitated TbNOT1 was reduced as expected (Figure 6A, lane 9). Thus, under the conditions used, TbCNOT10 appeared to be limiting for the TbCAF1–TbNOT1 association. Using the same extracts, the reciprocal pull-down of V5-TbNOT1 yielded TbCAF1-myc as expected (Figure 6B, lane 6), although again most TbCAF1-myc remained in the supernatant (Figure 6B, lane 8), consistently with the glycerol gradient result and the over-expression of TbCAF1-myc. Puzzlingly, when TbCNOT10-depleted trypanosomes were used for the V5-TbNOT1 pull-down, the amount of co-precipitated TbCAF1-myc was only slightly affected even though the amount of precipitated V5-TbNOT1 was lower (Figure 6B, compare lanes 6 and 9). This is difficult to explain because it is inconsistent with the other results; perhaps, it is connected with over-expression of TbCAF1-myc.
Figure 6.
Depletion of trypanosome CNOT10 reduces CAF1 association with the complex. (A) myc-beads were used to pull down myc-tagged TbCAF1 in normal trypanosomes (lanes 4–6) and cells with TbCNOT10 RNAi (lanes 7–9). Trypanosomes, expressing V5-tagged TbNOT1 without myc-tagged TbCAF1, were used as a negative control (lanes 1–3). One percent was loaded for input (I) and unbound fraction (U) and the rest was eluted (eluate, E). (B) The reciprocal experiment using V5-beads. (C, D) as in (A, B), respectively, but here the interaction between TbCAF1 and TbCAF40 was analysed with or without CNOT10 RNAi.
Because of this result, we also used the association of TbCAF40 to monitor complex integrity. In cells with normal amounts of TbCNOT10, precipitation of TbCAF1-myc resulted in co-precipitation of V5-TbCAF40 (Figure 6C, lane 6), but this no longer occurred in TbCNOT10-depleted trypanosomes (Figure 6A, lane 9). The results were confirmed by reciprocal pull down (Figure 6D, lanes 6 and 9).
Together, these results show that both TbCAF1 and TbCNOT10 interact directly with TbNOT1, and that the presence of TbCNOT10 enhances the interaction between TbCAF1 and TbNOT1. Depletion of TbCNOT10, with the accompanying reduction in the TbCAF1–TbNOT1 interaction, reduces the cellular level of TbNOT1.
CAF1 deadenylation activity is not complex dependent
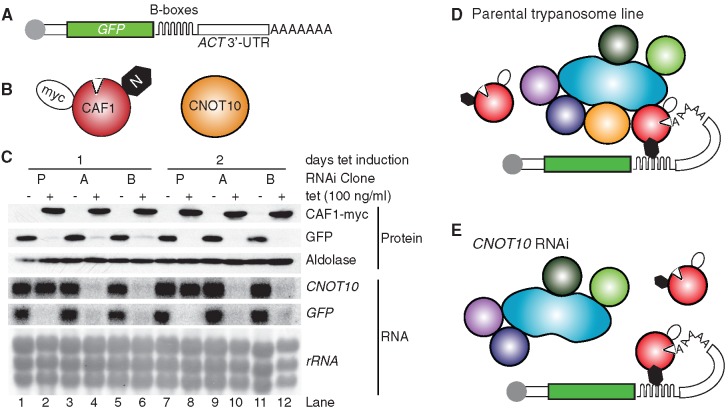
One possible interpretation of all observations so far was that when TbCAF1 is not in the NOT complex, it is unable to initiate mRNA degradation. However, we had previously shown that recombinant TbCAF1 is active in deadenylation in vitro (5). There are two possible explanations for this discrepancy. Either TbCAF1 enzyme activity in vivo is dependent on, or stimulated by, interactions in the complex, or TbCAF1 relies on the complex for attachment to substrates. To distinguish between these possibilities, we created a cell line containing a constitutively expressed mRNA reporter, which consisted of a GFP ORF followed by six boxB elements and an actin 3′-UTR (GFP-B-ACT). In addition, the cell line inducibly expressed a TbCAF1 fusion protein with a λN peptide at the N-terminus and myc at the C-terminus (λN-TbCAF1-myc). This hybrid protein should be ‘tethered’ to the GFP-B-ACT reporter RNA through the interaction between the B-boxes and the λN peptide. Upon induction of λN-TbCAF1-myc expression, GFP-B-ACT mRNA was destroyed (Figure 7, lanes 1, 2 and 7, 8). A control RNA with no boxB (GFP-ACT) was unaffected by λN-CAF1-myc expression (not shown).
Figure 7.
Tethered TbCAF1 destroys a reporter mRNA in trypanosomes. (A) Cartoon of the reporter RNA, with a cap (grey) GFP ORF and six Bbox loops at the start of the 3′-UTR, (B) λN-TbCAF1-myc is shown in red, and TbCNOT10 is in orange. (C) Western (above) and northern blots showing expression of GFP protein and GFP mRNA, as well as TbCAF1-myc, aldolase control and TbCNOT10 RNA. Equal loading of the RNA was shown using rRNA. Results are shown for the parental cell line with no RNAi (P) and two independent clones with CNOT10 RNAi, labelled A and B. (D) The situation in the parental cell line P—lanes 1, 2, 7 and 8 in (C). λN-TbCAF1-myc is shown interacting with the B-boxes on the reporter and also with the rest of the NOT complex; the tethered CAF1 can degrade the poly(A) tail. (E) The situations in cell lines A and B, after induction of RNAi against TbCNOT10 for 1 day (lanes 4 and 6 of (C)) or 2 days (lanes 10 and 12 of (C)). TbCAF1 is still tethered to the reporter RNA and can still degrade poly(A), even though association with the NOT complex has decreased.
So far the tethering result did not tell us whether TbCAF1 requires interaction with the NOT complex for its activity, since tethered λN-TbCAF1-myc might still interact with the NOT complex. To reduce the interaction, we depleted TbCNOT10 by RNAi. Now, λN-CAF1-myc alone should be tethered. Nevertheless, the GFP-boxB mRNA reporter was degraded as actively as before (Figure 7, lanes 3–6 and 9–12). This result suggests that TbCAF1 does not depend on NOT complex association in order to degrade a tethered substrate. In other words, TbCAF1 alone is indeed active in vivo, but it depends on the presence of TbCNOT10—and, by extension, perhaps the rest of the NOT complex—to be recruited to its substrate.
HsCNOT10 is probably not required for association of HsCAF1 with the NOT complex
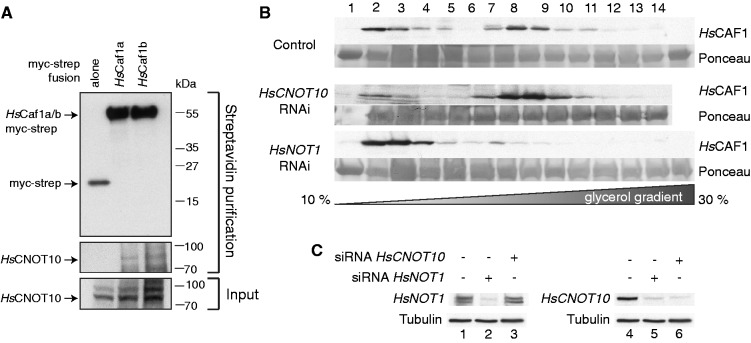
Knowing the importance of TbCNOT10 in mRNA degradation, we were interested whether this function was conserved in mammalian cells. Therefore, we first confirmed the interaction of HsCAF1a and HsCNOT10 in HEK 293 cells by co-IP (Figure 8A). Using myc-strep-tagged HsCAF1a or HsCAF1b, we were able to pull down endogenous HsCNOT10 and could confirm the interaction previously shown by TAP (38). To determine whether the association of HsCAF1a with core components of the human NOT complex is HsCNOT10-dependent, we knocked down HsCNOT10 by siRNA and analysed the migration of HsCAF1a in a glycerol gradient. As controls we used a scrambled siRNA and a siRNA against HsNOT1. Figure 8B shows that HsCNOT10 depletion did not influence the migration of HsCAF1a, whereas knock-down of HsNOT1 led to a shift of HsCAF1a towards lighter fractions. The knock-down efficiency of HsCNOT10 and HsNOT1 was verified by western blot analysis (Figure 8C). We also attempted to determine the effect of HsCNOT10 depletion on Caf1b complex association, using a Caf1b-specific antibody. Results suggested that Caf1b also might have remained complex associated after HsCNOT10 knock-down, but since Caf1b was extremely difficult to detect no firm conclusion could be drawn.
Figure 8.
HsCNOT10 is not required for association of CAF1 with the human NOT complex. (A) Myc-strep-tagged HsCAF1a or HsCAF1b were purified from HEK293 cells, and association of HsCNOT10 was assessed by western blot analysis. Cells expressing the tag alone served as the control. (B) Control, human CNOT10 and NOT1 RNAi extracts were run on 10–30% glycerol gradients and 15 fractions were collected. Western blots of the gradient fractions were analysed for the migration of HsCAF1a. The Ponceau-stained band is BSA, which was added to every fraction to serve as a control for equal precipitation efficiency. (C) HsNOT1 and HsCNOT10 depletion was confirmed by western blot analysis.
Notably, the knock-down of HsCNOT1 reduced the amount of HsCNOT10, whereas the reverse was not the case. These results suggested that HsCNOT10 is not required for association of HsCAF1a with HsNOT1 in HEK293 cells.
DISCUSSION
In this study, we have shown that in the relatively simple trypanosome NOT complex, association of TbCAF1 with TbNOT1 is enhanced by the presence of TbCNOT10. Further, our results indicate that in vivo, this association is required for deadenylation of mRNAs by TbCAF1.
Our yeast two-hybrid experiments revealed various interactions between TbCNOT10, TbCAF1, TbNOT5 and the N- and C-terminal parts of TbNOT1 (Figure 4). The two-hybrid interactions of TbCAF1, TbNOT5 and TbCNOT10 with TbNOT1 seem likely to be direct, as they parallel those seen in other species. The interaction between TbCAF1 and TbNOT1 most likely resembles that seen in opisthokonts (16,17). The relevant region of NOT1 is highly conserved across evolution and the most important CAF1-interacting residues of yeast and trypanosome NOT1s are identical. In CAF1, somewhat less conservation is evident in the interaction domain, but the overall hydrophobic character is conserved. In contrast, we cannot currently be sure that the apparent interaction between TbCAF1 and TbCNOT10 is direct, since we cannot rule out the possibility that they interact via yeast Not1p. Given the sequence conservation, TbCAF1 might indeed interact with yeast Not1p. In contrast, TbCNOT10 bears no resemblance to any yeast protein (including Caf130p) so interaction with Not1p would not be expected.
TAP of CNOT10, bearing an N-terminal protein-A-calmodulin-binding-peptide tag, did not result in co-purification of TbCAF1; exactly the same result was obtained for HsCNOT10 bearing an N-terminal FLAG-HA tag (9). This was not caused by the buffer conditions since TbCNOT10 was obtained after TAP of the complex using TAP-TbCAF1. It was probably not caused by the tag either: N-terminally TAP-tagged TbCNOT10 was functional, since trypanosomes expressing only the TAP-tagged version were viable, and TbCNOT10 that was N-terminally fused with either the V5 peptide or yeast-two hybrid components, was able to interact with N- or C-terminally tagged TbCAF1. The only explanation we can think of is that the interaction was somehow specifically competed by the binding of the tag to the resins.
In trypanosomes, TbCNOT10 was required both for association of V5-TbCAF1 with the complex, and for the presence of normal amounts of V5-TbNOT1. The easiest explanation of this result is that TbCNOT10, by interacting with both TbCAF1 and TbNOT1, stabilizes TbCAF1–TbNOT1 binding; and that TbNOT1, which is not associated with TbCAF1, is preferentially degraded.
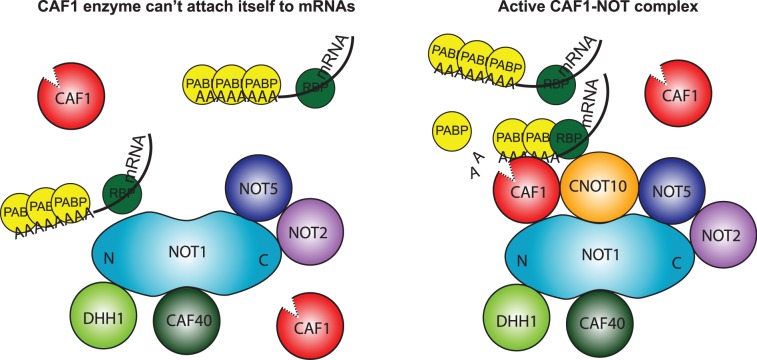
Glycerol gradient analysis for both trypanosomes and HEK cells showed that about half of V5-TbCAF1 or native HsCAF1 was not associated with the NOT complex. The role of this ‘free’ fraction is not known. It may have no function at all, or it could have a role independent of deadenylation, for example in repressing translation, as shown for Xenopus oocytes (39). In addition, it could also act as a reserve stock for newly formed NOT complexes. Since in trypanosomes, we saw a reduction in deadenylation and mRNA degradation in TbCNOT10-depleted cells, with concomitant loss of CAF1 association with the NOT complex, it seems likely that in vivo, free TbCAF1 is unable to digest PABP-coated poly(A) tails. In contrast, once TbCAF1 attaches to an mRNA, either via the complex or by tethering, digestion is possible (Figure 9). Similarly, in human cells, depletion of HsNOT1 or HsNOT2 destabilized the NOT complex and decreased deadenylation (14,19), and the interaction of HsCAF1a with HsNOT1 was required for Caf1-/Not-dependent degradation of an miRNA target (16). In budding yeast, disruption of the interaction between the deadenylases (Ccr4p and Caf1p) and Not1p, effected by expression of dominant-negative interaction-defective mutants, impaired growth and deadenylation (17).
Figure 9.
A model for the function of TbCNOT10 in trypanosomes. Left: in the absence of TbCNOT10, TbCAF1 is not able to bind PABP-covered mRNAs and initiate mRNA degradation. Right: upon association with the complex, TbCAF1 is ‘tethered’ to target mRNAs through other subunits.
In human cells, the NOT complex can be recruited to mRNAs through RNA-binding proteins (e.g. (13)). We expect the same to be true in trypanosomes, but so far there are no good candidate RNA-binding proteins that might fill this role. In our TAP of the trypanosome NOT complex, we did not identify any RNA-binding proteins, but this is not surprising. Eukaryotic NOT complexes probably interact with many different proteins, so that any one would be present at relatively low levels. Moreover, such interactions may be transient. Indeed, human TTP, which does interact with human NOT complex, was not detected in pull-downs with any human NOT complex component (9).
We do not know which NOT complex subunits are directly responsible for interactions with RNA or RNA-binding proteins. Potential candidates for recruitment of the complex to human mRNAs are HsCNOT3/5 and 9, both of which pulled down proteins with RNA-binding domains (9). In trypanosomes, recruitment of the NOT complex to different mRNAs through different subunits could at least partially explain why all of the subunits are required for cell growth. The role of CNOT10 in human cells remains to be elucidated, but one possibility is that both human and trypanosome CNOT10 play roles in the targeting of mRNAs for degradation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–3.
FUNDING
The Deutsche Forschungsgemeinschaft [CLl112/9-3, STO859/2-1 and STO859/3-1 to C.C. and G.S.]; a stipend from the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (to S.S.) and a bridging project grant from the ZMBH-DKFZ alliance. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Janine Philipp, Ute Leibfried and Claudia Hartman for technical assistance. We are also grateful to Ann-Bin Shyu (University of Texas-Houston Medical School) for the human Caf1a antibody.
REFERENCES
- 1.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem. Soc. Trans. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collart M, Panasenko O. The Ccr4-Not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 2009;422:443–453. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 10.Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4–NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 13.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito K, Takahashi A, Morita M, Suzuki T, Yamamoto T. The role of the CNOT1 subunit of the CCR4–NOT complex in mRNA deadenylation and cell viability. Protein Cell. 2011;2:755–763. doi: 10.1007/s13238-011-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azzouz N, Panasenko OO, Deluen C, Hsieh J, Theiler G, Collart MA. Specific roles for the Ccr4–Not complex subunits in expression of the genome. RNA. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4–NOT deadenylase complex. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks883. September 12 (10.1093/nar/gks883; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basquin J, Roudko VV, Rode M, Basquin C, Seraphin B, Conti E. Architecture of the nuclease module of the yeast Ccr4–Not complex: the Not1–Caf1–Ccr4 interaction. Mol Cell. 2012;48:207–218. doi: 10.1016/j.molcel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Russell P, Benson JD, Denis CL. Characterization of mutations in NOT2 indicates that it plays an important role in maintaining the integrity of the CCR4–NOT complex. J. Mol. Biol. 2002;322:27–39. doi: 10.1016/s0022-2836(02)00707-6. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Inoue T, Yokoyama K, Morita M, Suzuki T, Yamamoto T. CNOT2 depletion disrupts and inhibits the CCR4–NOT deadenylase complex and induces apoptotic cell death. Genes Cells. 2011;16:368–379. doi: 10.1111/j.1365-2443.2011.01492.x. [DOI] [PubMed] [Google Scholar]
- 20.Garces RG, Gillon W, Pai EF. Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 2007;16:176–188. doi: 10.1110/ps.062600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4–NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4–NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 24.Daniels J, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiol. Mol. Biol. Rev. 2010;74:552–569. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayton C, Michaeli S. 3′ processing in protists. Wiley Interdiscip. Rev. RNA. 2011;2:247–255. doi: 10.1002/wrna.49. [DOI] [PubMed] [Google Scholar]
- 26.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 27.Manful T, Fadda A, Clayton C. The role of the 5′–3′ exoribonuclease XRNA in transcriptome-wide mRNA degradation. RNA. 2011;17:2039–2047. doi: 10.1261/rna.2837311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clayton CE, Estevez AM, Hartmann C, Alibu VP, Field M, Horn D. Down-regulating gene expression by RNA interference in Trypanosoma brucei. Methods Mol. Biol. 2005;309:39–60. doi: 10.1385/1-59259-935-4:039. [DOI] [PubMed] [Google Scholar]
- 29.Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clayton CE. Import of fructose bisphosphate aldolase into the glycosomes of Trypanosoma brucei. J. Cell. Biol. 1987;105:2649–2653. doi: 10.1083/jcb.105.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93(Pt 3):491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 34.Zinoviev A, Akum Y, Yahav T, Shapira M. Gene duplication in trypanosomatids—two DED1 paralogs are functionally redundant and differentially expressed during the life cycle. Mol. Biochem. Parasitol. 2012;185:127–136. doi: 10.1016/j.molbiopara.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwede A, Manful T, Jha B, Helbig C, Bercovich N, Stewart M, Clayton C. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 2009;37:5511–5528. doi: 10.1093/nar/gkp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 39.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J. Biol. Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.