Abstract
Ribosomal RNA undergoes various modifications to optimize ribosomal structure and expand the topological potential of RNA. The most common nucleotide modifications in ribosomal RNA (rRNA) are pseudouridylations and 2′-O methylations (Nm), performed by H/ACA box snoRNAs and C/D box snoRNAs, respectively. Furthermore, rRNAs of both ribosomal subunits also contain various base modifications, which are catalysed by specific enzymes. These modifications cluster in highly conserved areas of the ribosome. Although most enzymes catalysing 18S rRNA base modifications have been identified, little is known about the 25S rRNA base modifications. The m1A modification at position 645 in Helix 25.1 is highly conserved in eukaryotes. Helix formation in this region of the 25S rRNA might be a prerequisite for a correct topological framework for 5.8S rRNA to interact with 25S rRNA. Surprisingly, we have identified ribosomal RNA processing protein 8 (Rrp8), a nucleolar Rossman-fold like methyltransferase, to carry out the m1A base modification at position 645, although Rrp8 was previously shown to be involved in A2 cleavage and 40S biogenesis. In addition, we were able to identify specific point mutations in Rrp8, which show that a reduced S-adenosyl-methionine binding influences the quality of the 60S subunit. This highlights the dual functionality of Rrp8 in the biogenesis of both subunits.
INTRODUCTION
Eukaryotic ribosomes are large ribonucleoprotein complexes consisting of two unequal subunits, the 40S and 60S. The synthesis of these ribosomes is a complex multi-step process where 79 ribosomal proteins and 4 ribosomal RNAs (rRNAs) are assembled into mature ribosomal particles. In Saccharomyces cerevisiae, >200 non-ribosomal proteins and 75 snoRNAs are required to complete this sophisticated task (1,2). The four mature rRNAs that form the ribosome are transcribed from a single rDNA unit, which is organized in tandem repeats on chromosome XII in S. cerevisiae. The 18S, 25S and 5.8S are transcribed as a single pre-35S transcript by RNA pol I, whereas the 5S rRNA is transcribed by RNA pol III. Early cleavage events at sites A0–A2 within the primary 35S transcript separate the biogenesis pathways of the two ribosomal subunits (1,3,4). Although the pre-rRNA intermediates of the pre-60S complexes are processed in the nucleus before nuclear export, the final processing and modification of pre-rRNA of 40S completes in the cytoplasm (5,6). During the processing events, the rRNA of both subunits also undergoes various chemical modifications. The most common chemical modifications in rRNA are pseudouridylations (Ψ) and 2′-O methylations (Nm), performed by small ribonucleoprotein particles (snoRNPs). These snoRNPs consist of an RNA component that functions in the substrate binding and different proteins that have structural and catalytic functions. Most of the snoRNPs fall into one of two classes, the box H/ACA snoRNPs and box C/D snoRNPs responsible for the pseudouridylation and 2′-O methylation, respectively (7,8). Mapping of the modified nucleotide residues in eukaryotic and archaeal ribosomes clearly revealed that these modifications cluster in highly conserved areas of the ribosome like the peptidyltransferase centre, sites of A- and P-tRNA binding, the peptide exit tunnel and intersubunit bridges (9). Therefore, it is believed that the modification of ribonucleotides optimizes the rRNA structure and represents a way to expand the topological potentials of RNA molecules (10).
Apart from the pseudouridylation and 2′-O ribose methylation, the bases of rRNA are also known to undergo methylation, where a methyl group is added to the nitrogen atom. Interestingly, these base modifications are introduced by specific and snoRNA-independent enzymes. In yeast Dim1 catalyses the N-dimethylation of two neighbouring adenosines at the 3′-end of 18S (11). Bud23, a base methyltransferase (MTase), has been shown to catalyse the N7-methylation of guanosine 1575 (12). Recently, we discovered Nep1 to be responsible for the N1-methylation of the unique hyper modified m1acp3Ψ 1191 base of eukaryotic 18S rRNA, which is located in the decoding centre of the ribosome (13). In contrast to the mild growth defects observed for snoRNA deletion mutants, the specific enzymes for base modifications described so far (Dim1, Nep1, Bud23) are either essential enzymes or have a drastic effect on cell growth. However, similar to snoRNA-dependent modifications, there is increasing evidence that the base modification itself is not essential, suggesting that base modifying proteins play a bifunctional role in ribosome biogenesis. Similarly, the snoRNAs that are involved in both modification and processing of rRNA are also the essential snoRNAs (14–16). Therefore, the concurrence of modification and cleavage events are suggested to happen in a coordinated manner, implicating that the binding of these factors to pre-rRNA has possible chaperone-like functions. Recent observations with the interdependence of the functional and modification domains for the snR10 have corroborated this hypothesis (17). Crystal structures of the essential protein Nep1 and its complexes with RNA also suggest that Nep1 changes rRNA structure on binding and stabilizes a stem–loop structure, which may guide the assembly of Rps19 (18). Importantly, the enzymatic activity is not required for the essential function, as mutations in Nep1 that impair S-adenosyl-methionine (SAM) binding do not result in lethality (19). The minimal impact on cell growth on loss of this rRNA modification corresponds to the observation that most of the snoRNAs responsible for modifications are not essential (20).
Despite the fact that all snoRNPs causing pseudouridylation and 2′-O methylation of 18S and 25S rRNA and all snoRNA-independent enzymes modifying the 18S rRNA have already been described, no information is available about proteins that catalyse the remaining base modifications of 25S rRNA in yeast. This may also argue for the involvement of non-essential proteins in these modification reactions. Only the essential protein Nop2, which is important for the processing and maturation of 27S pre-rRNA and large ribosomal subunit biogenesis, is predicted to be most likely an m5C MTase (21,22). Comparative with ribose modifications, the base modifications also tend to be in close vicinity to the highly modified regions, such as Helix 69, which forms the intersubunit bridge of the ribosome, or within the peptidyltransferase centre (9). Nevertheless, there is also a lack of information about the impact of these modifications on yeast cell growth. In contrast to this, a large number of base modifying enzymes for the 23S rRNA of Escherichia coli have been reported. However, the physiological relevance of most modifications for which the responsible proteins have been identified is still unknown (23–25). The current hypothesis is that these modifications could become important only under specific growth or stress conditions.
The identification of proteins responsible for the base methylations in yeast could help to unravel the physiological relevance of these modifications. Recent bioinformatic analyses have made it possible to predict the general substrates for several putative MTases (26). The ribosomal RNA processing protein ribosomal RNA processing protein 8 (Rrp8) has also been predicted to modify tRNA or rRNA. Rrp8 was previously shown to co-purify with snR190/U14 (boxG) as well as with rRNA corresponding to the A2 site of the ITS1 region (27). This is in accordance to its involvement in processing at site A2 and the genetic interaction with the essential H/ACA snoRNP protein, Gar1 (28). We have also reported a genetic interaction of RRP8 with the essential biogenesis factor NEP1, arguing for a specific role in 40S subunit biogenesis (29). Interestingly, Rrp8 was also found in early pre-60S complexes containing the snoRNA-independent MTase Spb1, suggesting that Rrp8 remains associated to the 27S A2 pre-rRNA directly after A2 cleavage (30). Moreover, together with snoRNPs and Spb1, it seems to dissociate from this precursor before processing proceeds, indicating that Rrp8 is part of an rRNA modification machinery. In this study, we showed for the first time that the Rossmann-fold-like protein Rrp8 is responsible for a base methylation of the 25S rRNA. In addition, we were able to identify specific point mutations in Rrp8 that showed a distinct effect on the 60S subunit. Furthermore, we elaborated the previously known genetic interaction of RRP8 with GAR1 emphasizing a yet unknown role of Rrp8 in the biogenesis of the 60S subunit. Our data highlight a dual functionality of Rrp8 in the biogenesis of both small and large subunits.
MATERIALS AND METHODS
Plasmid and yeast strain construction
Detailed descriptions are available in Supplementary Data. All yeast strains used and constructed are listed in Supplementary Table S1. Oligonucleotides used for plasmid constructions are listed in Supplementary Table S2.
Growth conditions and yeast media
Yeast strains were grown at 30°C in YEPD medium (1% of yeast extract, 2% of peptone, 2–4% of glucose) or in synthetic dropout medium (0.5% of ammonium sulphate, 0.17% of yeast nitrogen base, 2–4% of glucose). For selection on KanMX, G418 was added to the medium (0.2 mg/ml). The 5-FOA plates were prepared using synthetic dropout medium to which 0.05 mg/ml of uracil and 1 mg/ml of 5-FOA was added. For serial dilution growth assays, yeast cells were grown over night in YEPD medium and diluted to an OD600 of 1 followed by 1:10 serial dilutions. From the diluted cultures, 5 µl was spotted onto YEPD plates and was incubated at 30°C or 19°C. For growth analysis of tetracycline (tc) aptamer containing strains, serially diluted cells were spotted on YEPD plates containing 0 or 50 µM tetracycline. Growth curves with tc aptamer containing strains were performed in liquid YEPD media containing 0 or 250 µM tetracycline. To test for paromomycin sensitivity, 5 µl of a paromomycin solution (200 mg/ml) was spotted on filter discs, which were applied on YEPD plates containing the strains to be tested.
Protein expression and purification
DNA coding for the open reading frame of yeast RRP8 was cloned into the expression vector pPK591. The resulting plasmid pKO2 was transformed into E. coli strain BL21 (DE3) (Novagen) for protein expression. The cells were grown at 37°C to an OD600 of ∼0.6, and 0.1 mM isopropyl-ß-d-thiogalactopyranoside was used to induce protein expression for 18 h at RT. Purification of 6× His-tagged wild-type Rrp8 protein was performed with the HIS-Select® Nickel Affinity Gel (Sigma-Aldrich) according to the manufacturer’s protocol. Cells were disrupted in lysis buffer [300 mM of NaCl, 1 mM of ethylenediaminetetraacetic acid (EDTA), 50 mM of Tris–HCl pH 8.0] with 0.1 M of PMSF, 0.5 M of benzamidine and 0.015% of ß-mercaptoethanol (14.3 M) by extensive freezing and thawing. After binding the proteins, matrix was washed three times with wash buffer (50 mM of NaPO4 pH 8.0, 150 mM of NaCl and 10 mM of ß-mercaptoethanol). Elution was performed in wash buffer with the addition of 250–500 mM imidazol. Elution fractions were pooled and extensively dialysed with dialysis buffer (20 mM of Tris–HCl pH 7.5, 100 mM of NaCl, 0.1 mM of EDTA, 1 mM of DTT and 50% of glycerol).
In vitro SAM binding assay
Biochemical determination of SAM binding by ultraviolet (UV) cross-linking was performed as described previously (31). Recombinant 6× His–Rrp8 protein (25–30 µg) was mixed with 2 µCi [3H-methyl] SAM (80 Ci/mmol, Hartmann Analytic GmbH) in a buffer containing K3PO4 pH 7.0, 100 mM of NaCl, 2 mM of EDTA, 1 mM of DTT, 5% of glycerol in an open plastic 96-well plate. The reaction mixture was exposed to UV irradiation on ice in a Stratagene UV Stratalinker 1800 using the Autocrosslink function (120 000 μJ and 25–50 s). The products were run on a 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and stained with Coomassie blue. After destaining, the gel was treated for fluorography as previously described (32). The undried gel was exposed to X-ray film (GE Healthcare) for 72 h at −80°C.
Sucrose gradient analysis
For the separation of the subunits, sucrose gradient analysis was performed on a 10–50% gradient in ribosome buffer B (50 mM of Tris–HCl pH 7.6, 50 mM of NaCl, 1 mM of DTT). Lysates of exponentially growing yeast cells were prepared in 1 ml of buffer B with glass beads for 2 min at 4°C. The amount of RNA was determined at 254 nm, and 5-OD254 units were loaded on the gradients. Separation of the subunits was performed by gradient ultracentrifugation in a SW40 Ti rotor (Beckman Coulter Inc.) for 17 h at 24 500 r.p.m. and 4°C. Calculation of the 40S to 60S ratio was performed by using the ImageJ software (http://rsbweb.nih.gov/ij/). The area below the peaks was determined and multiplied by their respective concentrations. One OD254 unit of the 40S subunit corresponds to 54.92 pmol, whereas one OD254 unit of the 60S corresponds to 26.44 pmol. Preparation of polysome profiles was performed on 10–50% gradients in polysome buffer A (20 mM of HEPES pH 7.5, 10 mM of KCl, 2.5 mM of MgCl2, 1 mM of EGTA and 1 mM of DTT). Exponentially growing yeast cells were prepared as described, and 10-OD254 units were used for gradient ultracentrifugation in a SW40 Ti rotor (Beckman Coulter Inc.) for 17 h at 19 000 r.p.m. and 4°C.
RNA extraction and northern hybridization
For northern blot analysis, RNA was prepared by phenol/chloroform extraction as previously described (33). Ten microgram of RNA was separated on 1% of agarose gel in 1× TAE supplemented with 6.66% formaldehyde and transferred to a positively charged nylon membrane (Hybond N+, GE Healthcare) using capillary blotting. In all, 50 pmol of the corresponding oligonucleotides (Supplementary Table S2) were radioactively labelled at the 5′-end using 6 µl γ32P-adenosine triphosphate (∼3.3 pmol/µl, Hartmann-Analytik) and 1 µl T4 polynucleotide kinase (Roche) in the supplied buffer for 1 h at 37°C and purified with G-25 columns. Hybridization was done in 15 ml hybridization buffer (GE Healthcare) overnight at 42°C, and signals were visualized by phosphoimaging using a Typhoon 9100 (GE Healthcare).
Preparation of rRNA
For preparation of 25S rRNA, exponentially growing yeast cells from 200 ml of YEPD medium were harvested, and cell extracts were prepared using glass beads in ribosome buffer B. Ribosome subunits were separated as already described by gradient ultracentrifugation using a 20–50% sucrose gradient and 20-OD254 units. The 60S subunits were collected with the Density Gradient Fractionation System (Teledyne Isco) and precipitated with 2 vol of 100% ethanol at −20°C for 16 h. Precipitated 60S subunits were dissolved in water, and 25S rRNA was purified using the RNeasy Kit (QIAGEN) following the protocol for RNA cleanup. RNA was eluted in two steps with 65 µl water each.
High-performance liquid chromatography
For high-performance liquid chromatography (HPLC) analysis, 100 pmol 25S rRNA was digested with nuclease P1 and bacterial alkaline phosphatase (Sigma-Aldrich) according to the method of Gehrke and Kuo (1989). Nucleosides were analysed by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 µm) equipped with a pre-column (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. Buffer A (10 mM of NH4H2PO4, 2.5% of methanol at pH 5.3) and buffer B (10 mM of NH4H2PO4, 20% of methanol at pH 5.1) were used with gradient conditions described as follows. With a flow rate of 1.0 ml/min, the concentration was stable at 0% B for 12 min, changed to 10% B over 8 min, to 25% B over 5 min, to 60% B over 8 min, to 64% B over 4 min, to 10% B over 9 min and 0–100% A over 15 min. The applied gradient conditions provided highly reproducible elution profiles in the region before the first 20 min, elution conditions with increased methanol concentrations were less reproducible.
For mass spectrometry analysis, nucleosides were collected from four HPLC experiments and desalted twice with a Zorbax Eclipse XDB-C18 column (Agilent; 4.6 × 150 mm, 5 µm) using 5 mM of ammonium acetate pH 6.0 with a flow rate of 0.5 ml/min. After buffer evaporation, samples were re-solved in water and applied to MALDI mass spectrometry on a VG Tofspec (Fisons Instruments) in the negative ion mode.
Isolation of a defined rRNA sequence by specific hybridization and single-strand digestion
Specific sequence of the 25S rRNA was isolated by hybridization to complementary (25S–645 and 25S–2142) deoxyoligonucleotides following a protocol previously described with slight modifications (34). In all, 1000 pmol of the synthetic deoxyoligonucleotides complementary to C633–G680 or C2118–G2166 of yeast 25S rRNA was incubated with 100 pmol of rRNA and 1.5 µl of DMSO in 0.3 vol of hybridization buffer (250 mM of HEPES, 500 mM of KCl at pH 7). After hybridization, mung bean nuclease and 0.02 µg/µl RNase A (Sigma-Aldrich) were added to start the digestion. Before the separation of the samples on a 13% polyacrylamide gel containing 7 M of urea, they were extracted once with phenol/chloroform and precipitated as described. Bands were extracted using the D-Tube™ Dialyzers according to the manufacturer’s protocol for electroelution (Novagen®).
Western blot analysis of 3× HA–Gar1
Protein extracts from the HA-epitope tagged yeast strain were prepared using glass beads. Twenty micrograms total protein of each sample was separated with 12% SDS–PAGE and blotted on a PVDF membrane (Millipore). The membrane was blocked with 5% non-fat dry milk, and tagged proteins were detected with anti-HA monoclonal antibody (Roche; 1:1000 dilution) followed by anti-mouse IgG-conjugated horseradish peroxidase (BioRad; 1:10 000 dilution).
Protein localization
Yeast cells containing GFP–Rrp8 fusion encoding plasmids and a chromosomally integrated gene encoding for ScNop56–mRFP were grown to mid-logarithmic phase in synthetic medium lacking histidine. Protein localization was visualized using a Leica TCS SP5.
RESULTS
The C-terminus of Rrp8 is important for SAM binding and for Rrp8 function
Rrp8 was identified in S. cerevisiae as a result of its peculiar genetic interaction with the mutant gar1 allele. The accumulation of an aberrant 21S rRNA in the Δrrp8 deletion strain illustrated the role of Rrp8 in affecting rRNA processing at site A2 (28). In our previous study, we have also shown that Rrp8 plays a modest but yet significant role in 40S subunit biogenesis (29). Bioinformatics studies using distant homology detection and fold recognition predicted Rrp8 to be a class I MTase (Rossmann-like fold) with the MTase domain located in the C-terminal part of the protein (26). However, the substrate for the Rrp8 MTase remained unknown.
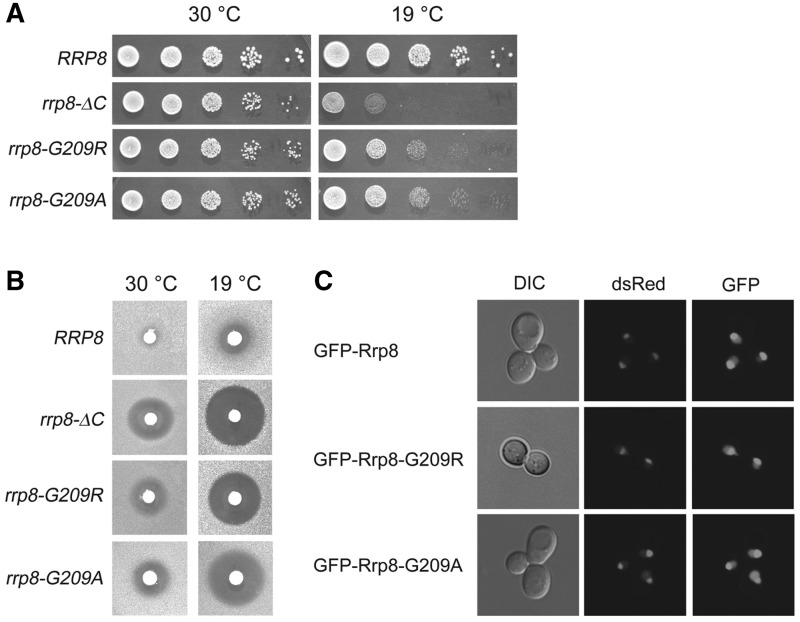
In this study, to unravel the function of the MTase domain, we used rrp8 mutant lacking the major part of the MTase domain (rrp8-ΔC). Based on previous experiments, we additionally generated a point mutation of a highly conserved glycine in motif I of the Rossmann-like fold. The mutation of this glycine to either arginine or alanine was previously shown to affect SAM binding and, therefore, protein function of the yeast arginine MTase Hmt1 (35). Consequently, we chose to mutate the equivalent residue and generated the substitution mutants, rrp8–G209R and rrp8–G209A. A serial dilution assay of the rrp8-ΔC mutant revealed the previously observed cold-sensitive phenotype and the hypersensitivity to paromomycin (28,29). The mutants expressing Rrp8–G209R and Rrp8–G209A also showed these phenotypes. However, the sensitivity to low temperature and paromomycin was not as strong as in the rrp8-ΔC mutant (Figure 1A and B). To test whether the mutant protein versions are not degraded and still localized to the nucleolus, we generated a GFP-fusion and compared the GFP signal of tagged wild-type Rrp8 with the tagged Rrp8–G209R and Rrp8–G209A proteins. Signal intensity and localization is comparable with the GFP-fusion construct with wild-type Rrp8, emphasizing that the mutant proteins are expressed to normal level and that the observed phenotypes are not because of a delocalization of the protein (Figure 1C). Taken together, these results demonstrated that the deletion of the MTase domain and a point mutation in a highly conserved SAM binding motif alters Rrp8 function.
Figure 1.
Phenotypic characterization of the strains used in this study. (A) Ten-fold serial dilutions of the indicated strains were spotted onto solid YEPD plates and were incubated at different temperatures. (B) Paromomycin sensitivity tests were performed by spotting 5 μl of paromomycin solution (200 mg/ml) on filter discs, which were then applied on YEPD plates containing the strains indicated. (C) To test the nucleolar localization of Rrp8 Rrp8–G209R and Rrp8–G209A, plasmids pKO1, pKO10 and pKO11 carrying GFP-fusion constructs were transformed into strain ScNop56-mRFP and were visualized with Leica TCS SP5.
Rrp8 binds to [3H-methyl] SAM in vitro
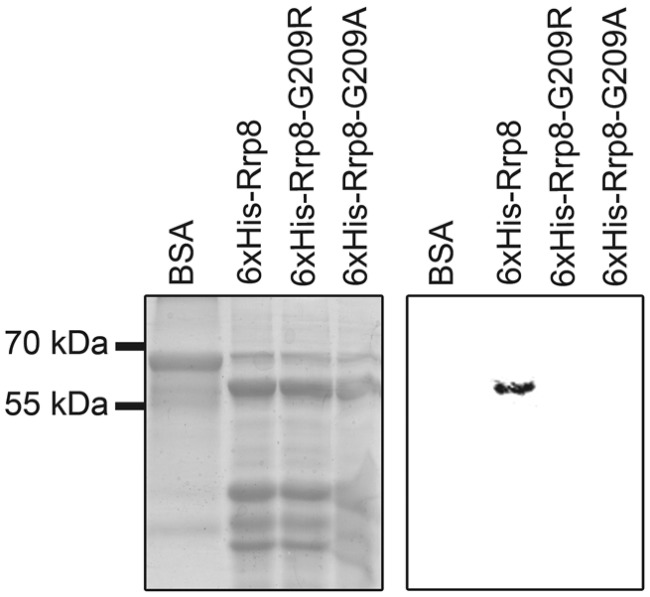
To biochemically verify the bioinformatic prediction and to test whether Rrp8–G209R and Rrp8–G209A affect SAM-binding, we used an in vitro SAM-binding assay. The N-terminal HIS-tagged Rrp8 proteins were incubated with [3H-methyl] SAM and were UV cross-linked. The co-migration of the [3H]-signal with the 6× His–Rrp8 protein illustrated that SAM binds to Rrp8 indeed. In contrast to this, no SAM binding of the respective Rrp8–G209R and Rrp8–G209A mutant proteins was observed (Figure 2). This result clearly demonstrated that yeast Rrp8 is able to bind SAM in vitro, and that the G209R and G209A mutations either abolish or strongly reduce the SAM-binding. This further supported the notion of Rrp8 being a SAM-dependent MTase. Therefore, loss of the Rrp8-mediated modification could either influence rRNA processing or ribosome assembly.
Figure 2.
In vitro SAM-binding of Rrp8. Recombinant 6× HIS–Rrp8, 6× HIS–Rrp8–G209R and 6× HIS–Rrp8–G209A were expressed in E. coli. The purified proteins (25–30 μg) were then incubated with [3H-methyl] SAM, and the reaction mixture was exposed to UV irradiation. The products were re-solved on a 12% SDS–PAGE gel and stained with Coomassie blue. After destaining, the gel was prepared for fluorography.
Hypomorphic expression of Gar1 in rrp8-ΔC affects the amount of mature 60S subunits
Previous observation, where Rrp8 was shown to be involved in 40S biogenesis, advocates its involvement in the modification of 18S rRNA. However, as the enzymes responsible for all known modifications of 18S rRNA had already been identified, the odds of Rrp8 being involved in 18S rRNA modification appeared nominal (11–13). Additionally, using a previously described protocol (13), we had already excluded the possibility of Rrp8 catalysing the acp-modification within the highly conserved helix 35 of 18S rRNA for which the responsible enzyme remains unknown (data not shown).
In our previous study, we made evident that the synthetic lethal interaction between a suppressed Δnep1 and Δrrp8 mutant is the result of defects in 40S biogenesis (29), which was the basis for our assumption of Rrp8 being a 40S MTase. Similarly, we next analysed another synthetic lethal interaction of the rrp8-ΔC mutant with a gar1 mutant. Gar1 is an essential structural protein of all H/ACA box snoRNPs and contains N-terminal and C-terminal GAR or RGG domains. Interestingly, these GAR domains are not essential for viability. However, as previously mentioned, it was shown that the expression of a mutant Gar1 protein lacking its two glycine/arginine-rich (GAR) domains (gar1ΔGAR) leads to inviability, when combined with a Δrrp8 deletion strain (28).
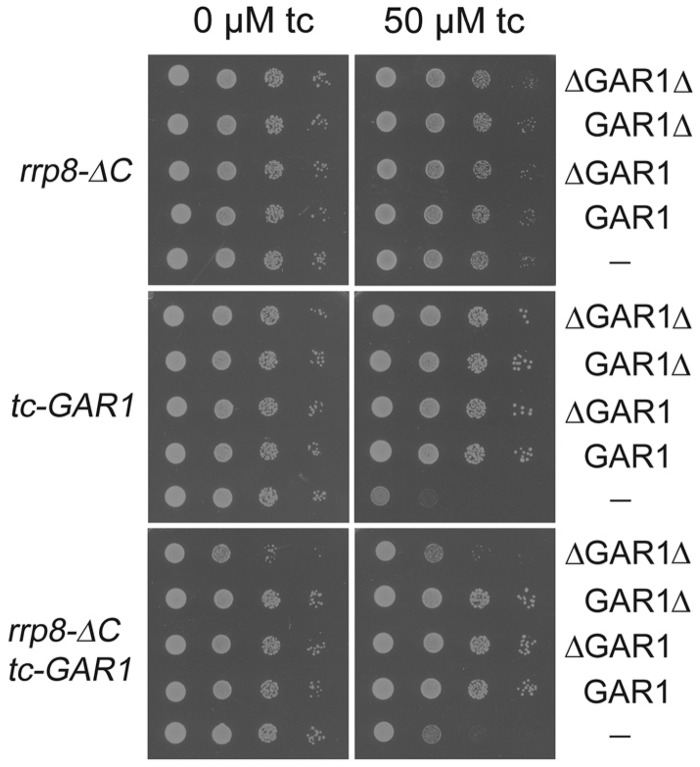
To analyse this interaction between rrp8-ΔC and gar1ΔGAR mutant, we regulated the expression of the essential Gar1 protein using the described tetracycline aptamer regulatory system, where addition of tetracycline prevents translation of the corresponding mRNA (36). In addition to this, we constructed plasmids expressing the whole Gar1 protein (GAR1), a Gar1 protein without the N-terminal GAR domain (ΔGar1), a Gar1 protein without the C-terminal GAR domain (Gar1Δ) and a protein without both GAR domains (ΔGar1Δ), to explore the specificity of the synthetic interaction. The tetracycline aptamer regulated TDH3p-tc3-3xHA-GAR1 allele (tc-GAR1 allele) and allowed gradual depletion of the Gar1 in the presence of tetracycline (Supplementary Figure S1). The tc-GAR1 allele was recombined with the various plasmids expressing the Gar1 variants. Addition of 50 µM tetracycline (tc) strongly inhibited the growth of the tc-GAR1 allele, and all Gar1 variants could complement this growth defect. No effect of tetracycline was observed for the rrp8-ΔC mutant (Figure 3). To analyse the effect of a reduced Gar1 expression on the rrp8-ΔC mutant, we combined the rrp8-ΔC allele with the tc-GAR1 allele and investigated the growth of this double mutant strain along with the Gar1 variants without and with tetracycline. As expected, the double mutant showed a synthetic growth defect. The plasmids expressing the Gar1, ΔGar1 and the Gar1Δ proteins complemented this deficiency, whereas the plasmid expressing the ΔGar1Δ protein failed to complement growth in the presence of tetracycline (Figure 3). This independently confirmed the previously described synthetic lethality of Δrrp8 with the gar1ΔGAR mutant (28). Additionally, as only the double mutant with the plasmid expressing the ΔGar1Δ protein showed the same growth phenotype as the double mutant with the empty vector, this indicated that only loss of both GAR domains impairs the functional interaction of Rrp8 and Gar1.
Figure 3.
Complementation assay of tc-GAR1 with different GAR1 constructs. After transforming the plasmids expressing the whole Gar1 protein (GAR1), a Gar1 protein without the N-terminal GAR domain (ΔGAR1), a Gar1 protein without the C-terminal GAR domain (GAR1Δ) and a protein without both GAR domains (ΔGAR1Δ), ten-fold serial dilutions of the single mutants (rrp8-_C, tc-GAR1) and the double mutant (rrp8-_C tc-GAR1) were spotted onto -ura plates with or without tetracycline. Plates were incubated at 30°C.
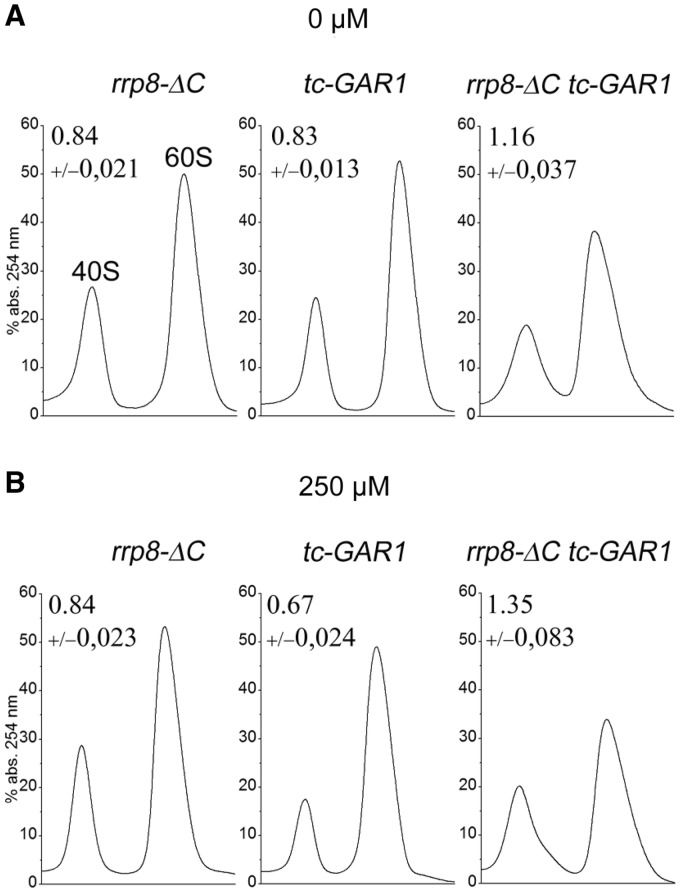
To investigate the influence of this impaired functional interaction on ribosome biogenesis, we then examined the relative abundance of ribosomal subunits in the rrp8-ΔC tc-GAR1 double mutant strain. Based on A254 absorbance estimates, the ratio of 40S to 60S subunits in the rrp8-ΔC mutant (0.84) was smaller than that of the wild-type cells (1.02, data not shown), which is in accordance to our previously described results. Importantly, also the pattern for cells expressing the tc-GAR1 allele was similar to previously published results, showing that a reduced level of Gar1 led to a strong 40S subunit decrease (37). Interestingly, the 40/60S subunit ratio increased in the rrp8-ΔC tc-GAR1 strain to a value of 1.35, emphasizing a decrease in the amount of 60S subunits (Figure 4). This decrease is also supported by the polysome profile of the double mutant, showing the formation of half-mers (data not shown). An influence of Gar1 on 60S production can be easily explained, as Gar1 being an essential structural protein of H/ACA box snoRNPs and is important for the modification of 25S, 5.8S and 5S rRNA. However, this phenotype occurs only with a concomitant loss of Rrp8, recommending that Rrp8, in addition to its function in 18S rRNA processing, might also play a role in 60S biogenesis. This was supported by a previously performed sedimentation profile of Rrp8 on a glycerol gradient and our recent ribosome profile analysis, which showed that the majority of Rrp8 localizes in fractions corresponding to the 60S subunits (28,29).
Figure 4.
Ribosome subunit analysis of tc-GAR1 rrp8-ΔC and rrp8-ΔC tc-GAR1. Sucrose gradient centrifugation was performed to analyse the influence of conditional expression of Gar1 in combination with rrp8-ΔC on ribosomal subunits. Cells were grown in liquid media without (A) or with 250 μM (B) tetracycline to an OD600 of 1. The volume of cell extracts containing 5-OD254 units was layered on 10–50% sucrose gradients and was centrifuged for 17 h at 24 500 r.p.m. and 4°C with a SW40 Ti rotor.
The rrp8–G209R and rrp8–G209A mutations led to half-mer formation
To elucidate that the decrease of 60S subunits in the rrp8-ΔC tc-GAR1 strain is because of loss of the MTase activity rather than to the loss of whole Rrp8, we next analysed the rrp8–G209R and rrp8–G209A mutants in combination with the tc-GAR1. Growth analysis of the corresponding double mutants (rrp8–G209R tc-GAR1, rrp8–G209A tc-GAR1) showed similar synthetic growth defects as observed with the rrp8-ΔC tc-GAR1 double mutant (Supplementary Figure S2). This observation made it apparent that it should be the SAM-dependent function of Rrp8 leading to lesser 60S subunits in the rrp8-ΔC tc-GAR1 strain.
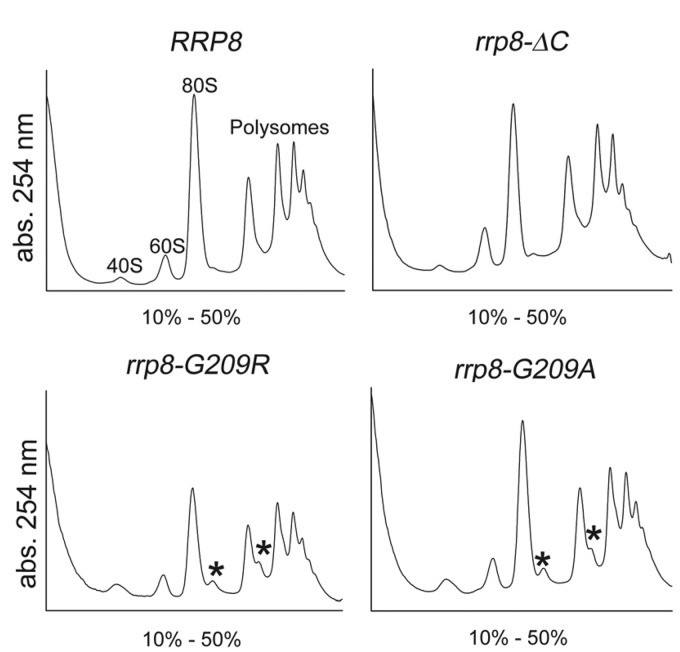
This became even more lucid with the polysome profiles of the rrp8–G209R and rrp8–G209A mutants. Cells expressing Rrp8–G209R or Rrp8–G209A exhibited half-mer polysomes (Figure 5). Half-mer polysomes are representatives of defects in the 60S subunit. Taken into account that the rrp8-ΔC mutant alone shows a normal polysome profile, and that a Δrrp8 mutant has no detectable influence on 60S biogenesis (28), these results seem to be counter-intuitive. However, comparable results were obtained using a mutant version of snR10 (snr10ΔC) that has no influence on 40S biogenesis but lacks the pseudouridylation of U2923 (38). A deletion of SNR10 showed no half-mers, whereas snr10ΔC results in the formation of these. Because loss of snR10 leads to a low level of free 40S subunits, the yield of half-mers was suggested to be low and perhaps below detection. Therefore, in the snr10ΔC mutant, the effects of snR10 on 40S subunit production can be separated genetically from its role in Ψ modification, which seems to affect the rate of 80S ribosome formation. According to this, half-mer production in the rrp8–G209R and rrp8–G209A mutants could also account for a defect that merely occurs when Rrp8 is present but is not able to perform its normal SAM-dependent function.
Figure 5.
Polysome profile analysis of the point mutants. Polysome profile analysis was performed to detect the translational status of rrp8–G209R and rrp8–G209A in comparison with the wild-type (RRP8) and the rrp8-ΔC mutant. Half-mers are indicated by asterisks.
Rrp8 performs m1A methylation at position 645 of the 25S rRNA
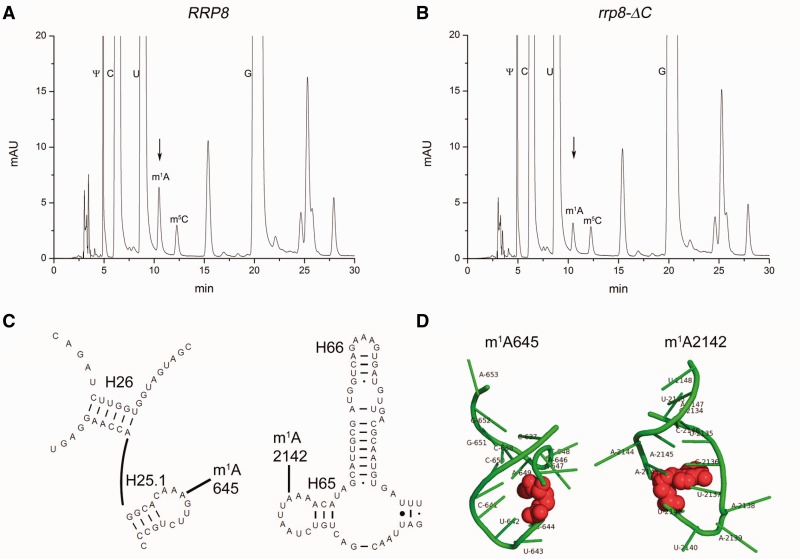
The described results prompted us to analyse a possible influence of Rrp8 on 25S rRNA modifications. Despite the already known 2′-O ribose methylations that are performed by snoRNPs and Spb1, seven base methylations are reported for which no corresponding proteins are known in yeast (9). To test for the loss of a base methylation, RP-HPLC analysis of nucleosides derived from yeast 25S rRNA was conducted. After sucrose gradient ultracentrifugation, 60S fractions of the wild-type and the rrp8-ΔC mutant were pooled separately, and RNA was extracted. The nucleoside composition of the 25S rRNA was analysed using a modified gradient of a previously described protocol on a Supelcosil LC-18-S HPLC column at 30°C (39). Surprisingly, we observed the reduction of a peak with a retention time of 10.48 min, which was close to the described retention time of 10.6 min for m1A under these elution conditions (Figure 6). MALDI mass spectrometry confirmed that the respective peak corresponds to the m1A modification (Supplementary Figure S3). As two such base methylations exist in the 25S rRNA, it became likely that a reduction of this peak corresponds to the loss of one m1A modification. This suggests Rrp8 to be responsible for the methylation of one adenine.
Figure 6.
Loss of Rrp8 influences the amount of 25S rRNA m1A modification. After sucrose gradient centrifugation, 60S subunits were collected with the Density Gradient Fractionation System (Teledyne Isco), and 25S rRNA was isolated. The 25S rRNA was digested with nuclease P1 and bacterial alkaline phosphatase (Sigma-Aldrich). Nucleosides from the wild-type (A) and the rrp8-_C mutant (B) were analysed by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 μm) equipped with a pre-column (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. (C) Localization of m1A modifications of 25S rRNA is shown in the secondary structure of Helix 25.1 and Helix 65 (http://www.rna.icmb.utexas.edu/). (D) 3D structure of the corresponding helices was made using PyMol software (PyMOL Molecular Graphics System, Version 1.2r3pre, Schröinger, LLC) using the recent 80S ribosome structure (3U5D.pdb and 3U5E.pdb). RNA is coloured in green, whereas the modified bases are shown in red spheres.
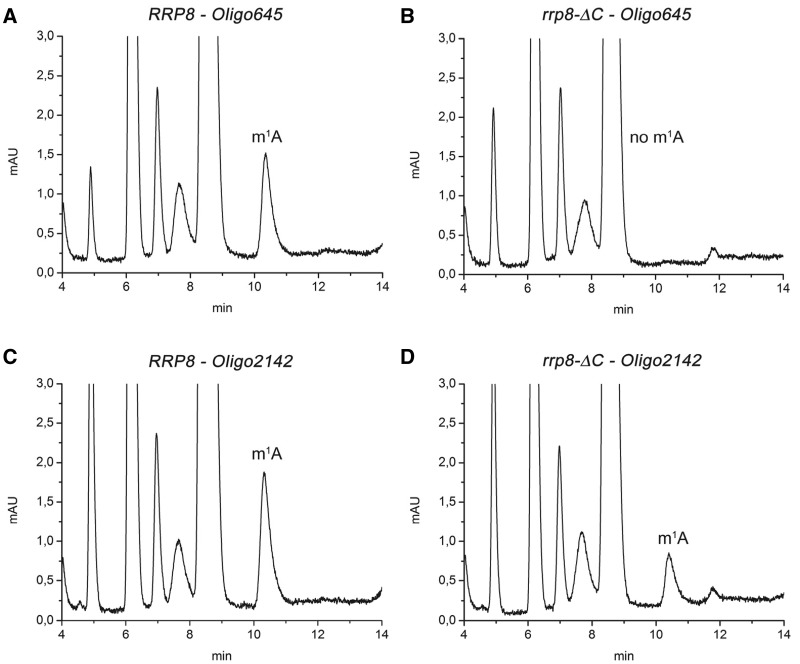
To determine which of the two m1A modifications is performed by Rrp8, we next isolated defined rRNA sequences using synthetic deoxyoligonucleotides complementary to nucleotides 633–680 (25S-645) and 2118–2166 (25S-2142) of yeast 25S rRNA. Isolated 25S rRNA from the wild-type and the rrp8-ΔC mutant was hybridized with the oligonucleotide 25S-645 or 25S-2142 to protect the respective rRNA sequence from nuclease digestion. Single-stranded DNA and RNA were, subsequently, removed by nuclease digestion, and the protected RNA fragment was gel-purified. The resulting RNA fragment was prepared for HPLC analysis again using the Supelcosil LC-18-S column. Analysis of the protected nucleotides within region 2118–2166 of wild-type and mutant 25S rRNA revealed a specific peak for the m1A modification, whereas no peak for this modification was observed in the nucleotide composition of region 633–680 from rRNA of the mutant (Figure 7). The results clearly showed that Rrp8 is responsible for the methylation at position 645 in yeast 25S rRNA.
Figure 7.
RP-HPLC analysis of mung bean digested RNA fragments. Specific sequences of the 25S rRNA from wild-type (A and C) and the rrp8-_C mutant (B and D), corresponding to Helix 25.1 and Helix 65, respectively, were isolated by hybridization to complementary deoxyoligonucleotides (Oligo645 and Oligo2142) followed by mung bean digestion. RP-HPLC analysis with these fragments was then carried out on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 μm) equipped with a pre-column (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. The following amounts of digested RNA were loaded: (A) 1.9 μg, (B) 1.3 μg, (C) 2.9 μg and (D) 1.8 μg.
Together with the 2′-O methylations Am649 and Cm650, which are caused by the U18 snoRNP, the modified nucleotide A645 is located in loop 25.1 between domain I and domain II of the 25S rRNA. Interestingly, sequences that are positioned in between these two elements are in close proximity to the association sites of 5.8S rRNA with 25S rRNA. These sites consist of two regions of extended base pairing between the 5′-region of 5.8S and 25S rRNA and between the 3′-region of 5.8S and 25S rRNA.
The Rrp8–G209R and Rrp8–G209A mutant proteins reveal a reduced m1A methylation state
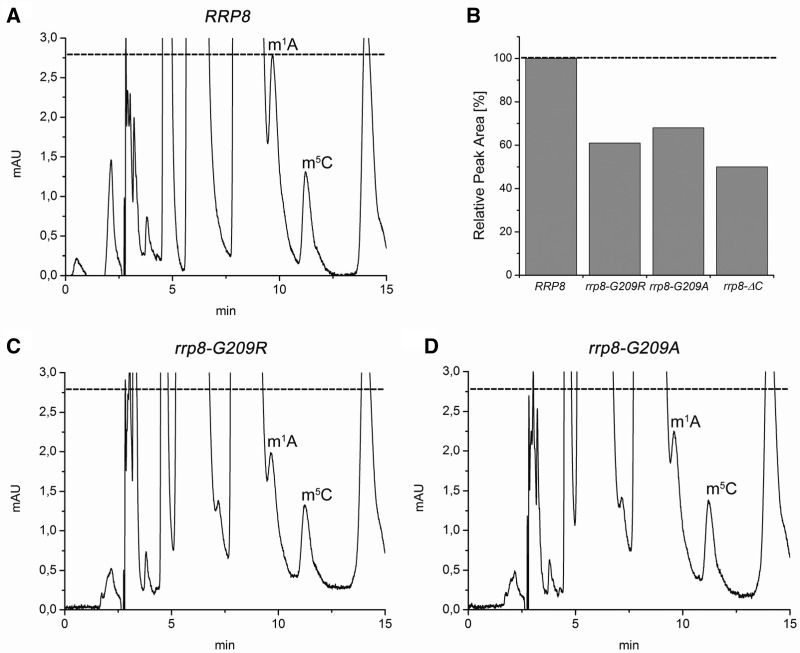
In accordance to our hypothesis that loss of a modification in the mutants expressing Rrp8–G209R and Rrp8–G209A might functionally influence the 60S subunit, we, consequently, investigated the methylation state of 25S rRNA in these mutants. Comparison of the peak height corresponding to the m1A peak in the HPLC runs of the wild-type and the respective point mutant suggested a reduction of the modification reaction in the mutant strains. We, therefore, estimated the peak areas, and calculated that in the point mutants, the m1A peak is reduced to ∼30% (rrp8–G209A) and to 40% (rrp8–G209R), whereas the rrp8-ΔC mutant showed reduction to one half (Figure 8). These results emphasize that the amino acid exchange in motif I of Rrp8 affects the enzymatic activity of the protein but does not totally abolish it. Our observations are in good agreement with previously performed experiments. Corresponding amino acid exchanges in motif I of the MTase Hmt1p also retain some activity (35).
Figure 8.
RP-HPLC analysis of 25S rRNA nucleosides from rrp8–G209R and rrp8–G209A mutant cells. After sucrose gradient centrifugation, 60S subunits were collected with the Density Gradient Fractionation System (Teledyne Isco), and 25S rRNA was isolated. The 25S rRNA was digested with nuclease P1 and bacterial alkaline phosphatase (Sigma-Aldrich). Nucleosides from the mutants (C and D) together with the respective wild-type strain (A) were analysed by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 μm) equipped with a pre-column (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. (B) Estimation of m1A peak areas in different rrp8 mutants. The m1A peak areas detected in the RP-HPLC analysis of the corresponding strain were estimated using the Agilent ChemStation software. The value for the wild-type was set to 100%.
Taken together, these results propose that the loss of m1A645 modification affects the function of the 60S subunit and that half-mer formation in the point mutants is because of defect in 60S subunits. The fact that no half-mers can be observed in the rrp8-ΔC mutant might be explained by the previously mentioned example of the snr10ΔC mutant. However, this would implicate that the observed growth phenotypes of the point mutants are not because of impaired processing of 18S rRNA.
G209R and G209A substitutions do not affect A2 cleavage
To test whether the A2 cleavage is affected in the mutants expressing Rrp8–G209R or Rrp8–G209A, we performed northern blot analysis. Both mutants showed normal A2 cleavage and no accumulation of the aberrant 21S pre-rRNA, indicating that the modifying function of Rrp8 can be separated from its role in small subunit synthesis and that m1A645 is not required for normal production of 40S particles (Supplementary Figure S4). In addition to this, we were not able to detect any accumulation of 25S rRNA precursors. This indicates that both point mutants do not effect the processing of rRNA species involved in 60S subunit biogenesis and that half-mer formation should result from functional or assembly defects in the large subunit. Because of the normal processing of 18S rRNA, the 60S defect is more pronounced in the point mutants, whereas in the rrp8-ΔC mutant, there seems to be reduction of both mature 40 and 60S subunits, which in turn prevents the half-mer formation. The half-mers formation depends on the relative amount of two subunits in the cytoplasmic pool, and the mutual decrease of both subunits should prevent any half-mer formation, as evident in the rrp8-ΔC mutant.
A mutation in 25S rRNA at position 645 affects 60S biogenesis
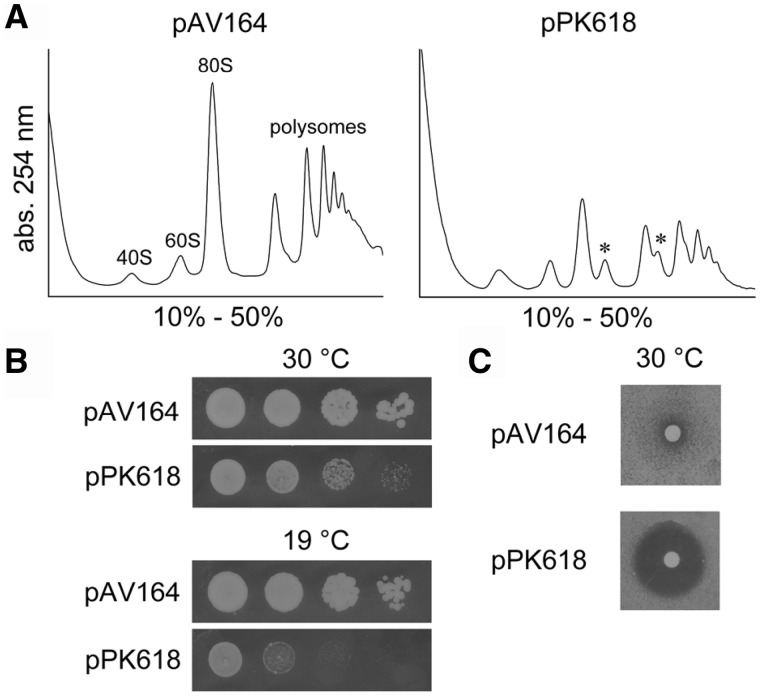
To gain further insights into the function of the m1A 645 modification, we decided to analyse the effects of a nucleotide exchange at position 645. Therefore, we exchanged A645 to the nucleotide T and episomally expressed the mutated 25S-A645T rRNA in a strain lacking the wild-type rDNA locus. In addition to our RNA protection assay, the HPLC analysis of the expressed mutant rRNA again clearly confirmed that the reduction of the peak at 10 min corresponds to the loss of the m1A modification (Supplementary Figure S5). The A to T substitution revealed strong formation of half-mers, which seemed to be even stronger than compared with the point mutants (Figure 9). Additionally, the growth defect of the strain along with the observed cold-sensitivity and hypersensitivity to paromomycin seemed to be much stronger than compared with the rrp8-ΔC mutant, suggesting that the nucleotide substitution leads to a stronger defect than the loss of the modification alone. This is most likely because of a disturbed rRNA structure that might affect processing or assembly of 60S subunits. By reason of these differences, we are not able to ascertain the precise function of the modification. However, because of the strong growth defect of the rRNA mutant, it is tempting to speculate that the structural integrity of Helix 25.1 plays an important role for 60S biogenesis.
Figure 9.
Polysome profiles and growth analysis of the rRNA mutant strain. (A) Polysome profile analysis was performed to detect the translational status of the rRNA mutant A645U. Mutated rRNA was expressed from the plasmid pPK618 in a strain where genomic rDNA was deleted. Wild-type rRNA was expressed from pAV164. (B) Thermosensitivity of strains carrying these plasmids was analysed on solid YEPD plates incubated at different temperatures. (C) Paromomycin sensitivity tests were performed as previously described.
Taken together, our results provide decent evidences that Rrp8 is responsible for the m1A645 modification of 25S rRNA and is involved in the biogenesis of both 40S and 60S subunits. The reduction of the 60S peak in the rrp8-ΔC tc-GAR1 strain asserts that the functional relationship of Rrp8 and Gar1 might be because of structural rearrangements during ribosome biogenesis. This could be supported by the fact that GAR domains have RNA-helix-destabilizing properties in vitro (40) and are implicated in non-specific protein RNA interactions (41). Furthermore, the importance of the physical presence of wild-type Rrp8 for the assembly of the 60S subunit is a new and interesting feature of Rrp8 that is valuable and has to be considered in futures studies.
DISCUSSION
Rrp8 was long thought to be a protein MTase involved exclusively in the late pre-40S biogenesis. Gar1 and histone H2 were considered to be the most favourable substrates for Rrp8, by virtue of their genetic and physical interactions (42). Also, as the human homologue of yeast Rrp8 was already shown to epigenetically regulate the rRNA transcription and exhibits a physical interaction with methylated histone H3, the histone H2 in yeast was considered to be the obvious target for the yeast Rrp8. In contrast, here, we demonstrated for the first time that yeast Rrp8 is an rRNA MTase responsible for the N1-methylation of A645 of 25S rRNA. In addition, we also provide evidences that Rrp8 is not only a part of the 40S biogenesis machinery but also plays an active role in the 60S subunit synthesis. As mentioned before, Rrp8 was considered to be a factor involved in the 40S biogenesis. Interestingly, although investigating the genetic interaction of rrp8-ΔC with the tc-GAR1 allele, we came across with the evidences that this genetic interaction is most likely the result of 60S biogenesis defects. The Rrp8 point mutants could show that it is indeed the alteration of the SAM-dependent function of Rrp8 that leads to the synthetic interaction with the tc-Gar1 allele. Unfortunately, we are not in a position to comment on the precise role of Gar1 in this interaction, except that both N- and C-terminal GAR or RGG domains are essential for this interaction.
The analysis with the specific Rrp8 point mutants helped us to further explain the importance of Rrp8 and its SAM-dependent function in the 60S biogenesis. With the in vitro SAM binding and RP-HPLC analysis, we could show that an amino acid exchange in motif I of Rrp8 leads to alteration in the SAM binding activity and affects the m1A modification. The polysome profiles with these point mutants displayed that, in contrast to 40S biogenesis defects in the rrp8-ΔC mutant, the rrp8–G209R and the rrp8–G209A mutant exhibit a specific defect in 60S subunits. Considering that Rrp8 is a MTase, and the point mutation alters the SAM binding, the decrease in mature 60S subunits could be explained with the two assumptions, (i) the decreased m1A modification in Rrp8 point mutants affects the processing or assembly of 60S subunits that leads to lesser mature 60S subunits or (ii) the point mutations in this region of Rrp8 alter the native conformation of the protein, which then impair its yet known function in 60S biogenesis. Interestingly, the first hypothesis seemed more preferable, as the rDNA point mutant where the A645 was exchanged with T displayed a similar decrease in the mature 60S subunits. Therefore, it is tempting to speculate that the loss or reduction in the modification of this A645 somehow influences the assembly of the 60S subunit. We could exclude the processing defects in the point mutants, as no accumulation of the 25S precursors was observed in northern blotting. As far as the alteration of the native conformation of Rrp8 is concerned in these point mutants, we could provide compelling evidences that the mutant rrp8 was expressed and translocated to the nucleolus just like the wild-type Rrp8. In addition, lack of 21S rRNA accumulation and any 40S biogenesis defects also made it clearer that theRrp8 point mutants were more or less still functional in terms of their SAM-independent function at the A2 site. Collectively, our results present convincing evidences for Rrp8 to be involved in the biogenesis of both large and small subunits.
To our knowledge, Rrp8 is the first example of a eukaryotic MTase that performs a m1A modification on ribosomal RNA. Despite the highly conserved 1-methyladenosine modification in the T-loop of tRNAs in all three domains of life, only few examples of the N1-methylation of adenosine in ribosomal RNAs are described to date (9). Bacterial rRNAs normally do not contain any m1A modification, with the exception of those pathogenic bacteria and antibiotic producers that have evolved mechanisms of resistance to antibiotics. Methylation of the N7 position of G1405 or the N1 position of A1408 in the 16S rRNA of pathogenic bacteria and antibiotic producers provide high-level of resistance to most aminoglycosides used in clinical practice and is carried out by specific MTases (43–45). For ribosomal RNAs in archaea, a m1A modification is reported in the case of A628 for Haloarcula marismortui 23S rRNA (46), and human 28S rRNA carries a m1A modification at position A1309 (47,48).
Generally, such nucleotide modifications have impact on the local structure of the RNA. Methyl groups can increase base-stacking because of their hydrophobicity, and they may also induce structural changes by increasing steric encumbrance or by blocking hydrogen bonds (49,50). The formation of m1A modulates the electron density in the aromatic purine system and, therefore, influences the strength of hydrogen bonds, which block a Watson–Crick position in the adenosine, and introduces a positive charge to the nucleoside (51). The 2′-O methylations, which are performed by the U18 snoRNP and are located in close proximity to the m1A645 modification, may help to establish a hydrophobic environment that could be important for the region of Helix 25.1. Therefore, it is possible that the loss of the modification affects fine-tuning of ribosome function that could give rise to the pronounced cold-sensitivity or decreased resistance to paromomycin.
The analysis of the 3D structure of the region around the modification site shows that the interaction of the 5′-end of 5.8S rRNA with 25S rRNA is in close proximity (Supplementary Figure S6C). A previous study suggests that helix formation in the region between domain I and II is a prerequisite for a correct topological framework for 5.8S rRNA to interact with 25S rRNA (52). The observed genetic interaction data support a function of Rrp8 in this region. Processing at cleavage sites A2 and A3 is an important step for the maturation of 5.8S rRNA, and the CRAC analysis from Tollervey’s laboratory has recently shown that the A3 cluster proteins influence the pre-rRNA folding and 5.8S rRNA maturation (53). This study also revealed the binding of the essential 60S biogenesis factor Nop4 to Helix 26, which is close to the Rrp8 modification site, raising the possibility that Rrp8 is also involved in structural rearrangements on binding to its target site. Consequently, this could also disturb the precise interaction of Nop4 with the rRNA leading to a processing defect. Supportive to this is the fact that the 60S biogenesis factors NOP12, NOP15 and NOP16, which belong to the A3 cluster, have been reported to show a positive genetic interaction with RRP8 (54). The predicted binding sites for Nop12 and Nop15 are within ITS2, the processing of which is also influenced by ITS1 (53). In addition to this, our study provides a strong evidence for a coordination of A2 cleavage with early maturation events of 27S A2 pre-rRNA as proposed recently.
Together with snR10 and Gar1, Rrp8 also turned out to be an interesting example for the robust coordination of 40S and 60S subunit biogenesis. Snr10 is responsible for a modification in the 25S rRNA, and the deletion of SNR10 leads to similar defects in 40S synthesis as observed in a Δrrp8 mutant. These examples again highlight the structural complexity of ribosome biogenesis and show that the physical presence of the protein and the catalytic activity affect the processing and assembly of pre-rRNA in different ways.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–6, Supplementary Methods and Supplementary References [13,36,55–59].
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft (En134-9); Excellence Cluster: Macromolecular Complexes.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Martin Held and Alexander Heckel for their excellent support with ESI-MS measurement. and Britta Meyer for her assistance with HPLC experiments. S.S. thanks DAAD (Deutscher Akademischer Austauschdienst) for the award of PhD scholarship.
REFERENCES
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Fatica A, Oeffinger M, Dlakic M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 2003;23:1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 8.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 9.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS One. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 12.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang XH, Liu Q, King TH, Fournier MJ. Strong dependence between functional domains in a dual-function snoRNA infers coupling of rRNA processing and modification events. Nucleic Acids Res. 2010;38:3376–3387. doi: 10.1093/nar/gkq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leulliot N, Bohnsack MT, Graille M, Tollervey D, Van Tilbeurgh H. The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of alpha/beta knot fold methyltransferases. Nucleic Acids Res. 2008;36:629–639. doi: 10.1093/nar/gkm1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 21.King M, Ton D, Redman KL. A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem. J. 1999;337(Pt 1):29–35. [PMC free article] [PubMed] [Google Scholar]
- 22.Hong B, Wu K, Brockenbrough JS, Wu P, Aris JP. Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:2927–2937. doi: 10.1093/nar/29.14.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwalla S, Kealey JT, Santi DV, Stroud RM. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- 24.Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S. Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003;31:4738–4746. doi: 10.1093/nar/gkg657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 26.Wlodarski T, Kutner J, Towpik J, Knizewski L, Rychlewski L, Kudlicki A, Rowicka M, Dziembowski A, Ginalski K. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS One. 2011;6:e23168. doi: 10.1371/journal.pone.0023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 28.Bousquet-Antonelli C, Vanrobays E, Gelugne JP, Caizergues-Ferrer M, Henry Y. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA. 2000;6:826–843. doi: 10.1017/s1355838200992288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schilling V, Peifer C, Buchhaupt M, Lamberth S, Lioutikov A, Rietschel B, Kotter P, Entian KD. Genetic interactions of yeast NEP1 (EMG1), encoding an essential factor in ribosome biogenesis. Yeast. 2012;29:167–183. doi: 10.1002/yea.2898. [DOI] [PubMed] [Google Scholar]
- 30.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subbaramaiah K, Simms SA. Photolabeling of CheR methyltransferase with S-adenosyl-L-methionine (AdoMet). Studies on the AdoMet binding site. J. Biol. Chem. 1992;267:8636–8642. [PubMed] [Google Scholar]
- 32.Bonner WM, Laskey RA. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur. J. Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- 33.McEntee CM, Hudson AP. Preparation of RNA from unspheroplasted yeast cells (Saccharomyces cerevisiae) Anal. Biochem. 1989;176:303–306. doi: 10.1016/0003-2697(89)90313-8. [DOI] [PubMed] [Google Scholar]
- 34.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride AE, Weiss VH, Kim HK, Hogle JM, Silver PA. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem. 2000;275:3128–3136. doi: 10.1074/jbc.275.5.3128. [DOI] [PubMed] [Google Scholar]
- 36.Kotter P, Weigand JE, Meyer B, Entian KD, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bousquet-Antonelli C, Henry Y, G'Elugne JP, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 39.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 40.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 41.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 42.Grummt I, Ladurner AG. A metabolic throttle regulates the epigenetic state of rDNA. Cell. 2008;133:577–580. doi: 10.1016/j.cell.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 43.Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 2007;45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 44.Koscinski L, Feder M, Bujnicki JM. Identification of a missing sequence and functionally important residues of 16S rRNA:m(1)A1408 methyltransferase KamB that causes bacterial resistance to aminoglycoside antibiotics. Cell. Cycle. 2007;6:1268–1271. doi: 10.4161/cc.6.10.4231. [DOI] [PubMed] [Google Scholar]
- 45.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob. Agents Chemother. 2007;51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirpekar F, Hansen LH, Rasmussen A, Poehlsgaard J, Vester B. The archaeon Haloarcula marismortui has few modifications in the central parts of its 23S ribosomal RNA. J. Mol. Biol. 2005;348:563–573. doi: 10.1016/j.jmb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 48.Ofengand J, Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- 49.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. Structural and functional roles of the N1- and N3-protons of psi at tRNA's position 39. Nucleic Acids Res. 1999;27:3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 51.Helm M, Giege R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- 52.Poll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL, Jr, Tschochner H, Milkereit P. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One. 2009;4:e8249. doi: 10.1371/journal.pone.0008249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Granneman S, Petfalski E, Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Entian K-D, Kötter P. In: Methods in Microbiology. Stanfield I, Michael JRS, editors. New York: Academic Press; 2007. pp. 629–666. [Google Scholar]
- 56.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 58.Orr-Weaver TL, Szostak JW. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl Acad Sci. USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.