Abstract
Reverse gyrase is an ATP-dependent topoisomerase that is unique to hyperthermophilic archaea and eubacteria. The only reverse gyrase structure determined to date has revealed the arrangement of the N-terminal helicase domain and the C-terminal topoisomerase domain that intimately cooperate to generate the unique function of positive DNA supercoiling. Although the structure has elicited hypotheses as to how supercoiling may be achieved, it lacks structural elements important for supercoiling and the molecular mechanism of positive supercoiling is still not clear. We present five structures of authentic Thermotoga maritima reverse gyrase that reveal a first view of two interacting zinc fingers that are crucial for positive DNA supercoiling. The so-called latch domain, which connects the helicase and the topoisomerase domains is required for their functional cooperation and presents a novel fold. Structural comparison defines mobile regions in parts of the helicase domain, including a helical insert and the latch that are likely important for DNA binding during catalysis. We show that the latch, the helical insert and the zinc fingers contribute to the binding of DNA to reverse gyrase and are uniquely placed within the reverse gyrase structure to bind and guide DNA during strand passage. A possible mechanism for positive supercoiling by reverse gyrases is presented.
INTRODUCTION
Regulation of DNA topology is central to a multitude of cellular events, including gene expression, DNA replication, recombination and repair (1,2). DNA topoisomerases inter-convert topoisomers by introducing or removing positive or negative supercoils. A number of topoisomerases are capable of relaxing DNA supercoils in an ATP-independent reaction but the introduction of supercoils is ATP-dependent. Bacterial gyrases introduce negative supercoils at the expense of ATP hydrolysis (3), thus relieving topological stress ahead of the replication fork. Conversely, reverse gyrases (4) which are unique to thermophiles and hyperthermophiles (5) catalyze the ATP-dependent introduction of positive supercoils into DNA and are thought to protect the genome at high temperatures, also via their DNA chaperone and renaturase activities (6,7). Reverse gyrases are usually single-chain molecules of >1000 amino acids, although exceptions are known from Nanoarchaeum equitans (8) and Methanopyrus kandleri (9), which harbor hetero-dimeric reverse gyrases. Early sequence alignments showed that reverse gyrases share a modular structure with an N-terminal cysteine-rich region (a putative zinc finger) preceding a helicase domain that is followed by a C-terminal topoisomerase domain (10). The helicase/topoisomerase domain arrangement was later confirmed by the crystal structure of Archaeoglobus fulgidus reverse gyrase (11). The zinc fingers were not resolved, but a so-called latch domain was identified within the helicase domain and was speculated to reversibly connect the helicase and topoisomerase domains during supercoiling (11) (Figure 1). The helicase domain is subdivided into H1 and H2 domains that are flexibly linked (12,13). H1 and H2 contain degenerated signature motifs of superfamily 2 (SF2) helicases. Among members of SF2, these motifs are necessary for ATP- and nucleic acid binding, and for duplex separation [reviewed in (14)]. A critical role of some of these motifs for positive DNA supercoiling by reverse gyrase has been demonstrated (15). In SF2 helicases, additional domains or domain insertions can modulate the helicase function (14). In reverse gyrase, apart from the N-terminal zinc finger and the latch domain, a so-called insert region is present in H1 (16) that may be involved in DNA binding (13). Some reverse gyrase topoisomerase domains also include a cysteine-rich region that has been proposed to form a second zinc finger (17).
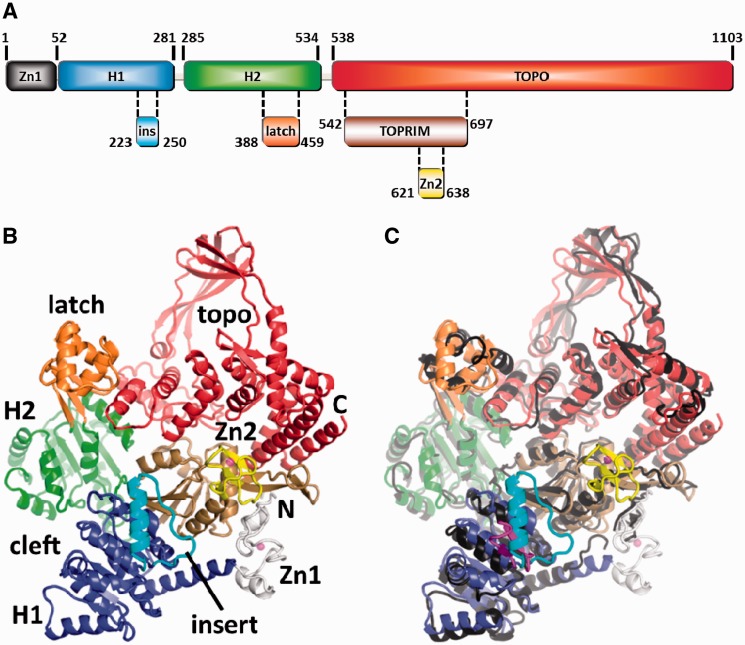
Figure 1.
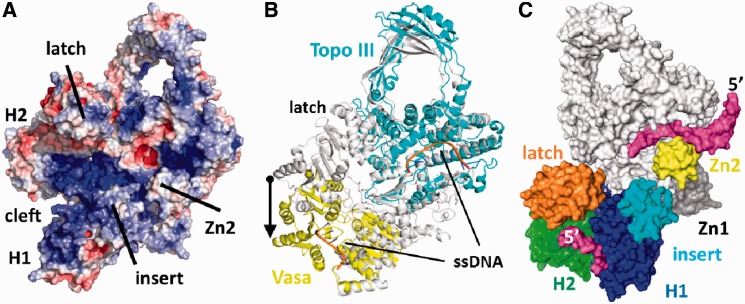
Overview of reverse gyrase. (A) Domain architecture of reverse gyrase. This color scheme is followed throughout the manuscript. (B) Structure of T. maritima reverse gyrase. The helicase subdomains H1 and H2 are shown in blue and green, respectively. H1 carries an insertion (cyan) that shares similarity with UvrD. The latch region (orange) is inserted in H2. The topoisomerase domain is colored red with the exception of the Toprim subdomain colored in brown. The Toprim domain (from topoisomerase/primase, a doubly wound Rossman fold) serves as a dedicated single-strand DNA-binding site (18). Zinc fingers are drawn in gray (Zn1) and yellow (Zn2). (C) Superposition of the T. maritima and A. fulgidus (PDB-ID 1GKU, dark gray) (11) structures highlights several differences, e.g. the zinc fingers (not resolved in A. fulgidus), the different orientations and folds of the latch and insert domains and a tilt in the topoisomerase domains. The much smaller insert domain of T. maritima reverse gyrase is colored magenta.
Previously, only the structure of A. fulgidus reverse gyrase was available, showing the overall composition of the enzyme (11). The helicase domain is tethered to the topoisomerase domain and the H1/H2 subdomains adopt an open conformation. It remains unclear whether this domain arrangement constitutes an intermediate during catalysis. From the A. fulgidus structure, large movements of the latch domain during catalysis were predicted but currently no experimental evidence supports this hypothesis. Also, the putative zinc fingers that are present in most reverse gyrase sequences were not resolved in the A. fulgidus structure but instead were proposed to become ordered upon DNA binding and to maybe contribute to DNA strand passage (11).
Here, we describe five structures of the entire Thermotoga maritima reverse gyrase that include the zinc fingers. The N-terminal zinc finger firmly attaches the H1 domain to the topoisomerase domain and may contribute to double-strand DNA (dsDNA) binding. The second zinc finger locates to the topoisomerase domain at a position close to the single-strand DNA-binding site. Comparison of all structures shows that the latch domain is a structurally plastic and flexible entity. Further, the insert region in H1, a putative DNA-binding helix–loop motif (Figure 1), displays rigid body flexibility. The latch and the insert helix contribute to binding of single-stranded DNA (ssDNA) to reverse gyrase (18). The zinc fingers, the latch and the insert region face the same side of reverse gyrase, allowing delineation of possible DNA-binding modes. We have shown previously that the isolated helicase module switches into a closed conformation during its catalytic cycle (12). Inferring that such a conformational change also occurs in the catalytic cycle of reverse gyrase, a closed form of reverse gyrase can be constructed that provides insight into possible conformational changes during positive supercoiling.
MATERIALS AND METHODS
Protein purification
Reverse gyrase, the helicase domain (rgyr_hel) and the helicase domain lacking the latch were constructed and purified as described and stored in 50 mM Tris/HCl pH 7.5, 1 M NaCl, 10 mM MgCl2, 0.1 mM ZnCl2 and 2 mM β-mercaptoethanol (16,20,21). Zinc finger deletions were Δ1-58 (Zn1)/Δ620-643(Zn2), and zinc finger mutations were Cys11Ala/Cys14Ala (Zn1_C2A2) and Cys635Ala/Cys638Ala (Zn2_C2A2). The helicase domain lacking the zinc finger comprises amino acids 59–541. In the helicase domain lacking the insert helix amino acids Ile224-Lys249 are deleted. Both proteins were purified as the wild-type helicase domain (21).
Positive DNA supercoiling
Positive supercoiling reactions were performed with 15 nM negatively supercoiled pUC18 and 1 µM reverse gyrase in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 100 µM Zn(OAc)2, 2 mM β-mercaptoethanol and 10% (w/v) PEG 8000 at 75°C as described (20). The reaction products at indicated time-points were analyzed as described (20).
Steady-state ATPase activity
Steady-state ATPase activity of 0.1 µM rgyr_hel in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 100 µM Zn(OAc)2, 2 mM β-mercaptoethanol containing 2 mM ATP was monitored in a coupled enzymatic assay and analyzed according to the Michaelis–Menten equation as described (20,21).
Determination of dissociation constants
Dissociation constants of rgyr_hel/DNA complexes were determined in fluorescence anisotropy titrations of 25 nM 5′-fluorescein-labeled 30mer or 60mer ssDNA or 60-bp dsDNA with rgyr_hel in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 100 µM Zn(OAc)2 and 2 mM β-mercaptoethanol as described (21).
Crystallization and structure determination
Crystals for full length reverse gyrase were obtained at 25°C from the PEG/Ion or Index screens (Hampton). Primitive monoclinic crystals (PDB-ID 4DDX) were obtained at 20°C in the micro-batch setup by mixing 26 µM reverse gyrase 1:3 by volume (v/v) with reservoir containing 20 mM citric acid, 80 mM Bis–Tris propane/HCl pH 8.8 and 16% PEG 3350. All other crystal forms were derived from 67 µM reverse gyrase mixed 1:5 (v/v) with reservoir and incubated at 20°C for 1–2 weeks. The C-centered orthorhombic form (PDB-ID 4DDW) was obtained from 0.2 M Mg(OAc)2, 20% PEG 3350 after addition of 0.15 M sodium citrate to clear drops. The first C-centered monoclinic form (PDB-ID 4DDU) was obtained from 0.2 M MgCl2, 0.1 M HEPES/NaOH pH 7.5 and 25% PEG 3350, whereas the second form (PDB-ID 4DDT) was obtained from 0.2 M K/Na tartrate pH 7.4 and 20% PEG 3350. The triclinic form (PDB-ID 4DDV) was obtained from 0.2 M sodium citrate and 20% PEG 3350. Crystals were vitrified by hyperquenching (22) without additional cryoprotectant. Data were collected at Swiss Light Source beamline PX-II, integrated with XDS (23), scaled with SADABS (Bruker) and are summarized in Table 1. High resolution limits for the data were selected based on I/σ(I) = 1 and CC1/2 > 0.3 (24) in the outer shell. A. fulgidus reverse gyrase (11) lacking the latch domain and with the helicase domain replaced by the T. maritima counterpart was used as the search model for phasing of the 3.2 Å resolution data with PHASER (25). This refined model was used for molecular replacement of the other four crystal forms. Models were built in COOT (26) and refined with BUSTER (PDB-ID 4DDV) (27) or PHENIX (28) using automatically determined translation, libration, screw-rotation (TLS) descriptions and secondary structure restraints. Grouped B-values were refined for resolutions >3.2 Å and tight non-crystallographic symmetry restraints were applied where available (PDB-IDs 4DDV, 4DDW and 4DDX). Data for PDB-ID 4DDU were collected and merged from two crystals to ensure high multiplicity (∼13) at a wavelength of 1.28 Å where calculated f”(Zn2+) = 3.4 e−. Data merging led to a significant increase in Rsym (27.4/94.1% for overall data/high resolution shell instead of 18.7/86.7% for the first crystal) but also improved I/σ(I) from 6.1/0.9 to 7.1/1.1 and increased CC1/2 from 0.988/0.306 to 0.988/0.414, indicating that the high resolution data benefited from merging, despite worse R-factors. Use of the merged data in refinement also yielded higher quality electron density maps and lower Rfree-values compared with data from only one crystal while keeping the same test set. The zinc ions were confirmed in maps calculated using the phases from the final model and the anomalous differences from the 4DDU data as Fourier coefficients (Supplementary Figure S1D). Statistics for the refinements are summarized in Table 1. Figures were created with Pymol (www.pymol.org).
Table 1.
Data collection and refinement statistics
| Dataset | 4DDU | 4DDT | 4DDV | 4DDW | 4DDX |
|---|---|---|---|---|---|
| Data collection | 44.9–3.0 | 48.2–3.2 | 49.1–3.5 | 49.0–3.9 | 49.2–4.2 |
| Resolution range (Å)a | (3.1–3.0) | (3.3–3.2) | (3.6–3.5) | (4.0–3.9) | (4.3–4.2) |
| Wavelength (Å) | 1.28 | 1.00 | 1.00 | 1.28 | 1.00 |
| Space group | C2 | C2 | P1 | C2221 | P21 |
| Cell dimensions (Å/°) | a = 170.0, b = 73.9, | a = 186.9, b = 104.4, | a = 95.7, b = 101.3, | a = 174.3, | a = 125.8, b = 104.9, |
| c = 111.4, β = 116.6 | c = 96.7, β = 116.6 | c = 104.4, α = 113.6, | b = 175.6, c = 104.1 | c = 126.2, β = 91.7 | |
| β = 97.5, γ = 110.4 | |||||
| Unique reflections | 24 946 (2311) | 27 506 (2398) | 39 606 (3186) | 14 481 (1024) | 24 000 (1652) |
| Multiplicity | 13.2 (13.8) | 3.6 (3.6) | 2.0 (2.0) | 5.5 (5.5) | 2.9 (2.9) |
| Completeness (%) | 99.8 (99.7) | 99.6 (99.1) | 98.4 (97.5) | 96.9 (95.2) | 96.8 (97.7) |
| Rsym (%)b,c | 27.4 (94.1) | 19.7 (68.3) | 14.9 (54.9) | 16.2 (82.7) | 28.4 (63.1) |
| CC1/2d | 0.988 (0.414) | 0.980 (0.458) | 0.981 (0.604) | 0.996 (0.343) | 0.937 (0.491) |
| Average <I/σ(I) | 7.1 (1.1) | 4.6 (1.1) | 4.0 (1.0) | 7.7 (1.2) | 3.1 (1.2) |
| Refinement | 44.2–3.0 | 48.2–3.2 | 49.1–3.5 | 48.7–3.9 | 48.4–4.2 |
| Resolution range (Å) | (3.2–3.0) | (3.3–3.2) | (3.6–3.5) | (4.2–3.9) | (4.5–4.2) |
| Rcryst (%)c | 21.5 (37.0) | 21.9 (34.4) | 23.5 (22.2) | 20.9 (30.7) | 24.5 (31.9) |
| Rfree (%)c | 27.8 (45.6) | 27.0 (40.0) | 27.8 (24.3) | 28.9 (36.8) | 30.5 (36.3) |
| No. of residues | 1102 | 1102 | 2204 | 2204 | 2203 |
| rmsd bonds/angles (Å/°) | 0.004 / 0.72 | 0.004 / 0.75 | 0.010 / 1.12 | 0.005 / 0.60 | 0.003 / 2.35 |
| Ramachandran plot (%)e | 87.1/12.6/0.1/0.2 | 85.3/14.2/0.2/0.3 | 87.6/11.5/0.6/0.2 | 87.4/11.9/0.3/0.4 | 83.5/15.4/0.8/0.3 |
aValues in parentheses correspond to the highest resolution shell.
bcalculated with XPREP (Bruker).
cR-factor definitions as summarized in (29). Rfree (30) is Rcryst with 5 % of test set structure factors.
dcorrelation coefficient between two random halves of the dataset as described in (24) and calculated using PHENIX (28).
ecalculated using PROCHECK (31). Numbers reflect the percentage of amino acid residues of the core, additionally allowed, generously allowed and disallowed regions, respectively.
RESULTS
Full-length T. maritima reverse gyrase was crystallized in five different space groups and data were collected to a maximum resolution of 3.0 Å (Table 1). Phasing was achieved by molecular replacement using a hybrid search model of the topoisomerase domain from the A. fulgidus structure (PDB-ID 1GKU) and the T. maritima H1 and H2 domains (16) (PDB-ID 3OIY). The obtained T. maritima model contained novel features that were absent in the search model. Most importantly, clear electron density for the two cysteine-rich regions, the putative zinc fingers, was present in the first maps calculated from molecular replacement (Supplementary Figure S1A–C). The highest peaks in the difference electron density maps corresponded to the zinc/cysteine cluster (>5 rmsd, root mean square deviation), and the presence of zinc was confirmed by its anomalous signal (see ‘Materials and Methods’ section and Supplementary Figure S1D). Neither of the zinc-binding regions was resolved in the structure of A. fulgidus reverse gyrase. In addition, electron density for the latch in T. maritima reverse gyrase was observed (Supplementary Figure S1A and B), which has a very different structure compared with the A. fulgidus enzyme. All residues of T. maritima reverse gyrase were traced and the models were refined with excellent R-values and stereochemistry (Table 1). Three of the five crystal structures have resolutions ≥3.5 Å (PDB-IDs 4DDV, 4DDW and 4DDX), yielding clear density for the subdomains but no details on side-chain conformations. These structures are included for the discussion of domain movements and exclusion of crystal packing effects, whereas side-chain orientations and more subtle conformational differences between reverse gyrases are evident from the higher resolution structures (PDB-IDs 4DDU and 4DDT).
Overall structure and domain organization
The overall padlock-like shaped structure of T. maritima reverse gyrase (1104 residues) shows a similar arrangement of the topoisomerase and helicase domains as observed for A. fulgidus reverse gyrase (1054 residues; Figure 1B and C). The enzymes exhibit 41% sequence identity (60% similarity) and share an overall rmsd of 2.1 Å over 889 residues, indicating that large parts of the protein structures (>200 residues) diverge significantly (for rmsd values of individual domains, Supplementary Table S1). The diverging regions include the tip of the topoisomerase domain, the latch domain and the insert region (Supplementary Table S1 and Figure 1C), which are further discussed below. The RecA-like H1 and H2 domains forming the helicase-module (32,33) are oriented similarly to each other as in the A. fulgidus structure (Figure 1C). In all five T. maritima reverse gyrase structures, H1 and H2 adopt an open conformation and their relative orientation with respect to the topoisomerase domain varies only slightly, indicating that this ‘open’ conformation is a stable state of the apo-enzyme. The interface connecting the H1/H2 helicase and topoisomerase modules appears to be plastic; superposition of the helicase domains of T. maritima and A. fulgidus reverse gyrases (Supplementary Table S1) reveals different positions of their topoisomerase domains. As a result of a tilt and shift, positional differences of equivalent residues exceed 10 Å at the apex of the topoisomerase domains. The H1 and H2 domains deviate from the canonical RecA fold by insertions. H1 contains an insert region (cyan in Figure 1) that in T. maritima reverse gyrase forms a lid-like loop-helix structure on the surface. This motif adopts the same conformation as in structures of the isolated helicase domain and has been suggested to play a role in DNA binding (13). In A. fulgidus reverse gyrase, the corresponding region forms a β-hairpin (magenta in Figure 1C) and was proposed to interact with DNA and promote strand separation (11). In Thermoanaerobacter tengcongensis reverse gyrase, deletion of this region reduces DNA affinity (34). The insertion in H2, called the latch (orange in Figure 1 and elsewhere), serves a crucial function in communication between the helicase and topoisomerase domains (16,35). In the isolated helicase domain, it is critical for cooperative binding of ATP and DNA (16) and for ATP-dependent DNA unwinding (18). The T. maritima reverse gyrase structure is completed by two zinc fingers, one at the N-terminus and a second inserted into the topoisomerase domain (see below). These zinc fingers were proposed to contribute to DNA binding and strand passage (11,36).
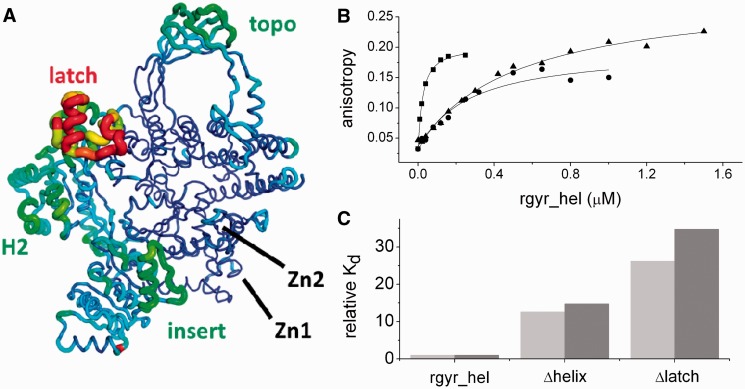
Comparison of the T. maritima revere gyrase crystal structures reveals elements that appear flexible or display rigid body movements (Figure 2A). Due to different packing arrangements in the five crystal forms, the observed structural diversity is not affected by crystal contacts. The latch, several α-helices in H2, the insert region and the apex of the topoisomerase domain show the largest variations, whereas other parts such as the zinc fingers and the core of the topoisomerase domain are invariant among these structures. The latch and insert region are directly involved in DNA binding (Figure 2B and C; see below).
Figure 2.
Role of the insertions in the helicase-domain. (A) Conformational variations in T. maritima reverse gyrase crystal structures. The mean rmsd values of the five structures were calculated per residue and placed into the B-value column of one of the models. For dimers, only the first chain was selected due to close similarity imposed by NCS restraints. The backbone was drawn as a tube with the radius correlating with the average rmsd. The scale of the rainbow coloring is from blue for rmsd <0.5 Å to red for rmsd >3.5 Å. Large variations in the structures are apparent for the latch and parts of the H2 domain (left), the insert region (bottom right) and the topoisomerase domain (top). In contrast, the core part of the topoisomerase domain and the zinc fingers appear structurally invariant. In some of the structures the latch is involved in crystal contacts, whereas in others it is devoid of external interactions. Averaging of the structures should thus produce a faithful picture of its plasticity. (B) Anisotropy titrations of a 30mer ssDNA with the helicase domain of reverse gyrase (squares), the helicase domain lacking the insert helix in H1 (Δhelix, shown as circles) and the helicase domain lacking the latch inserted into H2 (Δlatch, shown as triangles). The determined Kd-values of the DNA complexes are 20 ± 1 nM (helicase domain), 252 ± 65 nM (Δhelix) and 524 ± 74 nM (Δlatch). The 60mer ssDNA bound more tightly to all proteins, with Kd-values of 10 ± 2 nM (helicase domain), 147 ± 19 nM (Δhelix) and 347 ± 67 nM (Δlatch). (C) Comparison of relative ssDNA affinities for the 30mer (light gray) and the 60mer ssDNA (gray). Deletion of the insert helix reduced the DNA affinity 13- to 15-fold. Deletion of the latch reduces the affinity 26- to 35-fold, consistent with contributions from both elements to ssDNA binding.
Structural diversity and plasticity of the latch domains
The latch is inserted into H2 at Gly389 (T. maritima numbering). Its 70 residues (Arg390–Pro460) form an independent domain, as evidenced by the unaltered structure of H2 upon deletion of the latch and the fact that it can be produced as a separate entity (16). Although a Domain ALIgnment (DALI) (37) search for the A. fulgidus latch domain had indicated structural homology to the RNA-binding domain of transcription factor Rho (11), no homology to any known structure was found for the T. maritima latch domain. Aligning the latch domains by a superposition of T. maritima and A. fulgidus H2 reveals very different structures (Figure 3A and B). Based on the poor sequence conservation in this region (16) and the observation that the latch in different reverse gyrases can vary between 60 and 120 residues (Supplementary Figure S2), this lack of structural similarity is not unexpected. The folds of the latches in T. maritima and A. fulgidus reverse gyrases differ greatly due to a different arrangement of helical elements (Figure 3A and B). As a consequence, the structural similarity found between the latch of A. fulgidus reverse gyrase and the RNA-binding domain of transcription factor Rho is not upheld with the latch domain of T. maritima reverse gyrase. Thus, the T. maritima reverse gyrase latch should be viewed as a novel structure. The considerable sequence variability of the latch region among reverse gyrases (16) (Supplementary Figure S2) predicts different structures for this region in other reverse gyrases as well.
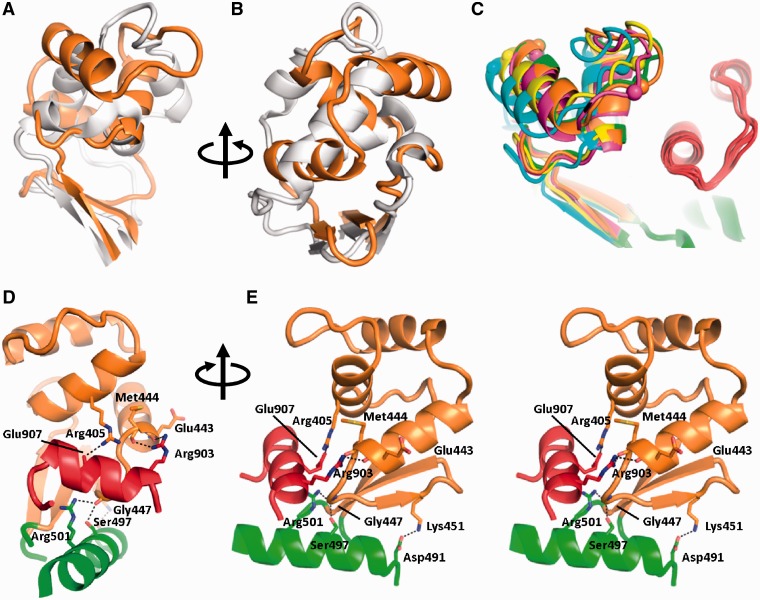
Figure 3.
Comparison of the latch structures. (A) Superposition of A. fulgidus (gray) and T. maritima (orange) reverse gyrases. The latch domains are connected to the H2 domains by a short β-sheet but have very different structures and orientations. (B) View rotated 90° around the vertical axis. (C) Superposition of the five T. maritima reverse gyrase structures (yellow, orange, magenta, cyan, green) on H2 (green, bottom right) reveals structural plasticity of the latch domain while the topoisomerase domain remains immobile (also compare Figure 2A). Spheres serve as markers for the magnitude of the structural differences in the latch domains. The orientation is the same as in (A). The part of the topoisomerase domain interacting with the latch is shown in red. (D) Thermotoga maritima reverse gyrase latch/topoisomerase interactions. Possible hydrogen bonds are drawn as dashed lines. (E) Stereo view rotated 90° about the vertical axis.
The latch has been proposed to mediate inter-domain communication between the helicase and topoisomerase parts (35,38). In A. fulgidus reverse gyrase, it suppresses DNA relaxation in the absence of ATP (35) and inhibits ATP hydrolysis in the absence of supercoiling (38). Although it seems to participate in DNA supercoiling during cleavage and re-ligation (38), deletion mutants still positively supercoil DNA (35). In T. tengcongensis reverse gyrase, the latch is not involved in DNA binding but required for positive DNA supercoiling (34). In contrast, the latch in T. maritima reverse gyrase has little effect on ATP hydrolysis but contributes to ssDNA and dsDNA binding to the helicase module (16). The isolated latch also weakly binds ssDNA and dsDNA (16). The latch is critical for the positive thermodynamic linkage between ATP and DNA binding to the helicase domain (16), and for ATP-dependent duplex separation (18). In contrast to A. fulgidus reverse gyrase, the latch does not seem to be involved in repressing relaxation in the absence of nucleotide but is required for positive supercoiling of DNA by T. maritima reverse gyrase (16). Although the different effects of deleting the latch in different enzymes have been puzzling, the different structures of the corresponding regions in A. fulgidus and T. maritima reverse gyrase could rationalize that the latch is a variable entity and may serve different purposes in different reverse gyrases.
Superposition of all T. maritima reverse gyrase structures on domain H2 shows large differences in the latch domains of up to 7 Å within residues 400–430, which were clearly visible even in the electron density for the lower resolution structures. An α-helix (left in Figure 3C) is displaced laterally as a rigid body, whereas the preceding loop region adopts a variety of conformations. These differences demonstrate that the latch is flexible, displaying both rigid body movements and structural plasticity (Figure 2A). Specific interactions of the latch with the topoisomerase domain include only 14 van der Waals interactions, hydrogen bonds between the carbonyl groups of Glu443 and Met444 with the side-chain of Arg903, the carbonyl group of Gly447 with the side-chains of Ser497 and Arg501 and two salt bridges (Arg405/Glu907 and K451/Asp491; Figure 3D and E; PDB-ID 4DDT). The contact surface of the latch and the topoisomerase domain is a mere 745 Å2. Residues involved in interactions with the topoisomerase domain are clustered at the base of the latch such that a tilting movement of the latch would not immediately result in loss of contact to the topoisomerase domain (Figure 3E). In line with this notion, these interactions are retained but for the Gly447/Arg501 hydrogen bond in the 3.0 Å resolution structure 4DDU, that displays a significant tilt of the latch. The small interface between the latch and the topoisomerase domains and the predominantly hydrophilic nature of their interactions would indicate a weak overall connection that can be overcome during the catalytic cycle. Thus, in principle, the structure is consistent with the previous suggestion (11) that the latch may transiently release parts of the topoisomerase domain during the supercoiling reaction.
Zinc fingers in reverse gyrase
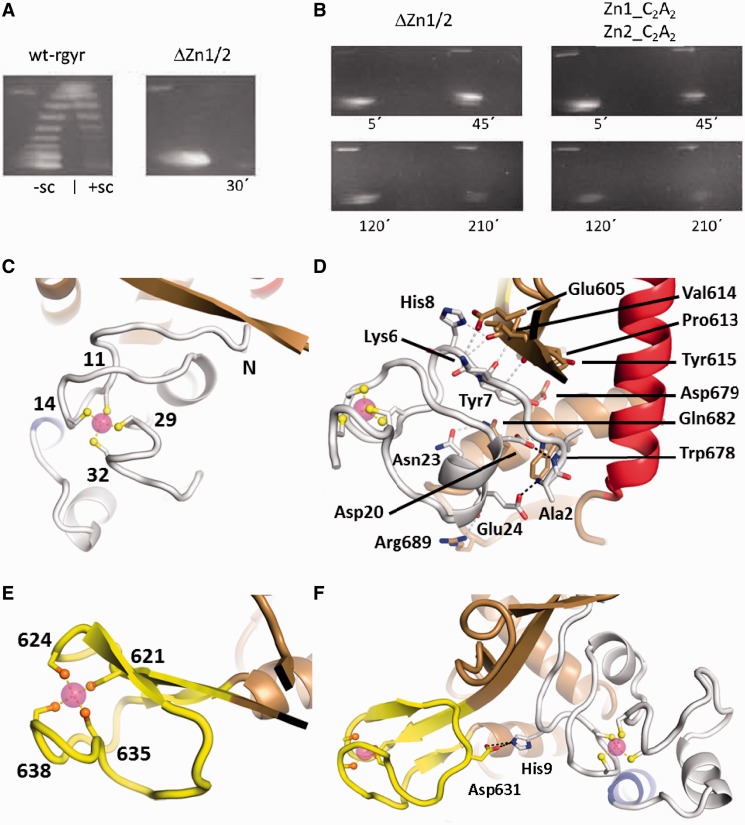
All sequences of reverse gyrases contain a cysteine-rich region within the first 40 N-terminal residues, and in most reverse gyrases a second cysteine-rich region is present within the topoisomerase domain (Supplementary Figure S3). These regions have been suggested to form zinc fingers involved in DNA binding (17). In T. tengcongensis reverse gyrase, both regions contribute to DNA binding (33). The N-terminal putative zinc finger in T. maritima reverse gyrase also contributes to DNA binding and deletion reduces the supercoiling efficiency (36). When we deleted both putative zinc fingers, or mutated two cysteines in each region to alanine, positive DNA supercoiling by T. maritima reverse gyrase was abolished (Figure 4A and B), underlining the critical function of these regions for the supercoiling reaction. Interestingly, reverse gyrase lacking the putative zinc fingers also loses the capability to relax DNA in the presence of ATP, suggesting that these regions may be critical for strand passage in reverse gyrases in general.
Figure 4.
The zinc fingers and their interactions. (A) Deletion of the zinc fingers abolishes supercoiling activity of reverse gyrase. Left panel: distribution of topoisomers generated by wild-type reverse gyrase (wt-rgyr) after 30 min and separated by 2D agarose gel electrophoresis. Negatively and positively supercoiled topoisomers are indicated as −sc and +sc, respectively. Right panel: deletion of the zinc fingers (ΔZn1/2) abolishes positive supercoiling and leads to almost no relaxation. (B) Both deletion of zinc fingers and mutation of individual cysteine residues in the zinc fingers impair supercoiling activities. Left panel: even after 3.5 h the zinc finger deletion construct displays no residual supercoiling activities. Right panel: mutation of two cysteine residues to alanine in both zinc fingers is sufficient for abolishment of supercoiling activities. For constructs, see ‘Materials and Methods’ section. (C) Structure of the N-terminal zinc finger. Zinc (magenta sphere) is coordinated by four cysteine residues (labeled) in an approximately tetrahedral geometry. (D) Possible hydrogen bonds between the N-terminal zinc finger and the Toprim domain are drawn as gray dashed lines. Two more hydrogen bonds in black show how the N-terminus is fixed by zinc finger residues Asp20 and Glu24. (E) The C-terminal zinc finger forms a zinc ribbon that is connected to the Toprim domain via a β-sheet. (F) Single interaction between the zinc fingers via a charged hydrogen bond.
The structure of T. maritima reverse gyrase now offers the first glimpse on the fold of these regions and their potential role in DNA binding and supercoiling. The N-terminal cysteine-rich region folds into a compact domain that harbors a Gag-knuckle zinc finger (39). A β-hairpin called a zinc knuckle carries two cysteine residues (Cys11 and Cys14 in reverse gyrase) at its tip and two more zinc ligands, Cys29 and Cys32, are provided by a short α-helix or loop region (Figure 4C). The charged N-terminus at Ala2 of reverse gyrase is incorporated into the small domain by a hydrogen bond to the side-chain of Glu24 and further fixation is achieved by a hydrogen bond of the first peptide group to the side-chain of Asp20 (Figure 4D). According to a DALI search, this domain has no structural precedent. The zinc finger is intricately connected to the topoisomerase domain. Contacts involve an extension of a β-sheet of the Toprim domain by two short strands (Figure 4C and D). Apart from these main-chain contacts, a total of 119 van der Waals contacts and several more hydrogen bonds stabilize the interface. The side-chains of Lys6, Tyr7 and His8 contact residues Glu605, Asp679 and Val614 (main chain), respectively (Figure 4D). Further hydrogen bonds exist between the side-chains of Asp22/Trp678 and Asn23/Gln682 and between both main-chain carbonyl groups of Glu24 and Arg25 with the side-chain of Arg689. A surface area of 1300 Å2 is buried on reverse gyrase with a relatively high surface complementarity coefficient of 0.7 (40), arguing in favor of a tethering function for the zinc finger that stands in contrast to the plasticity seen with the latch domain (see above). In the isolated helicase domain, this zinc finger can be removed without loss of structure (Supplementary Figure S4) or ATPase activity (Table 2). The N-terminal zinc finger has a peculiar effect on the isolated helicase domain of reverse gyrase. If present, the DNA-stimulated ATPase activity of the helicase module is 3-4-fold reduced, and the KM,app-values for ssDNA and dsDNA are 10- and 20-fold increased, respectively (Table 2). Consistent with these findings, Kd-values of the ssDNA and dsDNA complexes are also increased in the presence of the N-terminal zinc finger, regardless of the nucleotide present (Table 3). Together, these results indicate a detrimental effect of the N-terminal zinc finger to DNA binding in the absence of the topoisomerase domain.
Table 2.
Effect of Zn1 on the steady state ATP hydrolysis
| Enzyme + substrate | kcat (10−3s−1) | KM,app (µM) |
|---|---|---|
| rgyr_hel + ssDNAa | 1160 ± 188 | 0.07 ± 0.04 |
| rgyr_hel + dsDNAa | 1450 ± 64 | 0.18 ± 0.03 |
| rgyr_hel_Zn1 + ssDNA | 470 ± 50 | 0.7 ± 0.3 |
| rgyr_hel_Zn1 + dsDNA | 330 ± 70 | 4 ± 1 |
aFrom previous study (21).
Table 3.
Effect of Zn1 on equilibrium dissociation constants from fluorescence anisotropy titrations (µM)
| Enzyme + substrate | −nucleotide | +ADPNP | +ADP |
|---|---|---|---|
| rgyr_hel + ssDNAa | 0.2 ± 0.01 (n = 1.4)b | 0.46 ± 0.03 (n = 1.4)b | 0.28 ± 0.01 (n = 1.6)b |
| rgyr_hel + dsDNAa | 3.9 ± 0.6 | 0.19 ± 0.03 | 3.7 ± 0.5 |
| rgyr_hel_Zn1 + ssDNA | 1.3 ± 0.4 | 1.3 ± 0.3 | 3.4 ± 0.4 |
| rgyr_hel_Zn1 + dsDNA | not detected | 3.1 ± 0.2 | 10 ± 2 |
aFrom previous study (21).
bBinding curves for ssDNA were analyzed using the Hill equation with Hill coefficient n.
The C-terminal cysteine-rich region in reverse gyrase forms a zinc finger of the ribbon type (39) with two zinc knuckles clamping onto the zinc ion (Figure 4E). The zinc knuckles are joined by an additional β-strand to form a three-stranded anti-parallel β-sheet (Figure 4F). The zinc ribbon in reverse gyrase is inserted into the Toprim domain at the base of the DNA-binding site and protrudes from the surface of the topoisomerase domain (Figures 1B and 4E). No interactions of the zinc ribbon with the Toprim domain are present but the three-stranded β-sheet appears quite rigid, being structurally invariant in all T. maritima reverse gyrase structures (Figure 2A). The two zinc fingers in reverse gyrase are located next to each other on the surface of the topoisomerase domain. The zinc fingers are connected by a single interaction, a salt bridge between His9 and Asp631 (Figure 4F), which may allow for independent and coordinated movements of the zinc fingers at different stages during catalysis.
The zinc fingers are essential for both positive supercoiling and relaxation activities of reverse gyrase. Deletion of the zinc fingers abolishes either activity (Figure 4A), even after extended incubation times of 3.5 h (Figure 4B). More subtle alterations such as mutation of zinc-binding cysteine residues to alanine have the same effect, underscoring the importance of the zinc fingers for either DNA binding, supercoiling or both. Given that the C-terminal zinc finger is not conserved in reverse gyrases (Supplementary Figure S3B), it is probable that impairment of the N-terminal zinc finger is responsible for the observed lack of enzymatic activities. A negative effect on DNA binding after mutating cysteine residues in the N-terminal zinc finger was also noted in T. maritima reverse gyrase (36).
Interactions between the helicase and topoisomerase domains
We have previously shown that the isolated helicase domain of T. maritima reverse gyrase is a nucleotide-dependent conformational switch (21) that unwinds duplex DNA in an ATP-dependent reaction (18). Its DNA-stimulated ATPase activity is increased 10-fold compared with the authentic enzyme where its activity is modulated by the topoisomerase domain (12,21). Although the isolated helicase domain can adopt many different conformations, its conformational space is restricted by the topoisomerase domain in authentic reverse gyrase (12,13,16). This restriction is achieved by direct interactions between H1 and H2 with the topoisomerase domain in separate interfaces (excluding the latch, see above). Both interfaces extend over ca. 1200 Å2 with a comparatively low surface complementarity coefficient of 0.57. 18 residues of H1 form 13 hydrogen bonds, 10 salt bridges and 95 van der Waals contacts, most prominently with topoisomerase residues Ser540, Ser564, Ser565, Arg566, Lys567, Glu575 and Leu568. For H2, there are 17 residues involved that form 8 hydrogen bonds, 6 salt bridges and 94 van der Waals contacts with topoisomerase residues Arg701, Arg903, Glu907, Met908, Glu911 and Leu913 (detailed listing provided in Supplementary Table S2). Compared with H1, H2 has fewer interactions with the topoisomerase domain, which suggests that H2 is less tightly connected and could be released during catalysis. In contrast, H1 enjoys additional connections to the topoisomerase domain via the N-terminal zinc finger domain (Figure 4D), and should therefore remain firmly attached to the topoisomerase domain in the catalytic cycle.
DNA-binding sites on reverse gyrase
The electrostatic surface potential of T. maritima reverse gyrase has several positively polarized regions that could function as DNA-binding sites (Figure 5A). A continuous stretch of positive potential reaches from H1 underneath the latch, past the insert helix, over the zinc ribbon (Zn2), toward the ssDNA-binding site of the topoisomerase domain (Figure 5A). The DNA-binding site on the topoisomerase domain was also found for A. fulgidus reverse gyrase (11) and is exemplified by the crystal structure of Escherichia coli topoisomerase III in complex with ssDNA (19). A superposition of reverse gyrase with the DNA topoisomerase III/ssDNA complex places the ssDNA at an equivalent site on the reverse gyrase topoisomerase domain (Figure 5B). The ssDNA-binding site adjacent to the catalytic tyrosine in the topoisomerase domain of reverse gyrase is in a closed conformation, where the catalytic Tyr851 is inaccessible to solvent and the DNA-binding cleft is too narrow to accommodate ssDNA. Binding of ssDNA thus requires a subtle conformational change, such as a tilt of the topoisomerase domain that widens the DNA-binding cleft. To further dissect DNA binding to the helicase-domain and the contributions of the insertions in H1 and H2, we determined Kd-values of DNA complexes for the helicase domain and for deletion mutants lacking the latch or the helical insert (Figure 2B and C). The helicase domain bound a 30mer ssDNA with nanomolar affinities. Deletion of the insert helix in H1 leads to a 13-fold increase of the Kd-value, deletion of the latch causes a 26-fold increase. Similar effects of the insertions were observed with a 60mer ssDNA (Figure 2C). The latch and the insert helix thus contribute to binding of ssDNA to reverse gyrase, supporting the possible DNA path across reverse gyrase in its open conformation outlined by the electrostatic potential.
Figure 5.
Generation of a model for reverse gyrase with the helicase domain in the closed conformation. (A) Electrostatic surface potential calculated using APBS (41) and displayed at ±5 kbT. Positive potential (blue) suggests a continuous DNA-binding site from H1 to the topoisomerase domain. (B) Comparison of reverse gyrase with the closed conformation of the RNA helicase Vasa (42) in complex with Mg2+/ADPNP and oligo-U RNA (PDB-ID 2DB3; yellow) and with the E. coli DNA topoisomerase III/ssDNA (PDB-ID 1I7d; cyan) complex (19). The arrow depicts the magnitude of the projected H2 movement upon closure of the helicase domain. (C) Surface model of reverse gyrase with the helicase domain in the closed conformation. Large conformational changes of H2 and the latch relative to the topoisomerase domain are required. Bound DNA is shown in magenta. H1 and H2 are colored blue and green, respectively. Possible DNA interacting areas such as the latch, the insert region and the topoisomerase-based zinc fingers are colored separately. The topoisomerase domain is kept white and the directionality of the modeled ssDNAs is indicated by their 5′-end.
DISCUSSION
Here, we describe a set of T. maritima reverse gyrase structures that reveal two previously invisible zinc fingers and also include a novel structure for the latch domain. The structures represent the apo state where the helicase domain is in an open conformation and the topoisomerase domain is closed with respect to its ssDNA-binding site. This overall domain arrangement is similar to A. fulgidus reverse gyrase, lending support to the notion that this conformation is an intermediate during catalysis. We can map flexible regions in reverse gyrase by comparison of the independent T. maritima structures. For instance, structural plasticity is present in the apex of the topoisomerase domain. The latch domain inserted in H2 appears to constitute the most versatile part of reverse gyrase as judged by elevated B-values and differences among the individual structures. Rigid body flexibility is present in the helical insert in H1, which is well defined by electron density but may move somewhat independently of H1.
The two zinc fingers are essential for positive DNA supercoiling by T. maritima reverse gyrase. One is located at the N-terminus and another one is inserted into the topoisomerase domain. Both zinc fingers are loosely inter-connected by a single salt bridge. It has been suggested that the zinc fingers are natively unfolded and assume structure upon DNA binding (11) but in T. maritima, reverse gyrase DNA is not required for zinc finger folding. Instead, the N-terminal zinc finger extensively contacts the topoisomerase domain, providing a tight connection of the helicase and topoisomerase domains.
There are a number of inter-domain contacts between the helicase and the topoisomerase modules. They are predominantly centered at H1, whereas comparatively fewer contacts exist with H2, the latch domain being an exception. This intricate, yet plastic connection between the helicase module and the topoisomerase domain, particularly by the latch and the zinc fingers that are elements unique to reverse gyrase, supports a mechanism of positive DNA supercoiling that relies on a close communication between these two domains, yet involves structural rearrangements in the catalytic cycle. From the A. fulgidus structure, a model for DNA supercoiling by reverse gyrase has been put forward that predicted a conformational change in the helicase module as a first step, initiating a cascade of subsequent conformational changes that ultimately lead to strand passage and DNA supercoiling (11). We recently demonstrated that the isolated helicase module of T. maritima reverse gyrase is indeed a conformational switch that, in-line with its similarity to SF2 helicases, alternates between an open and a closed conformation in the nucleotide cycle when DNA is bound (12). In DEAD box helicases (14) closure of the cleft between the H1 and H2 domains creates a binding site for nucleic acids covering both domains (42). The bipartite binding site rationalizes the cooperativity between ATP and nucleic acid binding and the increase in nucleic acid affinity in the closed state (43). Strikingly, in the reverse gyrase helicase module, these conformational changes are coupled to changes in dsDNA affinity, but not to the affinity for ssDNA (12), suggesting that specific elements not shared with DEAD box proteins may contribute to ssDNA binding. Here, we have identified contributions of the insert helix and the latch to binding of reverse gyrase to ssDNA. The helicase module confers ATP-dependent DNA binding to reverse gyrase. In line with the observed closure of the reverse gyrase helicase domain in the presence of ATP and DNA, ATP-hydrolysis in authentic reverse gyrase only occurs upon binding of nucleic acid (12,21). The crystal structures presented here enable construction of a complete model for the DNA-bound state of T. maritima reverse gyrase, with the helicase module in the closed conformation (Figure 5). A closure of the helicase domain in reverse gyrase involves sizeable movements of the H1/H2 domains with respect to each other. The extent of the required domain movement becomes apparent upon alignment of the H1 domains of reverse gyrase and the closed conformation of a RNA helicase, e.g. Vasa (42) in complex with Mg2+/ADPNP and oligo-U RNA (Figure 5B). A movement of H2 toward H1 appears likely because the N-terminal zinc finger connects H1 tightly to the Toprim domain. An α-helical segment connects the zinc finger to H1. This arrangement could function as a lever arm that allows conformational cycling of the H1 and H2 domains between open and closed states without losing contact to the topoisomerase domain.
The helicase domain of reverse gyrase binds ssDNA and dsDNA in the absence of a nucleotide, with a strong preference for ssDNA, ss/dsDNA junctions and ssDNA bubbles (12). This high ssDNA affinity is retained in the closed conformation with ATP bound to it (12). We have recently demonstrated that the helicase domain, as well as full-length reverse gyrase unwind duplex DNA in an ATP-dependent reaction (18), suggesting that the helicase domain provides ssDNA regions for binding to and strand passage within the topoisomerase domain. A model of reverse gyrase in complex with ssDNA can be constructed by analogy with the E. coli topoisomerase III/ssDNA complex (Figure 5B) (19). Together with the model of the closed helicase module discussed above, an extended picture of the entire DNA-bound reverse gyrase emerges (20) (Figure 6). In this model, nucleic acid bound to the helicase and the topoisomerase domains is oriented with the same polarity, suggesting that they belong to the same strand of the dsDNA substrate (Figure 5C). This DNA strand becomes trans-esterified onto Tyr851, providing the gap through which the second strand must pass. The DNA is predominantly bound via the backbone phosphates in Vasa and in topoisomerase III, relying on electrostatic and hydrogen bond interactions. Importantly, the topoisomerase residues involved in DNA binding are conserved in reverse gyrase (Supplementary Table S3). Our model neatly juxtaposes the elements that contribute to DNA binding into the vicinity of the modeled DNA path. From the helicase domain, the DNA strands could stretch between the latch and the insertion in H1, and continue via the zinc ribbon to the ssDNA-binding site in the topoisomerase domain. The latch region is required for duplex unwinding by the helicase domain (18). The insert region in reverse gyrases is close to the suggested DNA path, contributes to ssDNA binding (13) and may even aid in strand separation (18). The conserved N-terminal zinc finger, is further away from the DNA-binding motifs but could be reached by the double-stranded part of substrate DNA, in agreement with the effect of this zinc finger on dsDNA binding (Table 3). In contrast, the topoisomerase-located zinc ribbon is expendable in some reverse gyrases (Supplementary Figure 3B). Although a zinc ribbon is present in T. maritima topoisomerase I (44) at a different location (Supplementary Figure S5), no zinc finger exists in E. coli topoisomerases I and III (19,45,46). Despite its dispensability, the prominent placement at the base of the topoisomerase DNA-binding site may point to an involvement of this zinc finger in DNA guidance during strand passage.
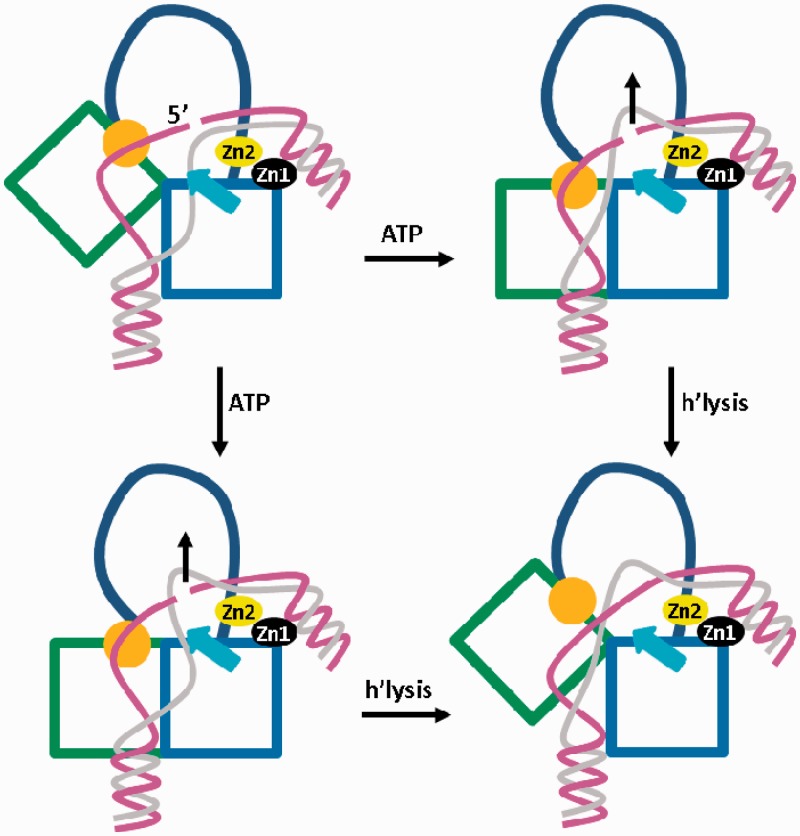
Figure 6.
Scheme for DNA strand passage coupled to closure of the helicase module. The H1 and H2 domains are drawn as blue and green squares, respectively. The latch domain in H2 is drawn in orange, the insert region in H1 is shown as a cyan arrow and the topoisomerase domain is simplified by an arc. The cleaved ssDNA strand is drawn in magenta and the ssDNA strand passing through the gap is shown in gray. The covalently fixed 5′-end is indicated. Zinc fingers are labeled. The latch and the insert region contact the single strands of the DNA substrate. Binding of ATP leads to closure of the helicase domain, and associated conformational changes of the lid, the insert and/or the zinc fingers direct the passing strand through the gap (black arrow), thus increasing the linking number. The insert region may disengage from the DNA before ATP hydrolysis (right wing) or after ATP hydrolysis (left wing). At the bottom right, the post-ligation state is shown.
The model illustrates a possible binding mode for the strands of the dsDNA substrate to reverse gyrase. The strand that is transiently cleaved by the topoisomerase domain is contacted by positively charged residues on the latch. The insert region might aid in fixing the opposite strand (Figure 6). After ATP binding and closure of the helicase module, a conformational change involving H2 and the latch may provoke a movement of the intact DNA strand upward through the gap in the opposite strand, created through cleavage by the topoisomerase domain. Most likely, the zinc fingers facilitate this DNA movement. As a result, the linking number would increase by one (Figure 6). Although the role of the insert region in H1 is not entirely clear, its transient interaction with ssDNA could rationalize facile disengagement at any point of the catalytic cycle, possibly coupled to strand passage. Alternatively, DNA may first be released from the latch domain depending on which interaction is the weakest. Distortion of the bound DNA by reverse gyrase may further contribute to release of DNA from the weakest interaction point. The model offers an explanation for our previous finding that the latch is required for positive supercoiling by T. maritima reverse gyrase, but is dispensable for (ATP-dependent) relaxation of negatively supercoiled dsDNA (16). Deletion of the latch results in a loss of DNA binding at this site of reverse gyrase and would abolish the coupling of the H2/latch movement to strand passage.
In conclusion, the structures of T. maritima reverse gyrase discussed here help complete a hitherto fragmented structural view on this enzyme class and allow delineation of a more detailed model for strand passage and supercoiling. This model is in agreement with present biochemical data and suggests specific roles for structural elements unique to reverse gyrase in nucleic acid binding and DNA supercoiling that will be addressed in future work.
ACCESSION NUMBERS
The coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 4DDU, 4DDT, 4DDV, 4DDW, 4DDX).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–5 and Supplementary References [48 and 49].
FUNDING
The Swiss National Science Foundation (to D.K.); DFG [SFB858, to D.K.]; European Union (Marie Curie Fellowship, to A.G.). Funding for open access charge: University of Muenster.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at SLS beamline PX-II for support during data collection, and Ines Hertel and Andreas Schmidt for purification of reverse gyrase. M.G.R. determined the crystal structures and wrote the manuscript. A.G., S.P.J. and Y.dT.D. performed enzymatic assays. D.K. designed the research and wrote the manuscript.
REFERENCES
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakasu S, Kikuchi A. Reverse gyrase; ATP-dependent type I topoisomerase from Sulfolobus. EMBO J. 1985;4:2705–2710. doi: 10.1002/j.1460-2075.1985.tb03990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 6.Kampmann M, Stock D. Reverse gyrase has heat-protective DNA chaperone activity independent of supercoiling. Nucleic Acids Res. 2004;32:3537–3545. doi: 10.1093/nar/gkh683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh TS, Plank JL. Reverse gyrase functions as a DNA renaturase: annealing of complementary single-stranded circles and positive supercoiling of a bubble substrate. J. Biol. Chem. 2006;281:5640–5647. doi: 10.1074/jbc.M513252200. [DOI] [PubMed] [Google Scholar]
- 8.Capp C, Qian Y, Sage H, Huber H, Hsieh TS. Separate and combined biochemical activities of the subunits of a naturally split reverse gyrase. J. Biol. Chem. 2010;285:39637–39645. doi: 10.1074/jbc.M110.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krah R, Kozyavkin SA, Slesarev AI, Gellert M. A two-subunit type I DNA topoisomerase (reverse gyrase) from an extreme hyperthermophile. Proc. Natl Acad. Sci. USA. 1996;93:106–110. doi: 10.1073/pnas.93.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc. Natl Acad. Sci. USA. 1993;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Toro Duany Y, Klostermeier D. Nucleotide-driven conformational changes in the reverse gyrase helicase-like domain couple the nucleotide cycle to DNA processing. Phys. Chem. Chem. Phys. 2011;13:10009–10019. doi: 10.1039/c0cp02859b. [DOI] [PubMed] [Google Scholar]
- 13.del Toro Duany Y, Klostermeier D, Rudolph MG. The conformational flexibility of the helicase-like domain from Thermotoga maritima reverse gyrase is restricted by the topoisomerase domain. Biochemistry. 2011;50:5816–5823. doi: 10.1021/bi200236a. [DOI] [PubMed] [Google Scholar]
- 14.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 15.Valenti A, Perugino G, D'Amaro A, Cacace A, Napoli A, Rossi M, Ciaramella M. Dissection of reverse gyrase activities: insight into the evolution of a thermostable molecular machine. Nucleic Acids Res. 2008;36:4587–4597. doi: 10.1093/nar/gkn418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganguly A, del Toro Duany Y, Rudolph MG, Klostermeier D. The latch modulates nucleotide and DNA binding to the helicase-like domain of Thermotoga maritima reverse gyrase and is required for positive DNA supercoiling. Nucleic Acids Res. 2011;39:1789–1800. doi: 10.1093/nar/gkq1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaxel C, Bouthier de la Tour C, Duguet M, Nadal M. Reverse gyrase gene from Sulfolobus shibatae B12: gene structure, transcription unit and comparative sequence analysis of the two domains. Nucleic Acids Res. 1996;24:4668–4675. doi: 10.1093/nar/24.23.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguly A, del Toro Duany Y, Klostermeier D. Reverse gyrase transiently unwinds double-stranded DNA in an ATP-dependent reaction. J. Mol. Biol. 2013;425:32–40. doi: 10.1016/j.jmb.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 20.Jungblut SP, Klostermeier D. Adenosine 5'-O-(3-thio)triphosphate (ATPγS) promotes positive supercoiling of DNA by T. maritima reverse gyrase. J. Mol. Biol. 2007;371:197–209. doi: 10.1016/j.jmb.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 21.del Toro Duany Y, Jungblut SP, Schmidt AS, Klostermeier D. The reverse gyrase helicase-like domain is a nucleotide-dependent switch that is attenuated by the topoisomerase domain. Nucleic Acids Res. 2008;36:5882–5895. doi: 10.1093/nar/gkn587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warkentin M, Thorne RE. A general method for hyperquenching protein crystals. J. Struct. Funct. Genomics. 2007;8:141–144. doi: 10.1007/s10969-007-9029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Evaluation of single crystal x-ray diffraction data from a position sensitive detector. J. Appl. Cryst. 1988;21:916–924. [Google Scholar]
- 24.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 28.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 29.Einspahr HM, Weiss MS. International Tables for Crystallography Vol. F, Crystallography of Biological Macromolecules. New Jersey: Wiley & Sons; 2011. Quality indicators in macromolecular crystallography: Definitions and applications. 2nd edn. pp. 64–74. [Google Scholar]
- 30.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 32.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 33.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl Acad. Sci. USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Liu J, Zhou J, Xiang H. Functional evaluation of four putative DNA-binding regions in Thermoanaerobacter tengcongensis reverse gyrase. Extremophiles:life under extreme conditions. 2011;15:281–291. doi: 10.1007/s00792-011-0356-5. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez AC. Studies of a positive supercoiling machine. Nucleotide hydrolysis and a multifunctional “latch” in the mechanism of reverse gyrase. J. Biol. Chem. 2002;277:29865–29873. doi: 10.1074/jbc.M202853200. [DOI] [PubMed] [Google Scholar]
- 36.Bouthier de la Tour C, Amrani L, Cossard R, Neuman KC, Serre MC, Duguet M. Mutational analysis of the helicase-like domain of Thermotoga maritima reverse gyrase. J. Biol. Chem. 2008;283:27395–27402. doi: 10.1074/jbc.M800867200. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez AC. Investigating the role of the latch in the positive supercoiling mechanism of reverse gyrase. Biochemistry. 2003;42:5993–6004. doi: 10.1021/bi034188l. [DOI] [PubMed] [Google Scholar]
- 39.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural Basis for RNA Unwinding by the DEAD-Box Protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 43.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl Acad. Sci. USA. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen G, Harrenga A, Wieland B, Schomburg D, Reinemer P. Crystal structure of full length topoisomerase I from Thermotoga maritima. J. Mol. Biol. 2006;358:1328–1340. doi: 10.1016/j.jmb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 46.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure. 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 47.Viard T, Lamour V, Duguet M, Bouthier de la Tour C. Hyperthermophilic topoisomerase I from Thermotoga maritima. A very efficient enzyme that functions independently of zinc binding. J. Biol. Chem. 2001;276:46495–46503. doi: 10.1074/jbc.M107714200. [DOI] [PubMed] [Google Scholar]
- 48.Viard T, Cossard R, Duguet M, de La Tour CB. Thermotoga maritima-Escherichia coli chimeric topoisomerases. Answers about involvement of the carboxyl-terminal domain in DNA topoisomerase I-mediated catalysis. J. Biol. Chem. 2004;279:30073–30080. doi: 10.1074/jbc.M309692200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.