Abstract
The conservation of hox genes as well as their genomic organization across the phyla suggests that this system of anterior–posterior axis formation arose early during evolution and has come under strong selection pressure. Studies in the split Hox cluster of Drosophila have shown that proper expression of hox genes is dependent on chromatin domain boundaries that prevent inappropriate interactions among different types of cis-regulatory elements. To investigate whether boundary function and their role in regulation of hox genes is conserved in insects with intact Hox clusters, we used an algorithm to locate potential boundary elements in the Hox complex of mosquito, Anopheles gambiae. Several potential boundary elements were identified that could be tested for their functional conservation. Comparative analysis revealed that like Drosophila, the bithorax region in A. gambiae contains an extensive array of boundaries and enhancers organized into domains. We analysed a subset of candidate boundary elements and show that they function as enhancer blockers in Drosophila. The functional conservation of boundary elements from mosquito in fly suggests that regulation of hox genes involving chromatin domain boundaries is an evolutionary conserved mechanism and points to an important role of such elements in key developmentally regulated loci.
INTRODUCTION
Hox genes are among the key regulatory factors that control developmental processes, in particular anterior–posterior (A–P) body axis across the phyla (1,2). In most cases, Hox genes are physically clustered and display temporal and spatial co-linearity (3–6). These genes have been widely studied in insects where they function to pattern ectodermal derivatives; determine the identity of appendages, such as mouthparts, antenna, wing, halteres and legs and pattern the mesoderm and gut endoderm along the A–P axis (7). The organization of Hox genes has been analysed in a number of insect species including several species of Drosophila (8,9), silk moth Bombyx mori (10,11), red flour beetle Tribolium castaneum (12,13), honey bee Apis mellifera (14), grasshopper Schistocerca gregaria (15) and mosquito Anopheles gambiae (16,17). Although the order and number of Hox genes is fairly conserved across insects, there are marked differences with respect to the size of the cluster, transcriptional direction and expression profile of individual genes.
Hox genes are best studied in Drosophila where the cluster is split at multiple locations in different species (9). In Drosophila melanogaster, the Hox cluster is split between Antp and Ubx to give rise to Antennapedia complex (ANT-C) and bithorax complex (BX-C). In Drosophila virilis, the cluster is broken between Ubx and abd-A, whereas in Drosophila buzzatii, an additional split has occurred between lab and pb. Outside Drosophila lineage, the split Hox cluster is only reported for B. mori (10). The splitting of Hox cluster in Drosophila is considered to be a derived condition acquired later in the evolution, as the Hox cluster has remained intact in most of the insect species, and more so in case of vertebrates (13,18). The persistence of Hox genes into clusters is widely believed to be due to multiple factors including the existence of overlapping regulatory elements that define the expression of these genes and the requirement to protect regulatory elements from position effects (3,19). The BX-C of Drosophila provides a well-studied example of the interplay of cis-regulatory elements to tightly regulate Hox genes. A series of cis-regulatory elements regulate the expression of Ubx, abd-A and Abd-B (the three homeotic genes of the BX-C) in a parasegment-specific manner (20). Critical to ensuring the functional autonomy of each domain are the chromatin domain boundary elements or insulators. Cis-regulatory elements are referred to as boundaries or insulators if they block enhancer–promoter interactions when positioned between the two and/or prevent reporter genes from position effects of the surrounding chromatin in transgenic assays. Some of the well-characterized insulators in the BX-C of Drosophila include Mcp, Fab-6, Fab-7 and Fab-8. Mutations that abolish the function of these boundary elements result in fusion of independent expression domains leading to misreguation of Hox genes and homeotic transformations (21–25).
While much of what is known about the organization and regulation of Hox genes have come from Drosophila which has a split Hox cluster, it is important to study the regulation of these genes in insects which possess an intact/ancestral Hox cluster. To this end, we chose mosquito, A. gambiae which represent a clad of lower dipterans, to search for chromatin boundary elements within the Hox complex. Mosquitoes belong to brachycera branch of Diptera, while Drosophila belong to a more advanced branch called nematocera. These two species are believed to have diverged around 250 Mya (26) and during this time have evolved variations on the common dipteran body plan. It is interesting to note that in contrast to Drosophila and B. mori, where the Hox clusters are split, the Hox genes in mosquito are arranged into a single cluster on chromosome 2R, an ancient feature of Hox gene organization (9,10,16,17,27,28). The mosquito Hox cluster contains all the canonical Hox genes including the Hox-derived genes zen and ftz and span a region of ∼1.2 Mb which is much larger than that of Drosophila (∼700 kb), Tribolium (∼700 kb) and Schistocera (∼700 kb), but smaller than that of Apis (∼1.37). In addition, the Hox cluster in Drosophila is interrupted by non-homeotic genes such as cuticle genes (between lab and pb) and amalgam gene (between bcd and Dfd), whereas in mosquito cuticle and amalgam gene homologues lie elsewhere on the chromosome 2R away from the Hox cluster (17,29). It is thought that the invasion of non-homeobox genes in the Hox cluster of fly is a relatively recent event and is correlated with the splitting of Hox genes. Despite these differences, the Hox genes in both the species are highly conserved and it is speculated that they might also share great degree of conservation in cis-regulatory elements (16,17).
In this study, we analysed the Hox complex of A. gambiae to identify and characterize chromatin domain boundaries. We searched for clusters of binding sites for boundary interacting proteins, GAF, dCTCF, BEAF, Su(Hw) and Zw5. All these proteins are also present in other insects including mosquito, except BEAF and Zw5 (30). Using our recently published chromatin domain Boundary Element Search Tool (cdBEST), we predicted several potential boundary elements in the Hox complex of mosquito, A. gambiae (31). Comparison of the distribution of boundary elements in mosquito Hox cluster with that of the Drosophila showed that most of the boundaries mark the domains of Hox genes as seen in the Drosophila BX-C. We assayed a subset of these (AgB1, AgB2, AgB7 and AgB18) for their enhancer-blocking activity in Drosophila and show that these elements function as enhancer blockers in both cultured cells and transgenic fly. Additionally, all the tested boundaries show GAF dependency for their function. This is the first study reporting identification of boundary elements from A. gambiae and their functional conservation in Drosophila. The functioning of mosquito boundary elements in Drosophila suggests that chromatin boundaries, like Hox genes themselves, may be conserved across insects.
MATERIALS AND METHODS
Genomic sequences and prediction of putative boundary elements
The A. gambiae Hox sequences (∼1.2 Mb) used in this study were downloaded from NCBI [accession No. NT_078266.2 (2R), 59191123–60432266, assembly AgamP3, dated 28 September 2011]. To precisely annotate the Hox genes on the current sequence assembly, we carried out blastp searches using the protein sequences from this region as a query, against the NCBI non-redundant database. Each blast search returned a reliable hit with a Drosophila Hox protein (Supplementary Table S1). Since the segment that contains genes lab, pb, zen and Dfd is inverted in the current assembly (against the expected and previously published version), we used the reverse complement sequence of this segment (59 191 123 to 59 514 080) to obtain the correct map (Supplementary Figure S1). We used cdBEST to predict boundary elements from Hox complex of mosquito (31). The tool was run with a set window size of 1000 bp and window slide of 10 bp across the Hox complex. All the parameters were set as default, except the score cut-off of Fab-7 type boundary which was set as 50. All the input and output files from the cdBEST program are included in Supplementary Zip file.
To map putative enhancers in the mosquito Hox complex, we wrote a Perl script that searches for the consensus binding motifs (derived from Drosophila) of KRUPPEL (Kr) and HUNCHBACK (Hb) proteins (32). Here a sequence segment is called as putative enhancer, if it contains a cluster of three or more motifs (1 HB, 1 KB, either 1 HB or 1 KB) with the set window size of 300 bp (32).
Transgene constructs
Primers were designed for four predicted boundary elements using online tools (Primer3 and OligoCal). The test fragments were amplified using DNA isolated from A. gambiae cell line (Suva4) with the following PCR primer pairs: AgHox-B1-F/R, AgHox-B2 F/R, AgHox-B7 F/R and AgHox-B18-F/R (Supplementary Table S4). The amplicons were cloned in TA-cloning vector (either pGEMT-Easy or TOPO-TA) following the manufacturer’s instructions. For enhancer-blocking assay in S2 cells, the test fragments were taken out and cloned in boundary assay vector, NPG (Neomycin–PE–GFP) (31) between PE and GFP as NotI fragment. For P-element transformation, the test fragments were excised out from TA-vector as EcoRI and inserted between two loxP sites of a modified pBlueScript vector, LML (33). From here the test fragments flanked by loxP sites were excised out and ligated into pCfhl (34) between the UPS/NE enhancers of fushi-tarazu (ftz) and hsp70 promoter as XhoI. Cloning of all the test fragments was confirmed by sequencing.
Cell culture, transfections and flowcytometry
Drosophila Schneider (S2) cells were cultured in Schneider’s complete media (Gibco) containing 10% inactivated FCS and 100 µg/ml PenStrep at 25°C. Before transfection, 106 cells in 1 ml medium were seeded in six-well plates and allowed to grow for 24 h. One microgram of column purified (Qiagen-Midi kit) plasmid was transfected using Effectene transfection reagent (Qiagen) as per the manufacturer’s instructions. For stable transfection, 1 mg/ml G418 was added to cells after 24 h of transfection. Transfected cells were selected on G418 for at least 6 weeks. For fluorescence activated cell sorting (FACS), around 40 000 cells were harvested, washed and resuspended in 1× PBS and analysed in FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems). Fluorescence was excited at 488 nm and the blocking of the enhancer by the test fragment was plotted as per cent GFP-positive cells.
P-element transformation
Transgenic flies were generated following standard procedures. Briefly, P-element constructs were injected into early embryos (pre-cellularization) of w1118 flies. Transgenic flies were identified by visual examination of rescue of the eye colour due to the presence of mini-white (MW) gene in the transformation vector and were outcrossed to w1118. Multiple independent lines were obtained for each construct. To generate balanced stocks and to determine the chromosome of insertion, each independent line was crossed to marked balancer chromosomes. All the fly stocks were maintained at 25°C.
Flipping out of test fragments from transgenic lines
To excise the test element from the integrated P-element, we used Cre recombinase expressing flies (35). Flies carrying loxP transgene were crossed to virgins of Cre expressing fly stock. From this, virgins of P-element and the Cre chromosomes were selected based on markers and balancers used and then crossed to Pin/Cyo;TM2/TM6B fly to make stocks (35). Excision was confirmed by PCR using the following specific primers: lacZ was amplified using primers LF 5′-ACTATCCCGACCGCCTTACT-3′ and LR 5′-GATGGCTGGTTTCCATCAGT-3′ and primers for AgB1, AgB2, AgB7 and AgB18 (listed in Supplementary Table S4).
β-Galactosidase assay
The 0- to 12-h-old embryos were collected from transgenic flies and dechorionated in 1:1 PBS–NaOCl (sodium hypochlorite). The embryos were washed thoroughly with PBS and fixed in saturated heptane (10 ml heptane was saturated by vigorously mixing with 5 ml PBS and 5 ml 25% glutaraldehyde, phases were then allowed to separate and the top phase of heptane was used) for 15 to 20 min rotating on a shaker at RT. The fixation solution was removed and the embryos were washed at least six to seven times with PBST (PBS+0.3% Triton X-100). Embryos were incubated in pre-warmed staining solution for 10 min. The staining solution was removed and the embryos were resuspended in 10 ml staining solution containing 0.1% X-gal (Sigma) and incubated from 4 h to overnight at 37°C in the dark. Staining solution was removed and the embryos were washed two to three times in PBS in order to remove the left over staining solution. For each construct, staining was done simultaneously in a grid along with controls [empty vector was used as negative control and five binding sites for Su(Hw) and Fab-7 were used as positive controls]. Staining was repeated at least four to five times to obtain a consistent pattern. The images of embryos were taken in a Leica Stereomicroscope.
Boundary activity in mutant background
To test the enhancer-blocking activity of AgB1, AgB2, AgB7 and AgB18 in mutant backgrounds, homozygous males carrying the test fragments were mated with the following mutations: TrlR85/TM3,Ubx-lacZ, dCTCFy+6/TM3,Ubx-lacZ, CP190H31-2/TM3,Ubx-lacZ and BEAFKO/Cyo,hb-lacZ. The embryos were stained for seven stripe and CNS expression pattern of β-galactosidase as described above.
Quantitative analysis of lacZ staining
For quantification of lacZ staining, an open source image processing tool, ImageJ 1.46r (http://rsbweb.nih.gov/ij/), was used. The intensity quantification of lacZ-stained embryos was performed as follows. Representative images for each boundary construct and control lines (under normal, flipped-out or mutant situations) were processed for quantification. The captured images were first converted to 8-bit grey-scale and then inverted in ImageJ software. Using the Oval Tool, a circular selection was placed over one of the seven lacZ stained stripes and the intensity was measured. The size of the area measured was kept the same in all the embryos analysed. The values were normalized by the background having no lacZ staining. At least five embryos were analysed for each case and graphically represented as mean ± SD.
Chromatin immunoprecipitation
Third-instar larvae containing AgB1, AgB2, AgB7 and Ag18 were collected, rinsed in ice-cold PBS and homogenized in homogenization buffer supplemented with DTT, PMSF and protease inhibitor cocktail (Roche). To remove debris, the homogenate was filtered through two layers of Mira cloth and centrifuged at 400g for 5 min at 4°C. Supernatant was transferred into a fresh tube and spun at 1100g for 10 min at 4°C. The pellet was resuspended in ice-cold homogenization buffer, cross-linked with 1% formaldehyde for 10 min at room temperature and quenched with 0.125 M glycine. The samples were washed with PBS and protease inhibitors and centrifuged at 1100g for 10 min at 4°C. The cells were lysed in lysis buffer supplemented with PMSF, DTT and protease inhibitors and incubated for 10 min. The chromatin was shared to an average size of 200–600 bp fragments by sonication using Bioruptor (Diagenode). The sonicated chromatin was pre-cleared using protein-A beads and incubated with anti-GAF (Home-raised). Alongside, chromatin was also incubated with IgG to serve as controls. After reversing the cross-linking, the DNA was purified using Isoamyl:Phenol:Chloroform. Relative abundance of GAF at test fragment (Supplementary Figure S2) and at control region was estimated using 7900 HT Fast real-time PCR system from Applied Biosystems. POWER SYBR GREEN PCR master mix, 2 pmol of each primer and 50 ng of template, was used per reaction with PCR conditions as follows: 5 min of 95°C, cycling conditions were 40 cycles of 94°C 15 s, 58°C 30 s and 68°C 30 s. Dissociation curves were analysed as a means to ensure quality of amplicon and to monitor primer dimers. Enrichment was determined based on the differences of the critical threshold (ΔCt) measurements of negative pull-down control versus pull down. One ΔCt unit corresponds to 2-fold enrichment. The primers used in the chromatin immunoprecipitation (ChIP) are given in Supplementary Table S4.
RESULTS
Sequence analysis to search for boundary elements in the Hox complex of mosquito
Despite poor sequence conservation, boundary elements share functional properties from flies to human. However, like other cis-regulatory elements, they contain small sequence motifs that serve as binding sites for proteins involved in boundary function. Drosophila uses the binding sites of multiple proteins, dCTCF, Su(Hw), BEAF, GAF, Zw5, Mod(mdg4) and CP190, to derive its boundary function. Majority of the Drosophila boundary interacting proteins are conserved in mosquito and other insects and thus may recognize similar DNA motifs and contribute towards boundary function in A. gambiae (30,36). Based on these facts, we used cdBEST that was originally developed for Drosophila, to identify potential boundary elements from the Hox locus in mosquito, A. gambiae (31).
Using cdBEST, we predicted 54 putative boundary elements across the Hox cluster of A. gambiae (Supplementary Figure S1). Among them 17 boundaries fall within the Ubx to Abd-B region, whereas 30 boundaries reside within the labial to Antp region. Seven boundaries are located in the intergenic region between Antp and Ubx. Most of the predicted boundaries fall in the intergenic regions and have multiple binding sites for GAF (Supplementary Table S2). To understand how boundary elements may define domains in A. gambiae Hox cluster, we mapped putative enhancers identified by a Perl script based on a recent study by Starr et al. (Supplementary Table S3) (32). Interestingly, the identified boundary elements flank the mapped enhancers across the Hox cluster (Figure 1B and Supplementary Figure S1). This arrangement of boundaries flanking the enhancers fits well with their proposed role in the formation of chromatin domains.
Figure 1.
Location and distribution of the boundary elements in the Hox complex of mosquito, A. gambiae. (A) The Hox cluster of A. gambiae is shown (not to scale) with tested boundary elements as red ovals. All the genes are shown schematically (not to scale) including the non-homeotic gene, ftz with a colored arrow. The direction of the arrow indicates orientation of the gene. The Hox complex of D. melanogaster (not to scale) with boundaries that may be counterparts of mosquito boundaries are also shown for reference. Abbreviations: labial (lb); proboscipedia (pb); Deformed (Dfd); Sex combs reduced (Scr); Antennapedia (Antp); Ultrabithorax (Ubx); abdominal-A (abd-A); Abdominal-B (Abd-B); fushi-tarazu (ftz), zerknullt (zen); bicoid (bcd). (B) The distribution of boundary elements across the BX-C of A. gambiae and Drosophila. Genes are shown as coloured arrows. The yellow lines within the genes indicate exonic regions. Broken line indicates domains/boundaries that may be functional equivalents in the two species. The shaded regions in the A. gambiae Hox cluster indicate domains that may correspond to iab-domains of the Drosophila. The domains shaded in orange (abx/bx, bxd/pbx) control the expression of Ubx, the grey domains (iab-2, iab-4) regulate abd-A and green domains (iab-5 to iab-8) are responsible for the expression of Abd-B in Drosophila.
Comparison of cis-regulatory elements revealed that like Drosophila, Hox cluster of A. gambiae also contains an extensive array of boundaries and enhancers organized into domains (Figure 1A, B and Supplementary Figure S1). It is interesting to note that although the Hox complex in A. gambiae is intact and 1.2 Mb long, the BX-C region is roughly of the same size as that of Drosophila. More interestingly, the putative boundaries and enhancers are organized into iab-type domains seen in the Drosophila BX-C (Figure 1B). There are two putative domains for mosquito Ubx, corresponding to abx/bx and bxd/pbx, one for abd-A corresponding to iab-2 and three for Abd-B corresponding to iab-6, iab-7 and iab-8. The Ubx domains are defined by boundaries, B38, B41 (AgB1) and B44 (AgB2), while abd-A domain is marked by B44 (AgB2) and B45. The Abd-B domains are demarcated by B52, B53 and B54. The region corresponding to iab-4 and iab-5 of Drosophila seems to be fused into a single domain in A. gambiae marked by B51 (AgB7) and B52. However, we find two additional putative domains between abd-A and mir-iab-4 which in Drosophila is organized into a single domain (iab-3). Furthermore, there are additional boundaries in the BX-C of A. gambiae between Ubx-abd-A and abd-A-mir-iab-4 (Figure 1B). These differences in boundary number and domain organization may be specific to the regulation of Hox genes in A. gambiae.
To study boundary elements in detail, we focused on four of the predicted boundary elements: AgB18 from the ANT-C, Scr-ftz region and three boundaries from the BX-C, AgB1 and AgB2 from Ubx-abd-A and AgB7 from abd-A-Abd-B region (Figure 1A). These boundaries were chosen due to their interesting location and correspondence with known boundaries of the Drosophila Hox cluster (Figure 1A and B). In Drosophila, Scr-ftz region contains a GAF-dependent boundary, SF1 (37). As most of the regulatory elements are located in the 5′-end of the Hox genes, AgB18, which reside in the first intron of Scr and contains GAF-binding sites, was the most likely candidate for SF1. In Drosophila, the chromatin domain regulatory elements in BX-C are very well characterized and each chromatin domain is marked by specific boundaries. AgB1, AgB2 and AgB7 are positioned strategically to demarcate the domains of Ubx, abd-A and Abd-B and may correspond to Drosophila boundaries 424, Fab-2 and Fab-4 on the basis of their position (Figure 1B).
Overall, the preferential localization of boundaries in regions largely devoid of protein-coding sequences between genes is in agreement with a role for these elements in organizing the genome into independent expression domains of gene expression.
Boundaries identified in the Hox complex of mosquito function as enhancer blockers in Drosophila S2 cells
We tested the boundary function of AgB1, AgB2, AgB7 and AgB18 using enhancer blocking assay in Drosophila S2 cells. Cell-culture-based assay has been widely used to analyse boundary function in vertebrates (38–40) and in Drosophila cells (41). We used GFP reporter transgene containing the test fragment between the PE (twist proximal element) enhancer and the hsp70 promoter that drives GFP expression (Figure 2A). The use of fluorescent reporters such as GFP allows a rapid and quantitative FACS assessment of the boundary function. The assay vector also contains neomycin which provides antibiotic resistance to transfected cells in the presence of neomycin drug G418.
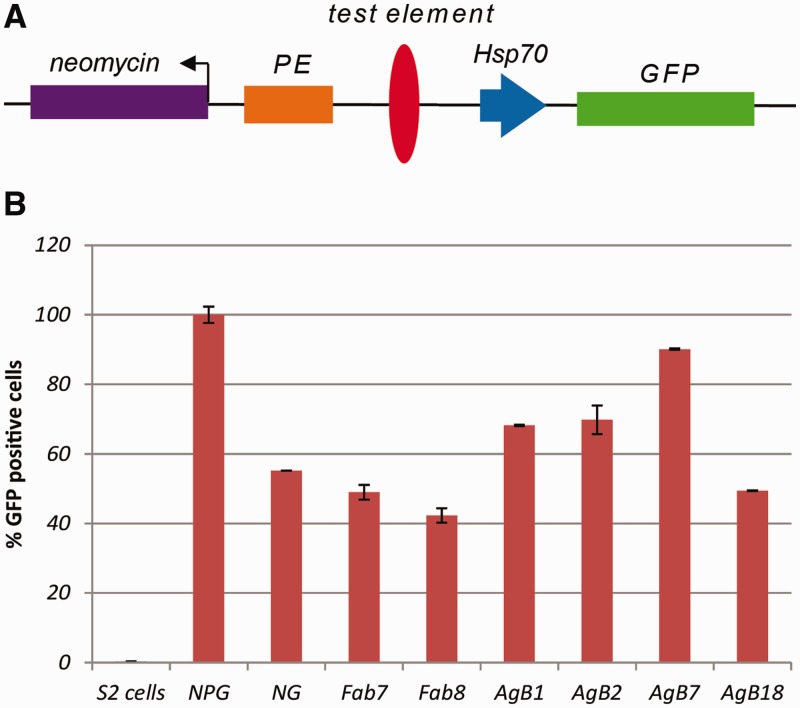
Figure 2.
Predicted mosquito boundary elements block enhancer (PE) mediated GFP expression in S2 cells. (A) Schematic of the boundary assay transgene containing the following components: neomycin gene for selection (purple), PE enhancer (orange), predicted boundary element (red oval) and hsp70-driven GFP reporter gene. (B) FACS analysis of GFP-positive cells selected for 6 weeks on G418 after transfection and plotted as GFP-positive cells. NPG; empty construct, NG; enhancer-less construct, Fab-7 and Fab-8 were used a positive controls and to compare the blocking of PE enhancer by mosquito boundaries.
S2 cells were transfected with constructs containing test boundary elements along with the empty vector (NPG) as a negative control and Fab-7 and Fab-8 as positive controls. We also used an enhancer-less construct (NG) to set the basal level of GFP expression. In stable transfections, the transgene integrates in a chromosomal environment and therefore provides a chromatin environment needed to study boundary function. After 6 weeks of selection on neomycin, we analysed cells for their enhancer-blocking activity using flowcytometry. We observed that cells transfected with transgenes containing AgB18 show a strong enhancer-blocking activity (50% reduction in GFP expression). The enhancer blocking by AgB18 was comparable to the blocking shown by Fab-7 and Fab-8 (50–60% reduction in GFP expression), two well-characterized boundaries of the BX-C of Drosophila (Figure 2B). Cells that were transfected with AgB1 and AgB2 showed a significant reduction in GFP expression (30% to 35%) indicating that they also act as enhancer blockers (Figure 2B). However, the enhancer-blocking activity of these two elements was not as strong as that of Fab-7 and Fab-8. Cells transfected with AgB7 showed a 10% reduction in GFP expression, thus behaving as a weak boundary. These results indicate that the predicted elements we analysed here from Hox complex of mosquito function as boundary elements of variable strength in Drosophila S2 cells.
Cell-based enhancer-blocking assay is a quick and easy way to analyse boundary function. However, it might not work with boundary elements whose function is tissue or developmental stage specific. In addition, cells over a period of time may lose physiological state and lack the relevant protein components necessary for boundary function. Therefore, cell-based boundary assay should be used to complement genetic and transgenic approaches to study boundary function. To complement our results obtained from cell-based enhancer-blocking assay, we used a transgenic approach to test these elements in Drosophila.
Identified boundary elements from mosquito function as enhancer blockers in transgenic Drosophila
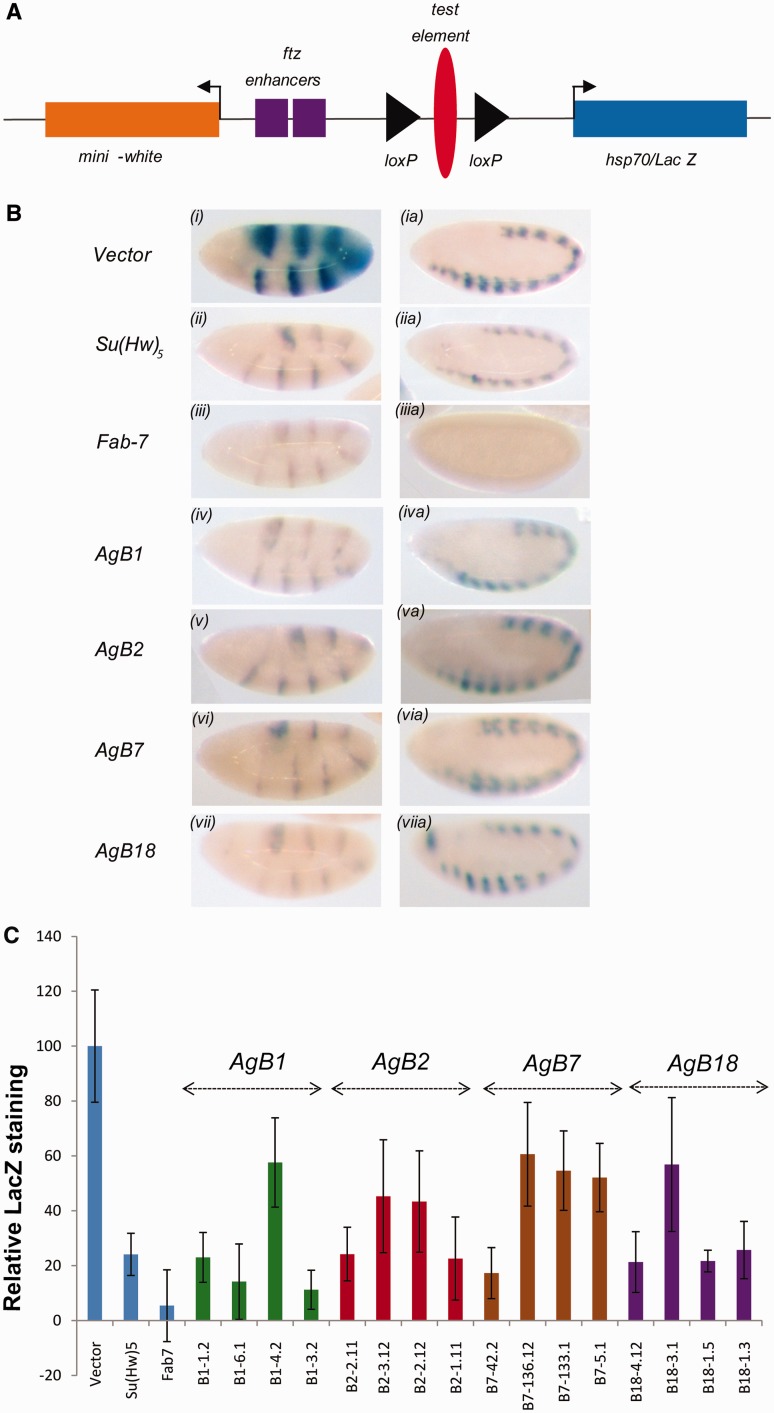
Cis-regulatory sequences taken from other Drosophila species or other distantly related Dipterans (Musa deomestica, Simulium vittatum) and even vertebrates have been successfully tested in Drosophila using transgenes (33,42). To test whether boundary elements from mosquito show functional conservation in Drosophila, we generated transgenic flies containing AgB1, AgB2, AgB7 and AgB18. The transgene construct to assay the boundary activity contains the test element inserted between the fushi-tarazu (ftz) enhancer and hsp70 promoter driving the lac-Z expression (34). Two ftz enhancers, UPS (seven stripes in early embryos) and NE (CNS pattern in late embryos), drive hsp70-lacZ gene in the transgenic flies carrying the assay construct (Figure 3A). In all the assay vectors, the test element was flanked by loxP sites to flip it out from the transgenic lines in order to rule out any position effect. We used an empty vector (pCfhl) as negative control and DNA fragment having five binding sites for SuHw, Su(Hw)5 and Fab-7 as positive controls. In transgenic lines carrying the empty vector, both UPS and NE enhancers of ftz drove a high level of lac-Z expression as determined by direct visualization and quantification of the staining intensity with ImagJ software (Figure 3B and C). In contrast, lac-Z expression driven by UPS enhancer was strongly reduced in three out of four lines carrying AgB1 or AgB18 (Figure 3B and C). The reduction in lac-Z staining in both the cases was comparable to control lines of Su(Hw)5 or Fab-7, both of which are known to have strong boundary activity (Figure 3B and C). These results indicate that AgB1 and AgB18 act as strong enhancer blockers in the early stages of Drosophila embryo. Although AgB2 transgene reduced the staining significantly in two lines, the blocking was seen to be slightly weaker than that of Fab-7 and Su(Hw)5 for other two lines (Figure 3B and C). In case of AgB7, one line showed strong blocking, whereas three others moderately blocked lac-Z expression when compared with Fab-7 or Su(Hw)5 (Figure 3B and C). Based on these results, we have referred AgB2 and AgB7 as moderate enhancer blockers.
Figure 3.
Mosquito boundary elements function as enhancer blockers in Drosophila embryos. (A) a diagram of the enhancer-blocking transgene pCfhl. The test element bracketed by loxP sites is cloned between ftz-enhancers and the hsp70-driven lac-Z reporter gene. (B) β-Galactosidase assay on transgenic embryos. All the embryos were stained in parallel to directly compare the level of lac-Z expression among different transgenic lines. On the left is shown lac-Z staining in germband-extended embryos of Stage 10 driven by UPS enhancer. On the right is shown NE-driven lac-Z staining of Stage 14 embryos in the central nervous system (CNS). Upper row is the representative embryos from the empty control construct (pCfhl) followed by five binding sites for Su(Hw), Fab-7 fragment, AgB1, AgB2, AgB7 and AgB18. (C) Quantification of lacZ staining using ImageJ software. Embryo images from four independent lines for each P element were analysed for lacZ staining. One of the seven stripes was taken as a representative for lacZ staining intensity and normalized to background. Relative lacZ staining is plotted along with the empty vector and positive controls, Su(Hw)5 and Fab-7. Error bars represent standard deviation from five embryos of the same line.
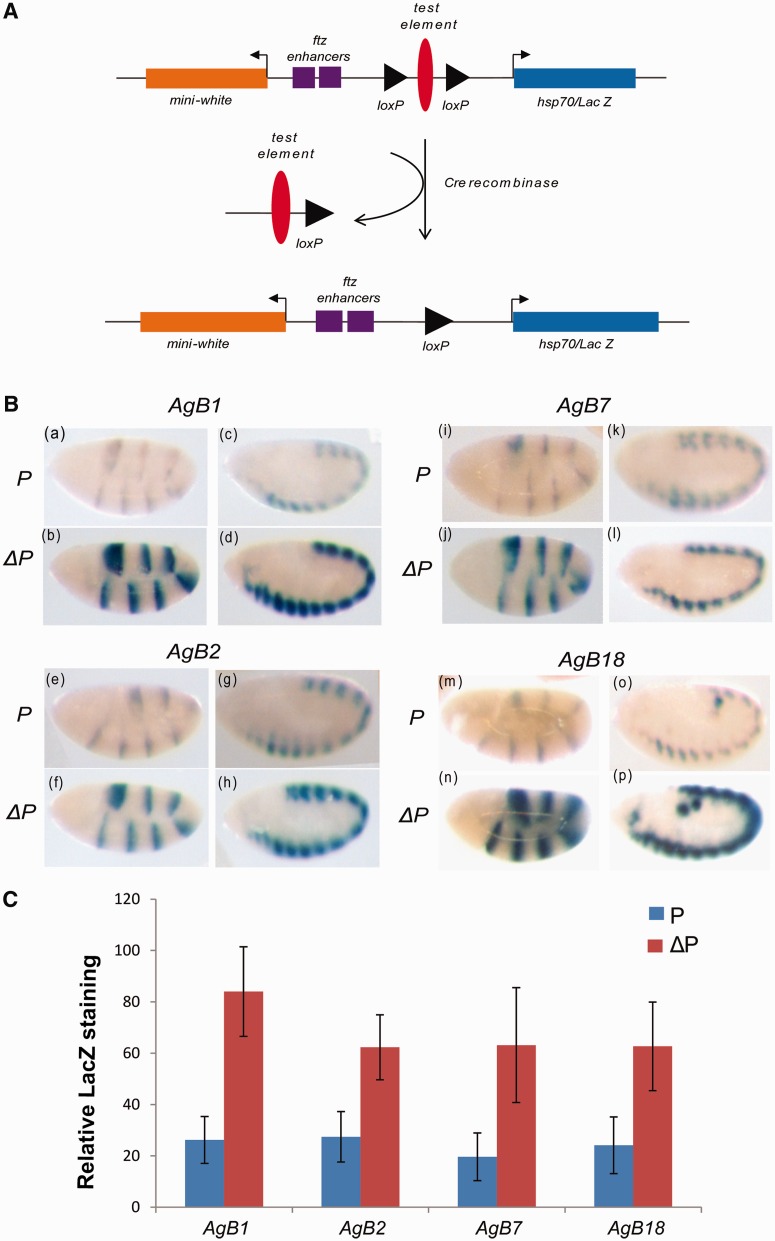
To check whether the above elements show enhancer blocking in the later stages of the Drosophila embryo, we also analysed lac-Z expression driven by NE enhancer. Interestingly, we noticed a significant reduction in lac-Z expression in the later stages of the embryo (Figure 3B) suggesting that these elements also act as enhancer (NE) blockers in the later stages of the embryonic development. Based on these results, we classify AgB1 and AgB18 as strong boundaries while AgB2 and AgB7 as moderate boundaries. Finally, to rule out position effect of the insertion site, we crossed our blocker lines to Cre-expressing fly to flip out the test elements (Figure 4A) (35). Several lines for each construct were analysed after excision of the test fragment. The level of staining was clearly restored to levels comparable to empty vector when AgB1, AgB2, AgB7 or AgB18 was flipped out (Figure 4B and C). These results clearly suggest that the reduction in lac-Z expression is indeed due to the boundary function of the mosquito elements that prevent enhancer from acting on the hsp70 promoter and not due to chromosomal environment of the transgene insertion.
Figure 4.
β-Galactosidase assay in embryos after flipping out of the test element. (A) The test element flanked by loxP sites is removed upon crossing the transgenic line to a Cre expressing fly. (B) Boundary activity of mosquito test elements is position independent as the removal of the test element restores the lac-Z staining. Top and bottom row of the lower panel represent the embryos of initial lines and flipped-out lines, respectively. All the genotypes were stained in a grid under identical conditions. (C) Quantitative representation of the relative staining of lacZ in transgenic and flipped-out lines. Embryo images for transgenic initial line and the flipped-out line were processed in ImageJ. P indicates transgenic initial line and ΔP indicates the flipped-out version of the same line. Error bars represent standard deviation from five embryos.
Genetic interaction of mutations in boundary interacting factors and mosquito boundary elements in Drosophila
In the previous sections, we established that mosquito boundary elements act as enhancer blockers in Drosophila. Next, we asked, what trans-acting factors are responsible for their boundary function? The test elements analysed here are predominantly rich in GAF-binding sites. (Supplementary Table S2 and Supplementary Figure S2). Binding sites for GAF are prevalent in the regulatory regions of Hox genes as well as elsewhere (43,44) and several of these have been implicated in boundary function. For example, GAGAG sites in the Drosophila eve promoter and in the SF1 boundary element are required for their insulator function (37,45). To test if GAF is required for the boundary activity of the test elements, we crossed TrlR85 heterozygous females to homozygous males containing the test element and analysed the enhancer-blocking activity in embryos resulting from the cross. We observed significant increase in the lac-Z staining in embryos of AgB1 and AgB2 in the TrlR85 mutant background (Figure 5A and B). These results indicate that GAF is important for the boundary activity of AgB1 and AgB2. Although not as pronounced as in case of AgB1 and AgB2, we also observed a significant increase in the staining of embryos from AgB7 and AgB18 lines (Figure 5A and B). When we brought flipped-out lines of the test fragments in TrlR85 mutant background, no increase in staining was observed, confirming our results (Supplementary Figure S3).
Figure 5.
(A) Boundary activity of AgB1, AgB2, AgB7 and AgB18 in mutant backgrounds. Males of initial transgenic line were crossed to virgin females of TrlR85, CTCFy+6, BEAFKO and CP190H31-2. The embryos resulting from the cross were analysed for enhancer-blocking activity by β-galactosidase assay. (B) Quantitative representation of the effect of mutations in Trl-GAGA, CP190 and dCTCF on mosquito boundary elements. Images of the embryos in various mutant backgrounds were analysed for lacZ staining using ImageJ software as described earlier. Noticeable increase is observed in Trl and CP190 backgrounds when compared with wild-type embryos (PxCS). Error bars represent standard deviation from five embryos. (C) ChIP analysis of GAF on transgenic larvae containing AgB1, AgB2, AgB7 and AgB18. Chromatin was pulled-down with anti-GAF and analysed by real time PCR using SYBR green mix. Fold enrichment at the target region was calculated as relative enrichment over the negative control (MW—mini-white coding region). Results shown are mean of two independent experiments with error bars.
To further confirm our results that GAF binds to AgB1, AgB2, AgB7 and AgB18, we performed ChIP experiments using GAF antibody on larvae of transgenic flies carrying the test elements. More than 10-fold enrichment was seen in GAF pull-down compared with the negative control region of MW gene which does not contain binding sites for GAF (Figure 5C). Known GAF-binding region of iab-PRE7 locus (46) was used as a positive control that showed >30-fold enrichment over negative control. These results indicate that GAF indeed interacts with the test elements in the context of transgenic Drosophila.
Although it has not been shown to directly bind to insulator sequences, CP190 is an essential co-factor for several boundary elements in Drosophila (47,48). Therefore, we were interested to test the effect of CP190 mutation on the function of above tested elements. We used CP190H31-2 and examined the boundary function in the mutant context. We found an appreciable increase in lac-Z staining in case of AgB1and AgB2, while AgB7 and AgB18 showed only a mild increase (Figure 5A and B). These observations suggest that CP190 acts as a co-factor for the boundary function of the test elements in Drosophila.
To test the functional relevance of BEAF sites in test elements, we tested boundary activity in BEAFKO background. No effect was seen on the boundary function as indicated by no change in lac-Z expression (data not shown). Although none of the tested elements has binding sites for dCTCF, nevertheless, we analysed boundary function in dCTCFy+6 backgrounds and found no effect on boundary activity. Taken together, it appears that the mosquito boundary elements tested in Drosophila use conserved protein components such as GAF and CP190 to carry out their function.
DISCUSSION
Many genomic loci in eukaryotes contain gene clusters that share regulatory elements. Such loci must be tightly regulated to prevent undesirable interaction among different regulatory elements. Chromatin boundary elements ensure proper regulation of such genomic loci by preventing the cross-talk between enhancers, silencers and promoters. Hox gene cluster is one such locus where boundary elements have been shown to be critical for the proper expression of Hox genes along the A–P axis. Due to the independent, yet co-ordinated control of multiple genes by a complex set of regulatory elements, Hox gene cluster is an interesting model to study the correlation between gene organization and gene regulation. Furthermore, Hox genes and their organization have remained conserved during evolution and it is envisaged that their regulatory elements may also be invariant and share appreciable similarities between closely related species. In this study, we searched for chromatin boundary elements from the Hox complex of malarial mosquito, A. gambiae, and analysed their function in D. melanogaster. We found that mosquito boundary elements (AgB1, AgB2, AgB7 and AgB18) function as enhancer blockers in Drosophila suggesting a conserved mechanism of boundary function across insect species.
In Drosophila, especially in the bithorax complex, a number of boundary elements have been identified that regulate individual Hox genes by defining cis-regulatory domain (49). Although mosquito Hox complex is not split, it has striking similarity with that of the fly Hox cluster with respect to its organization (Figure 1A and B). Given the faster divergence rate of Drosophila, splitting of its Hox cluster and an evolutionary distance of 250 million years, it is interesting to note that the BX-C region in mosquito and Drosophila is fairly of equal size. Moreover, it is also interesting to point out that mosquito BX-C region, like Drosophila, also contains a series of cis-regulatory elements organized into putative iab-type domains defined by boundary elements that include AgB1, AgB2 and AgB7 (Figure 1B). The fact that breaks in Drosophila Hox cluster have rarely occurred in the BX-C, it is tempting to assume that sharing of cis-regulatory regions may have kept the bithorax complex intact across insect species. The ANT-C in Drosophila does not contain as many boundaries and enhancers as the BX-C. However, the ANT-C as well as the Antp-Ubx intergenic region in mosquito contain a number of putative boundaries and enhancers suggesting that the overlapping nature of cis-regulatory elements may be a constraining force for the intact Hox cluster of mosquito. Additional experiments will be necessary to understand the function of regulatory elements to address the question whether Hox genes in A. gambiae are clustered because of the intermingling of regulatory elements of one Hox gene with the transcription unit of another or the intact Hox cluster of A. gambiae is simply a product of phylogenetic inertia.
One of the tested elements, AgB18, acts as a strong boundary and separates Scr from ftz. The Scr-Antp region in Drosophila Hox cluster contains two homeotic genes, Scr and Antp, and the segmentation gene ftz. Scr and ftz are divergently transcribed and separated by a 15-kb intergenic region that harbours enhancers (D and AE1) which specifically interact with ftz promoter and drive its expression. The Scr is protected from the effects of ftz enhancers by a boundary element, SF1, located in the intergenic region of Scr-ftz (37). The Scr gene has a complex expression pattern driven, in part, by the distal T1 enhancer located 3′ of the ftz gene which specifically interacts with Scr promoter. Scr-distal T1 enhancer is thought to bypass SF1 either by promoter-targeting sequence (PTS) or interaction of SF1 with other boundaries located downstream of ftz which may also shield ftz from T1 enhancer. Like Drosophila, the Scr and ftz genes in A. gambiae are also divergently transcribed. However, nothing is known about the regulatory elements like enhancers, boundaries or PTS in mosquito. In our analysis, we could neither locate a boundary nor any enhancer element in the intergenic interval of Scr-ftz in A. gambiae. However, we found putative boundaries and enhancers within the intronic region of the Scr (Supplementary Figure S1). Presence of enhancers and their regulation by boundary elements in the intronic region has been reported earlier (50). It is possible that the enhancers embedded in the Scr gene drive tissue-specific expression of Scr. If this is true, the promoter of ftz will need to be shielded from the influence of these intronic enhancers. In such context, AgB18 may restrict the effect of downstream enhancers located in the Scr to its promoter and prevent them from inappropriately interacting with the ftz promoter. The enhancer located in the 3′ of ftz (Supplementary Figure S1) may not activate Scr like T1 distal enhancer described above, but instead drive ftz expression. In this scenario, boundary element, B28, may be required to shield ftz transcription unit from enhancers located downstream in the Antp region. Although the regulation of Scr-ftz interval in mosquito may differ from that of Drosophila, we speculate that AgB18 may be related, at least in part, to SF1 to maintain the functional independence of Scr and ftz transcription in Antennapedia complex of A. gambiae. Interestingly, like SF1, AgB18 is GAF dependent for its enhancer-blocking activity.
For a detailed understanding of the boundary function, it is important to identify the protein components involved in boundary function. Thus far, the proteins that directly associate with boundary sequences include Zw5, BEAF-32 (51,52), GAF (37,45), Su(Hw) (53) and dCTCF (54). Although the presence of vertebrate GAF has recently been reported, CTCF is the only known boundary factor in vertebrates (55,56). Other factors that may not directly interact with boundary elements but are implicated in boundary function include CP190, Mod(mdg4), dTopors, Rm62, cohesion and Ago2 (47,57–60). Computational analysis of boundary interacting factors across species has revealed that, with the exception of BEAF and Zw5, all the boundary interacting factors are conserved in mosquitoes and other insect species (30,36). It has also been shown that mosquito CTCF interacts with known boundary elements from Drosophila (Fab-8) and chicken (5′HS4) in vitro (36). Binding site analysis of boundary elements analysed in this study revealed that these elements are predominantly GAF dependent. Using genetic approach, we show that GAF is a key component mediating the enhancer-blocking function of AgB18, AgB1, AgB2 and AgB7. Upon the loss of a single copy of GAF, we found that the boundary function of all these elements is compromised. We also show that GAF directly associates with the tested boundary elements in vivo (Figure 5B). These results suggest that GAF may be the major player mediating the boundary function in mosquito Hox complex. Apart from being a transcriptional activator and recruiting remodelling complexes (61,62), GAF is known to be an important factor for boundary elements in the Hox complex of Drosophila (37,63). We have earlier shown that the Drosophila GAF can mediate the boundary function of a mouse boundary element located in the Hoxd13-Evx2 region (33), suggesting that GAF may be one of the major protein component of evolutionarily conserved aspect of boundaries, specifically in the developmentally regulated loci.
We also found that CP190 is required for boundary function of the tested elements as loss of a single copy reduced the enhancer-blocking activity of these elements. It is not surprising that the mosquito boundary elements also require CP190 as a co-factor for their boundary function because CP190 is known to be required for dCTCF and Su(Hw)-dependent boundary function in Drosophila (47,48,64). In addition, CP190 has been shown to associate with BEAF on a genome-wide scale (65). Although AgB1, AgB2 and AgB18 contain binding sites for BEAF, no effect was seen on the boundary function in BEAFKO background. It is possible that these sites represent non-functional BEAF sites, because BEAF is believed to be absent from all the species other than Drosophilids (30). Moreover, none of these boundaries contains palindromic BEAF sites as frequently seen in BEAF-dependent boundaries in Drosophila (66). Taken together, this study shows that mosquito boundary elements identified here employ GAF and CP190 for their enhancer-blocking activity. We propose that the boundary elements identified here from the Hox complex of mosquito may share common components and mechanisms with Drosophila and belong to a class of conserved boundary elements that regulate enhancer–promoter interactions in the Hox complexes.
Finally, isolation of boundary sequences from mosquito lays a strong foundation to probe further into the regulation of Hox genes in insects with intact Hox clusters such as mosquito. In addition, the comparative analysis of regulation of Hox genes in mosquito and Drosophila will significantly help to increase our knowledge about characteristics, evolutionary conservation and location of functional elements within the complex genetic loci. With the improvement in molecular and transgenic tools available for mosquito, it will be possible to explore these elements in their native chromosomal context. Furthermore, efforts should be made to identify such elements from other species in order to better understand their distribution and evolutionary conservation. Additionally, this study provides promising avenues to flank transgenes with endogenous boundary elements which could greatly improve the expression levels of transgenes in this medically important mosquito species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–3 and Supplementary zip file.
FUNDING
Department of Biotechnology (DBT), India (to Y.S.S. and R.K.M.); Indian counsel of Medical Research (ICMR), India, and Deutscher Akademischer Austausch Dienst (DAAD), Germany (to S.H.A.); Council of Scientific and Industrial Research (CSIR) (to R.K.M.). Funding for open access charge: Centre for Cellular and Molecular Biology, Hyderabad, India.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Francois Karch, Craig Hart and Victor G Corces for mutant stocks of Drosophila used in the study and Paul Schedl for control enhancer-blocking test lines. They also thank Navneet K Matharu for help with ChIP experiments and critical reading of the manuscript.
REFERENCES
- 1.Kaufman TC, Lewis R, Wakimoto B. Cytogenetic analysis of chromosome 3 in Drosophila melanogaster: the homoeotic gene complex in polytene chromosome interval 84a-B. Genetics. 1980;94:115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 3.Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 5.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 6.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence PA, Morata G. Homeobox genes: their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 8.Von Allmen G, Hogga I, Spierer A, Karch F, Bender W, Gyurkovics H, Lewis E. Splits in fruitfly Hox gene complexes. Nature. 1996;380:116. doi: 10.1038/380116a0. [DOI] [PubMed] [Google Scholar]
- 9.Lewis EB, Pfeiffer BD, Mathog DR, Celniker SE. Evolution of the homeobox complex in the Diptera. Curr. Biol. 2003;13:R587–R588. doi: 10.1016/s0960-9822(03)00520-7. [DOI] [PubMed] [Google Scholar]
- 10.Yasukochi Y, Ashakumary LA, Wu C, Yoshido A, Nohata J, Mita K, Sahara K. Organization of the Hox gene cluster of the silkworm, Bombyx mori: a split of the Hox cluster in a non-Drosophila insect. Dev. Genes Evol. 2004;214:606–614. doi: 10.1007/s00427-004-0441-1. [DOI] [PubMed] [Google Scholar]
- 11.Chai CL, Zhang Z, Huang FF, Wang XY, Yu QY, Liu BB, Tian T, Xia QY, Lu C, Xiang ZH. A genomewide survey of homeobox genes and identification of novel structure of the Hox cluster in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1111–1120. doi: 10.1016/j.ibmb.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Denell RE, Brown SJ, Beeman RW. Evolution of the organization and function of insect homeotic complexes. Semin. Cell Dev. Biol. 1996 527–538. [Google Scholar]
- 13.Shippy TD, Ronshaugen M, Cande J, He J, Beeman RW, Levine M, Brown SJ, Denell RE. Analysis of the Tribolium homeotic complex: insights into mechanisms constraining insect Hox clusters. Dev. Genes Evol. 2008;218:127–139. doi: 10.1007/s00427-008-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walldorf U, Binner P, Fleig R. Hox genes in the honey bee Apis mellifera. Dev. Genes Evol. 2000;210:483–492. doi: 10.1007/s004270000091. [DOI] [PubMed] [Google Scholar]
- 15.Ferrier DE, Akam M. Organization of the Hox gene cluster in the grasshopper, Schistocerca gregaria. Proc. Natl Acad. Sci. USA. 1996;93:13024–13029. doi: 10.1073/pnas.93.23.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devenport MP, Blass C, Eggleston P. Characterization of the Hox gene cluster in the malaria vector mosquito, Anopheles gambiae. Evol. Dev. 2000;2:326–339. doi: 10.1046/j.1525-142x.2000.00074.x. [DOI] [PubMed] [Google Scholar]
- 17.Powers TP, Hogan J, Ke Z, Dymbrowski K, Wang X, Collins FH, Kaufman TC. Characterization of the Hox cluster from the mosquito Anopheles gambiae (Diptera: Culicidae) Evol. Dev. 2000;2:311–325. doi: 10.1046/j.1525-142x.2000.00072.x. [DOI] [PubMed] [Google Scholar]
- 18.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 19.Randazzo FM, Seeger MA, Huss CA, Sweeney MA, Cecil JK, Kaufman TC. Structural changes in the antennapedia complex of Drosophila pseudoobscura. Genetics. 1993;134:319–330. doi: 10.1093/genetics/134.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 21.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- 22.Galloni M, Gyurkovics H, Schedl P, Karch F. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogga I, Mihaly J, Barges S, Karch F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell. 2001;8:1145–1151. doi: 10.1016/s1097-2765(01)00377-x. [DOI] [PubMed] [Google Scholar]
- 24.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 25.Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolshakov VN, Topalis P, Blass C, Kokoza E, della Torre A, Kafatos FC, Louis C. A comparative genomic analysis of two distant diptera, the fruit fly, Drosophila melanogaster, and the malaria mosquito, Anopheles gambiae. Genome Res. 2002;12:57–66. doi: 10.1101/gr.196101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Adv. Genet. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- 29.Seeger MA, Haffley L, Kaufman TC. Characterization of amalgam: a member of the immunoglobulin superfamily from Drosophila. Cell. 1988;55:589–600. doi: 10.1016/0092-8674(88)90217-6. [DOI] [PubMed] [Google Scholar]
- 30.Schoborg TA, Labrador M. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J. Mol. Evol. 2010;70:74–84. doi: 10.1007/s00239-009-9310-x. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan A, Mishra RK. Chromatin domain boundary element search tool for Drosophila. Nucleic Acids Res. 2012;40:4385–4395. doi: 10.1093/nar/gks045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starr MO, Ho MC, Gunther EJ, Tu YK, Shur AS, Goetz SE, Borok MJ, Kang V, Drewell RA. Molecular dissection of cis-regulatory modules at the Drosophila bithorax complex reveals critical transcription factor signature motifs. Dev. Biol. 2011;359:290–302. doi: 10.1016/j.ydbio.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasanthi D, Anant M, Srivastava S, Mishra RK. A functionally conserved boundary element from the mouse HoxD locus requires GAGA factor in Drosophila. Development. 2010;137:4239–4247. doi: 10.1242/dev.058701. [DOI] [PubMed] [Google Scholar]
- 34.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 35.Siegal ML, Hartl DL. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray CE, Coates CJ. Cloning and characterization of cDNAs encoding putative CTCFs in the mosquitoes, Aedes aegypti and Anopheles gambiae. BMC Mol. Biol. 2005;6:16. doi: 10.1186/1471-2199-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Recillas-Targa F, Bell AC, Felsenfeld G. Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc. Natl Acad. Sci. USA. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao S, Osborne CS, Bharadwaj RR, Pasceri P, Sukonnik T, Pannell D, Recillas-Targa F, West AG, Ellis J. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res. 2003;31:5317–5323. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Belozerov VE, Cai HN. Analysis of chromatin boundary activity in Drosophila cells. BMC Mol. Biol. 2008;9:109. doi: 10.1186/1471-2199-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittkopp PJ. Evolution of cis-regulatory sequence and function in Diptera. Heredity. 2006;97:139–147. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- 43.Poux S, Melfi R, Pirrotta V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leibovitch BA, Lu Q, Benjamin LR, Liu Y, Gilmour DS, Elgin SC. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol. 2002;22:6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chopra VS, Srinivasan A, Kumar RP, Mishra K, Basquin D, Docquier M, Seum C, Pauli D, Mishra RK. Transcriptional activation by GAGA factor is through its direct interaction with dmTAF3. Dev. Biol. 2008;317:660–670. doi: 10.1016/j.ydbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 51.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 53.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 54.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 56.Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate homologue of Drosophila GAGA factor. J. Mol. Biol. 2010;400:434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 58.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 60.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okada M, Hirose S. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol. Cell. Biol. 1998;18:2455–2461. doi: 10.1128/mcb.18.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sultana H, Verma S, Mishra RK. A BEAF dependent chromatin domain boundary separates myoglianin and eyeless genes of Drosophila melanogaster. Nucleic Acids Res. 2011;39:3543–3557. doi: 10.1093/nar/gkq1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.