Abstract
The mRNA export complex TREX (TREX) is known to contain Aly, UAP56, Tex1 and the THO complex, among which UAP56 is required for TREX assembly. Here, we systematically investigated the role of each human TREX component in TREX assembly and its association with the mRNA. We found that Tex1 is essentially a subunit of the THO complex. Aly, THO and UAP56 are all required for assembly of TREX, in which Aly directly interacts with THO subunits Thoc2 and Thoc5. Both Aly and THO function in linking UAP56 to the cap-binding protein CBP80. Interestingly, association of UAP56 with the spliced mRNA, but not with the pre-mRNA, requires Aly and THO. Unexpectedly, we found that Aly and THO require each other to associate with the spliced mRNA. Consistent with these biochemical results, similar to Aly and UAP56, THO plays critical roles in mRNA export. Together, we propose that Aly, THO and UAP56 form a highly integrated unit to associate with the spliced mRNA and function in mRNA export.
INTRODUCTION
The highly conserved TREX complex (TREX) plays key roles in mRNA export. The human TREX contains the multi-subunit THO complex (THO) as well as proteins UAP56, Aly and Tex1 (known as THO, Sub2, Yra1 and Tex1, respectively, in yeast) (1–3). In recent studies, DDX39 and CIP29 were also reported to be putative new human TREX components (4,5). The yeast THO complex contains Tho2, Hpr1, Mft1 and Thp2 (6). The Drosophila and human THO complexes contain homologs of yeast Tho2 and Hpr1 (Thoc2 and Thoc1), as well as Thoc5, Thoc6 and Thoc7 that do not have apparent counterparts in yeast (2,7). THO subunits tightly bind with one another. In humans, Thoc2 immunodepletion led to co-depletion of the whole THO complex (2). Moreover, in both Drosophila and mammals, THO proteins require one another for maintaining their stability (2,7,8). Although Tex1 is known to be an independent TREX component parallel to THO, Aly and UAP56, in both yeast and humans, stable association of Tex1 with THO has been reported (2,3). In yeast, a most recent study revealed that Tex1 is co-purified with THO even in 500 mM salt condition (9). In humans, Tex1 is always present in stoichiometric amount with other THO subunits in immunoprecipitates by THO antibodies (2).
Previous studies in humans showed that UAP56, which is an ATP-dependent RNA helicase, binds directly with Aly (10–12). More recent studies revealed that UAP56 interacts with Aly in an ATP-dependent manner, and the ATPase activity of UAP56 is stimulated by Aly (5,13). These studies suggest that Aly might play a regulatory role in TREX formation. Moreover, UAP56 was shown to be required for the interaction between Aly and THO, leading to the view that UAP56 is the core component of TREX (5). Consistent with this view, Aly and THO associate with different regions of UAP56 (2). However, it remains to be investigated whether Aly and THO function in TREX assembly.
Despite the high conservation in constitution and function of TREX, recent studies revealed that it is linked to different machineries in yeast and humans (14). The yeast THO associates with transcription machinery and consequently recruits and transfers Yra1 and Sub2 to nascent transcripts (15–19). A more recent study reported an alternative model for TREX recruitment in yeast (20). In this model, Yra1 is recruited by Pcf11, which is a component of CF1A complex, and is transferred to Sub2 and THO already associated with the mRNA. In both models, the yeast TREX is thought to be recruited in a stepwise way co-transcriptionally. In humans, the entire TREX is connected to the splicing machinery and is recruited to mRNA in a splicing-dependent manner (2,21,22). In addition to splicing, our previous study revealed that the 5′-cap is required for TREX recruitment and mRNA export (23). The cap-binding protein CBP80 associates with TREX and this association was thought to be mediated by the interaction between Aly and CBP80 (23). Thus, it is possible that Aly is required for efficient recruitment of TREX to the spliced mRNA. However, this possibility has not been investigated and the role of THO in recruitment of TREX remains to be determined.
In this study, we sought to investigate how each human TREX component functions in assembly of the complex and in association of TREX with the mRNA. We found that Tex1, a previously reported independent TREX component, is essentially a subunit of the THO sub-complex. Significantly, similar to UAP56, Aly and THO are also critical for TREX assembly. Moreover, both Aly and THO function in linking TREX to nuclear cap-binding complex (CBC) and are required for binding of TREX components with the spliced mRNA. Consistent with these roles of THO, we found that THO is required for the export of total mRNA.
MATERIALS AND METHODS
Plasmids and antibodies
Plasmids encoding AdML and Ftz pre-mRNA, GST-Aly, His-UAP56, GST-AlyΔC, GST-UAP56 and GST-eIF4A3 were described previously (11,24–26). Maltose-binding protein (MBP)-tagged Aly, UAP56, Tex1, Thoc6 and Thoc7 plasmids were constructed by inserting the corresponding coding sequence into pMAL-c2X plasmid. To construct His-Aly and His-AlyΔC plasmids, NcoI–BamHI fragment from GST-Aly and BamHI–SacI fragment from GST-AlyΔC were inserted into the same sites of pET 9d and pRSET A plasmids, respectively. The rabbit polyclonal Tex1 antibody was raised against MBP-Tex1 and affinity purified. Aly, UAP56, Thoc7 and Thoc2 antibodies were raised against full-length GST-Aly, GST-UAP56, MBP-Thoc7 and C-terminal part of Thoc2, respectively. Antibodies to Thoc5, CBP80 and eIF4A3 were described previously (2,23). The negative control antibody used for immunoprecipitations (IPs) is against fSAP130 (27). Tubulin and SC35 antibodies were purchased from Sigma.
Cell culture and RNAi
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Biochrom). For shRNA-mediated knockdown, pLKO.1 plasmid inserted with DNA oligonucleotide that encodes shRNA was introduced into 293 FT cells together with packaging plasmid psPAX2 and envelope plasmid pMD 2.G. After 48 h, the medium containing lentiviruses was filtered and added to HeLa cells. Two microgram per milliliter of puromycin (Sigma) was added to the medium after 24 h. Cells were used for fluorescence in situ hybridization (FISH), western and reverse transcriptase–polymerase chain reaction (RT–PCR) analyses after another 72 h. The shRNAs targeting sequences are shown in Supplementary Table S1.
Gel filtration
Gel filtration was performed on a 1.5-cm/50-cm Sephacryl S-500 gel filtration column (GE Healthcare). The gel filtration buffer contains 20 mM HEPES, 500 mM KCl, 2.5 mM ethylenediaminetetraacetic acid (EDTA), 0.1% Triton and 0.01% sodium azide. 250 μl of HeLa nuclear extract was mixed with the same volume of splicing dilution buffer (20 mM HEPES at pH 7.6 and 100 mM KCl) and loaded onto the column. Samples were separated at a flow rate of 0.1 ml/min and each 1 ml fraction was collected and analyzed by western analyses. The column was calibrated with Gel Filtration Calibration Kit HMW (GE healthcare). The void volume was 23 ml.
FISH and immunofluorescence
FISH was preformed as previously described using a high performance liquid chromatography-purified Alexa 548-conjugated oligo dT (70) probe (5). Immunofluorescence was performed by incubating fixed cells with the SC35 antibody 1:200 diluted in blocking buffer (1× phosphate buffered saline (PBS), 0.1% Triton and 2 mg/ml bovine serum albumin) for 30 min at the room temperature. Cells were then washed three times with PBS for 5 min each and incubated with the Alexa-488-labeled anti-mouse antibody 1:2000 diluted in blocking buffer for another 30 min at the room temperature, followed by DAPI staining and three washes in PBS for 10 min each.
Immunodepletions and protein IPs
For immunodepletions, antibodies were covalently cross-linked to protein A Sepharose beads (GE Healthcare) at 4:1 ratio (crude serum volume:packed beads volume) using dimethylpimelimidate (Sigma). One milliliter of high salt HeLa nuclear extract (350 mM) was mixed with 250 μl of beads and rotated for 1.5 h at 4°C. The procedure was repeated for three times to deplete Aly and THO, and for seven times to deplete UAP56. For protein IPs, antibodies were covalently cross-linked to protein A beads at 1:2 ratio (crude serum volume:packed beads volume). Seventy-five microliter of HeLa nuclear extract was incubated under splicing condition for 20 min in the presence of 50 ng/µl RNase A. The mixture was combined with 125 µl of IP buffer (1× PBS, 0.1% Triton, 0.2 mM PMSF and protease inhibitor) and 20 µl of beads cross-linked with antibodies. The mixtures were rotated overnight at 4°C, followed by six washes with 1.5 ml of IP buffer. Proteins were eluted with sodium dodecyl sulfate (SDS) loading buffer and separated by SDS–polyacrylamide gel electrophoresis (PAGE).
RNA IPs
RNA IPs were performed as previously described (23). Pre-mRNAs were incubated with HeLa nuclear extract for indicated times under splicing condition. Five microliters of splicing reaction and 100 µl of binding buffer (20 mM HEPES at pH 7.9, 200 mM KCl, 0.1% Triton, 2.5 mM EDTA and 5 mM DTT) were mixed with 10 µl of protein A Sepharose beads coupled with 5 µl of antibodies. IPs were performed at 4°C for 2 h, followed by six washes with 1.5 ml of binding buffer. RNAs were recovered by phenol/chloroform extraction and ethanol precipitation. RNA was analyzed on denaturing polyacrylamide gels and visualized by PhosphorImager. One-fourth of the input was loaded.
GST and MBP Pull downs
For each pull down, 8 µg of purified GST- or MBP-tagged proteins bound to 20 µl of Glutathione Sepharose 4B or Amylose resins were used. For pull downs from insect cell lysates, High Five cell lysate containing overexpressed THO proteins were mixed together with protein-bound beads in pull down buffer (1× PBS with 0.1% Triton, 0.2 mM PMSF, 50 ng/µl RNase A and protease inhibitor). The mixtures were rotated overnight at 4°C and beads were washed for five times. Proteins were eluted with SDS loading buffer, separated by SDS–PAGE, followed by western analyses. GST pull down for in vitro-translated proteins was performed as previously described (23). To pull down purified Thoc6, Thoc7 or Tex1, 8 µg of proteins were mixed with 20 µl of protein-bound beads in 1× PBS with 0.1% Triton, 0.2 mM PMSF and protease inhibitor. The mixtures were rotated overnight at 4°C and beads were washed with 1× PBS/0.1% Triton for five times and eluted with SDS loading buffer. Samples were separated by SDS–PAGE and stained with Coomassie blue.
RESULTS
Tex1 is a subunit of the THO complex
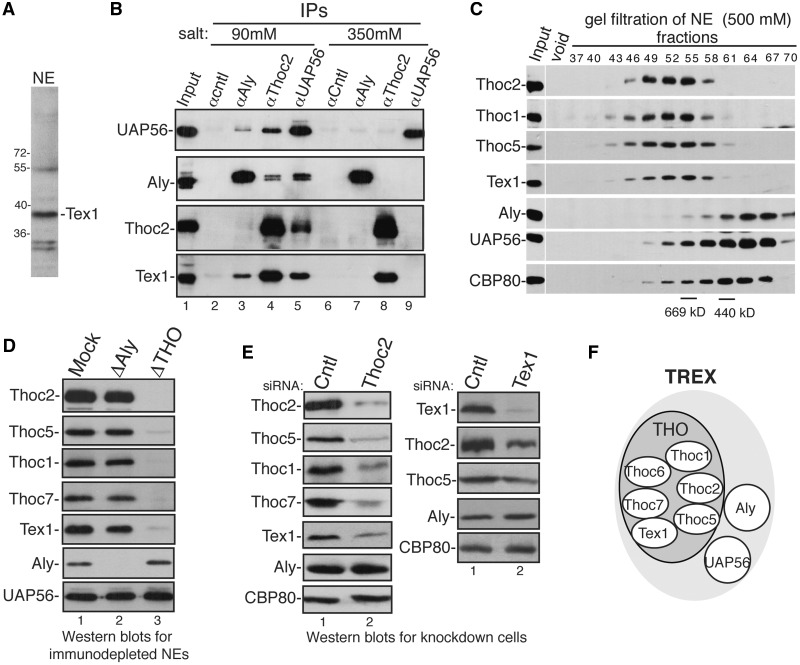
Tex1 is known to be a TREX component independent of Aly, THO and UAP56 (2). To study the interactions between Tex1 and other TREX components, we raised an antibody to Tex1. As shown in Figure 1A, this antibody detected a band of the appropriate size for Tex1 (39 kDa) in HeLa nuclear extract. We then examined whether Tex1 is immunoprecipitated (IP’d) from RNased HeLa nuclear extract by Aly, THO and UAP56 antibodies in normal and high salt conditions. In normal salt condition, as previously reported, Aly, UAP56 and Thoc2 were co-IP’d with one another, and Tex1 was also co-IP’d with them (Figure 1B, 90 mM) (2,5). In contrast, in high salt condition, only Tex1–Thoc2 interaction remained (Figure 1B, 350 mM). The control antibody to eIF4A3, which is a component of the exon–junction complex, did not IP any TREX protein under either salt condition. These results indicate that Tex1 tightly associates with THO. Consistent with these results, when the nuclear extract was size fractionated by gel filtration under high salt condition (500 mM), Tex1 was present in the same fractions as THO subunits that peaked in fractions (52 and 55) containing larger complexes. In contrast, Aly, UAP56 as well as CBP80 were eluted later from the gel filtration column (Figure 1C). We next examined whether Tex1 is co-immunodepleted with THO. As shown in Figure 1D, in the control ΔAly extract, Aly was absent without reducing the levels of other TREX proteins. In contrast, in ΔTHO extract, which was prepared with the Thoc2 antibody, neither THO proteins nor Tex1 was detected (Figure 1D). These results demonstrate that Tex1 is co-depleted with THO. Together, these data indicate that Tex1 forms a salt-stable complex with THO in nuclear extract and raise the possibility that Tex1 might be a subunit of THO.
Figure 1.
Tex1 is a subunit of the THO sub-complex. (A) Western analysis of nuclear extract with antibody against Tex1. Molecular size markers (in kilodaltons) are shown. (B) IPs from RNased nuclear extracts using indicated antibodies were performed under normal salt (90 mM) or high salt (350 mM) conditions followed by western analyses using indicated antibodies. (C) Nuclear extracts were separated on a gel filtration column followed by western analyses of the fractions using indicated antibodies. Molecular weight standards were indicated in the fractions. (D) Western analyses of Mock, ΔAly or ΔTHO nuclear extracts using indicated antibodies. (E) Western analyses of whole-cell lysates of control knockdown and Thoc2 knockdown cells (left panel) or control knockdown and Tex1 knockdown cells (right panel) using indicated antibodies. (F) Schematic of composition of the TREX complex.
Previous studies reported that THO proteins require one another for their stabilities (7,8). To investigate whether Tex1 is a THO subunit, we examined whether Tex1 is co-knocked down with THO. As shown in Figure 1E, in Thoc2 knockdown cells, the amount of THO proteins including Thoc2, Thoc5, Thoc1 and Thoc7 markedly went down. Significantly, the level of Tex1 decreased as well, whereas Aly and CBP80 were unchanged. Consistent with these results, in Tex1 knockdown cells, the levels of Thoc2 and Thoc5 were also reduced (Figure 1E, right panel). These results indicate that Tex1 and THO proteins require one another for their stabilities. Taking these data together, we conclude that Tex1 is a subunit of THO (see model in Figure 1F).
Interactions between Aly, THO and UAP56 are interdependent
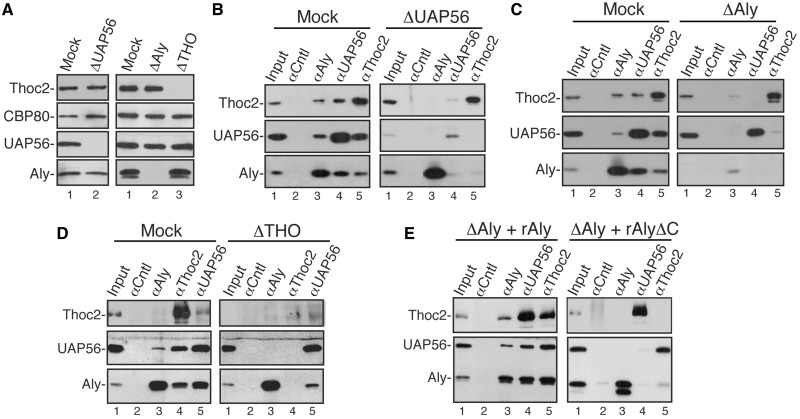
Previously, it was reported that UAP56 is required for TREX assembly, leading to the view that UAP56 is the core component of TREX (5). We sought to investigate whether Aly and THO play any role in TREX assembly. To do this, we used RNased ΔAly and ΔTHO extracts for IPs. Mock and ΔUAP56 extracts were used as controls. As shown in Figure 2A, Aly, UAP56 or THO was completely depleted from the corresponding nuclear extract without affecting the levels of others. As expected, in Mock extract, Aly, UAP56 and Thoc2 were co-IP’d with one another (Figure 2B–D, Mock), whereas in ΔUAP56 extract, no interaction was detected between Aly and Thoc2 (Figure 2B, ΔUAP56) (5). Significantly, in ΔAly extract, Thoc2 was not detected in the immunoprecipitate of the UAP56 antibody, and the amount of UAP56 co-IP’d with Thoc2 also decreased to background level (Figure 2C, ΔAly). Similarly, in ΔTHO extract, UAP56 was not present in the immunoprecipitate of the Aly antibody, and the level of Aly co-IP’d with UAP56 was dramatically reduced (Figure 2D, ΔTHO). These results indicate that like UAP56, Aly and THO are also required for TREX assembly.
Figure 2.
Interactions between Aly, THO and UAP56 are interdependent. (A) Western analyses of immunodepleted HeLa nuclear extracts with indicated antibodies. (B) RNased Mock (left panel) or ΔUAP56 nuclear extracts (right panel) were used for IPs with indicated antibodies followed by western analyses using Thoc2, UAP56 and Aly antibodies. (C and D) Same as (B), except that ΔAly and ΔTHO nuclear extract was used instead of ΔUAP56 nuclear extract, respectively. (E) Purified recombinant His-Aly or His-AlyΔC proteins were added to ΔAly extract and IPs were performed with indicated antibodies followed by western analyses.
The C-terminal region of Aly has been reported to be important for its association with UAP56 (11). To determine whether the C-terminus of Aly is required for TREX assembly, we added equal amount of recombinant His-Aly and His-AlyΔC to RNased ΔAly extract followed by IPs (Figure 2E and Supplementary Figure S1). Significantly, add-back of His-Aly, but not His-AlyΔC, completely restored Thoc2–UAP56 interaction. These results indicate that the C-terminus is critical for the role of Aly in TREX assembly.
Aly, but not UAP56, directly interacts with Thoc2 and Thoc5
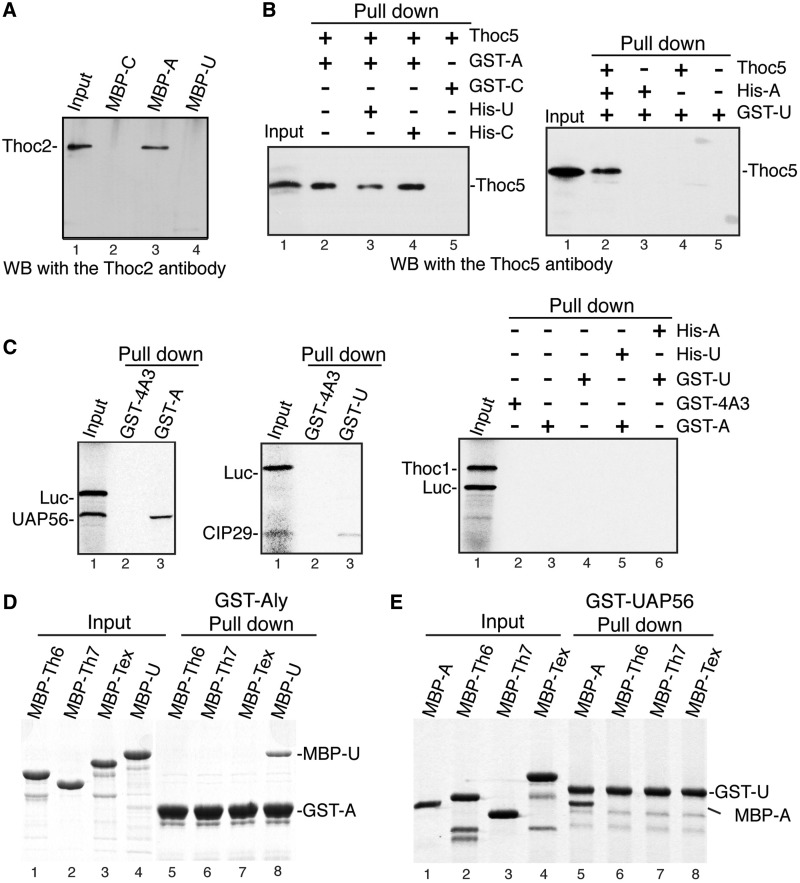
It has been reported that Aly can directly interact with UAP56 and the THO subunit Thoc5 (5,28). However, it remains unclear whether Aly directly interacts with other THO proteins and whether UAP56 makes direct contact with THO. To investigate these questions, we systematically examined whether Aly and UAP56 pull down each THO protein. As Thoc1, Thoc2 and Thoc5 were difficult to express using prokaryotic expression system in our hands, these proteins were expressed in insect cells. However, only Thoc2 and Thoc5 were soluble. When insect cell lysate containing overexpressed Thoc2 was used for pull down, MBP-Aly, but not equal amount of MBP-tagged UAP56 or the control protein, pulled down Thoc2 (Figure 3A). Insect cell-expressed Thoc5 was not pulled down by GST-UAP56, whereas it was pulled down by GST-Aly as previously reported (Figure 3B and Supplementary Figure S3) (28). The presence of UAP56, but not the control protein, in the pull down reaction reproducibly slightly inhibited the Aly–Thoc5 interaction (Figure 3B, left panel). One possible explanation of this inhibition is that UAP56 and Thoc5 share multiple binding sites on Aly. Consistent with this possibility, GST-UAP56 efficiently pulled down Thoc5 when Aly was present (Figure 3B, right panel). Together, these results indicate that Aly directly interacts with Thoc2 and Thoc5. It was also possible that these interactions we observed were results of chimeric THO complexes formed by human and insect proteins. To investigate this possibility, we used insect cell lysates containing overexpressed Tex1 (shown in below not to interact with Aly directly) as a control. Significantly, GST-Aly pulled down Thoc5, but not Tex1 (Supplementary Figure S3). We also used purified Tex1 and Thoc5 proteins for GST-Aly pull downs and obtained similar results (Supplementary Figure S3). Collectively, these data indicate that Aly, but not UAP56, interacts with Thoc2 and Thoc5 directly.
Figure 3.
Thoc2 and Thoc5 bind to Aly directly. (A) MBP pull downs were performed from insect lysate containing overexpressed Thoc2 using MBP-tagged Aly (MBP-A), UAP56 (MBP-U) and Mtr4N (MBP-C) followed by western analysis with the Thoc2 antibodies; 1% of the input was loaded. (B) Left panel, GST pull downs were performed from insect lysate containing overexpressed Thoc5 using GST-tagged Aly (GST-A) and eIF4A3 (GST-C). Purified His-UAP56 (His-U) or control protein (His-C) was added in pull down reactions where indicated. Western analysis was performed with the Thoc5 antibody. GST-eIF4A3 served as a negative control. Right panel, same as the left panel, but GST-UAP56 (GST-U) was used for pull downs; 50% of the input was loaded. (C) Left panel, GST pull downs were preformed from in vitro-translated 35S-labeled UAP56 and Luciferase (Luc) using GST-4A3 or GST-Aly. Proteins pulled down were visualized by autoradiography. Middle panel, same as left panel, except that GST-UAP56 was used for pulling down in vitro-translated 35S-labeled CIP29. Right panel, GST-tagged proteins as indicated were used for pulling down in vitro-translated 35S-labeled Thoc1 and Luc. His-tagged proteins were added in the pull down reaction where indicated; 20% of the inputs were loaded. (D) GST-Aly was used for pull down of purified MBP-tagged proteins followed by Coomassie staining; 6% of the inputs were loaded. MBP-Th6, MBP-Th7 and MBP-Tex indicate MBP-Thoc6, MBP-Thoc7 and MBP-Tex1, respectively. (E) Same as (D), except that GST-UAP56 was used for GST pull downs instead of GST-Aly; 12.5% of the inputs were loaded.
To test whether Aly and UAP56 interact with Thoc1, we performed GST pull downs using in vitro-translated Thoc1. In control experiments, GST-Aly and GST-UAP56 pulled down UAP56 and CIP29, respectively, but not Luciferase. In contrast, neither GST-Aly nor GST-UAP56 pulled down Thoc1, indicating that they do not interact with Thoc1 directly (Figure 3C). Finally, we examined whether GST-Aly and GST-UAP56 pull down purified MBP-tagged Tex1, Thoc6 and Thoc7. As shown in Figure 3D, although GST-Aly and GST-UAP56 efficiently pulled down MBP-UAP56 and MBP-Aly, respectively, neither of them pulled down Tex1, Thoc6 or Thoc7 (Figure 3D and E). These results indicate that neither UAP56 nor Aly directly associates with Tex1, Thoc6 or Thoc7. Taken together, our data indicate that Aly interacts with THO through Thoc2 and Thoc5, whereas UAP56 does not interact with THO directly.
Both Aly and THO are required for linking UAP56 to CBC
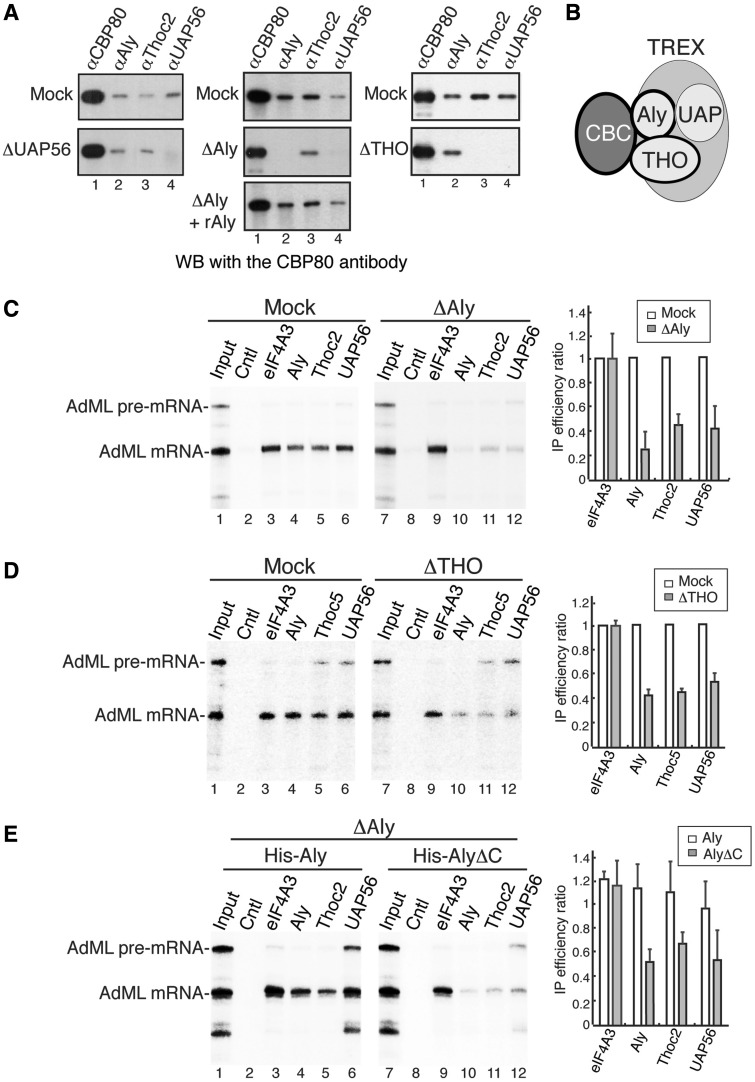
In a previous study, we have shown that recruitment of TREX to the spliced mRNA depends on the 5′-cap and this cap-dependent TREX recruitment occurs through TREX–CBC interaction (23). The finding that Aly, but not UAP56, directly interacts with CBC led to the view that Aly is the TREX component linking TREX to CBC (23). However, it remains to be investigated whether Aly is the sole TREX component mediating TREX–CBC interaction. To investigate this, we performed IPs with TREX antibodies from RNased ΔAly, ΔTHO, ΔUAP56 and Mock extracts, followed by western analysis with the CBP80 antibody. As expected, CBP80 was present in the immunoprecipitates of CBP80, UAP56, Aly and Thoc2 antibodies in Mock extract, and UAP56 depletion did not affect the amount of CBP80 IP’d by Aly and Thoc2 antibodies (Figure 4A, left panel) (23). In contrast, in ΔAly extract, CBP80 was not IP’d by the UAP56 antibody, and the amount of CBP80 IP’d by the Thoc2 antibody was reduced to ∼75% (Figure 4A, middle panel and Supplementary Figure S2). Add-back of recombinant Aly completely restored CBP80–UAP56 and CBP80–Thoc2 interactions (Figure 4A, middle panel). These results indicate that UAP56 requires Aly for interacting with CBP80, whereas THO can interact with CBP80 independent of Aly. In ΔTHO extract, CBP80 was not co-IP’d with UAP56, whereas the amount of that co-IP’d with Aly was unaffected (Figure 4A, right panel), indicating that THO is required for UAP56, but not for Aly, to interact with CBP80. These results are consistent with our data that Aly and THO depletions impaired the stable association of UAP56 with THO and Aly, respectively, as otherwise these associations would link UAP56 to CBC. Together, we conclude that both Aly and THO function in mediating TREX–CBC interaction (see model in Figure 4B).
Figure 4.
Aly and THO are required for UAP56–CBC interaction and stable association of TREX components with the spliced mRNA. (A) IPs were performed from RNased Mock, ΔUAP56, ΔAly and ΔTHO extracts as well as ΔAly extract supplemented with purified His-Aly using indicated antibodies. Western analyses were performed with the CBP80 antibody. (B) Schematic of the interaction between CBC and TREX. Aly and THO interact with CBP80 and mediate the interaction between TREX and CBC. (C) AdML pre-mRNA was incubated in Mock or ΔAly nuclear extract under splicing condition followed by mRNA IPs using indicated antibodies. The control antibody was an antibody against fSAP130; 25% of the input was loaded. Quantifications of three independent experiments are shown in the right panel. The bars indicate the ratio of IP efficiency in ΔAly extract relative to the corresponding IP efficiency in Mock extract. Error bars represent standard deviations (n = 3). (D) Same as (C), except that ΔTHO nuclear extract was used instead of ΔAly nuclear extract. (E) ΔAly nuclear extracts supplemented with purified recombinant His-Aly or His-AlyΔC proteins were used for mRNA IPs with indicated antibodies.
Aly and THO are required for association of TREX components with the spliced mRNA
Our data indicating that both Aly and THO are required for UAP56–CBC interaction raised the possibility that Aly and THO might be required for UAP56 associating with the mRNA. To test this possibility, we examined the effects of Aly and THO immunodepletions on association of UAP56 with the mRNA. First, we spliced AdML pre-mRNA in Mock and ΔAly extracts and used the spliced mRNA for IPs with antibodies to TREX components. eIF4A3 antibody was used as a control. In Mock extract, AdML spliced mRNA was efficiently IP’d by antibodies to eIF4A3, Aly, Thoc2 and UAP56 (Figure 4C, lanes 1–6). In striking contrast, in ΔAly and ΔTHO extracts, although AdML mRNA was efficiently IP’d by the eIF4A3 antibody, the amount of that associated with UAP56 significantly decreased (Figure 4C and D, lanes 6 and 12). Interestingly, AdML mRNA was barely associated with Thoc2 in ΔAly extract or with Aly in ΔTHO extract (Figure 4C, lanes 5 and 11; Figure 4D, lanes 4 and 10). These results indicate that both Aly and THO play key roles in stable binding of TREX components with AdML spliced mRNA. To investigate whether these roles of Aly and THO are general, we used Ftz pre-mRNA for the same experiment and obtained similar results (Supplementary Figure S4). We also sought to determine whether UAP56 is involved in association of TREX components with the spliced mRNA. However, to deplete UAP56, we had to carry out many rounds of depletion that destroyed the splicing activity of both Mock and ΔUAP56 extracts (Wang,Q. and Cheng,H., unpublished data). Thus, we were not able to test this role of UAP56. Together, we conclude that both Aly and THO are required for stable association of TREX components with the spliced mRNA.
We next examined whether the C-terminus of Aly is required for stable binding of TREX components with the spliced mRNA. As shown in Figure 4E, His-Aly, but not equal amount of His-AlyΔC, restored associations of Aly, THO and UAP56 with AdML mRNA. These results indicate that the C-terminus of Aly is important for its role in association of TREX components with the spliced mRNA.
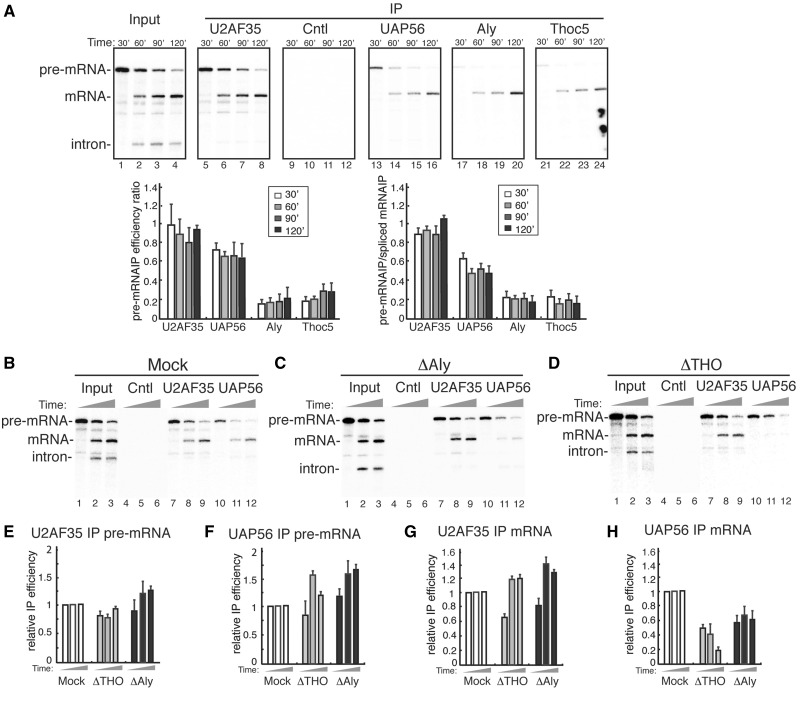
UAP56 associates with the pre-mRNA independent of Aly and THO
UAP56 has been implicated in multi-steps of splicing (12,25). Consistent with its role in splicing, UAP56 was shown to be present in mRNPs formed at early stages of splicing (11). However, it is not clear whether UAP56 associates with pre-mRNAs and whether this association requires Aly and THO (2). To investigate this, we performed mRNA IPs along a splicing time course. In vitro-transcribed Ftz pre-mRNA was spliced for 30, 60, 90 and 120 min, followed by IPs with antibodies to UAP56, Aly and Thoc5. As controls, antibodies to U2AF35 and an irrelevant protein were also used for IPs. Ftz spliced mRNA started to accumulate at 60 min and at 120 min most pre-mRNA has been spliced (Figure 5A). The U2AF35 antibody IP’d the pre-mRNA at 30 min, and IP’d both pre-mRNA and spliced mRNA at later time points with similar efficiencies (Figure 5A, lanes 5–8). Consistent with previous reports (2), antibodies to Aly and Thoc5 selectively IP’d the spliced mRNA (Figure 5A, lanes 17–24). In contrast, UAP56 associated with the pre-mRNA at 30 min (Figure 5A, lane 13). At subsequent time points, the UAP56 antibody IP’d both the pre-mRNA and the spliced mRNA (Figure 5A, lanes 14–16), and the IP efficiency for the spliced mRNA is higher than that for the pre-mRNA (Figure 5A, lower panel). Together, these results indicate that UAP56 is recruited to both pre-mRNAs and spliced mRNAs, whereas Aly and THO are only recruited to spliced mRNA.
Figure 5.
UAP56 associates with the pre-mRNA independent of Aly or THO. (A) Ftz pre-mRNA was incubated in HeLa nuclear extract under splicing condition for 30, 60, 90 or 120 min, followed by mRNA IPs using indicated antibodies; 25% of the input was loaded. Quantification of three independent experiments is shown below. In the left graph, bars indicate the relative pre-mRNA IP efficiency calculated by dividing the IP efficiency of pre-mRNA at each time point by that of spliced mRNA at 120 min. In the right graph, bars indicate the ratio of IP efficiency for the pre-mRNA to that for the spliced mRNA. Error bars represent standard deviations (n = 3). (B–D) Ftz pre-mRNA was incubated under splicing condition in mock (B), ΔAly (C) or ΔTHO (D) nuclear extract, respectively, for 30, 90 or 120 min followed by IPs using indicated antibodies; 25% of the input was loaded. (E) Bars indicate relative pre-mRNA IP efficiencies of U2AF35 at each time point that are calculated by dividing pre-mRNA IP efficiencies in ΔAly and ΔTHO extracts by those in Mock extract. Error bars represent standard deviations (n = 3). (F–H) Similar to (E), except that pre-mRNA IP efficiencies of the UAP56 antibody (F), spliced mRNA IP efficiencies of the U2AF35 antibody (G) and spliced mRNA IP efficiencies of the UAP56 antibody are indicated.
To investigate whether association of UAP56 with the pre-mRNA requires Aly and THO, we used Mock, ΔAly and ΔTHO extracts for UAP56 IPs along a splicing time course. Same as the results shown in Figure 4, Aly and THO depletion disrupted the association of UAP56 with spliced mRNA (Figure 5B–D and H). In contrast, the amount of pre-mRNA bound to UAP56 did not decrease, but even increased, in the absence of either Aly or THO (Figure 5B–D and F). This increase is consistent with our observation that UAP56 associates with spliced mRNA more efficiently than with pre-mRNA. As the control, in ΔAly and ΔTHO extracts, association of U2AF35 with the pre-mRNA was not affected, and the amount of spliced mRNA bound to U2AF35 increased for some unknown reason (Figure 5B–E and G). Together, our data indicate that UAP56 is recruited to the pre-mRNA independent of Aly and THO.
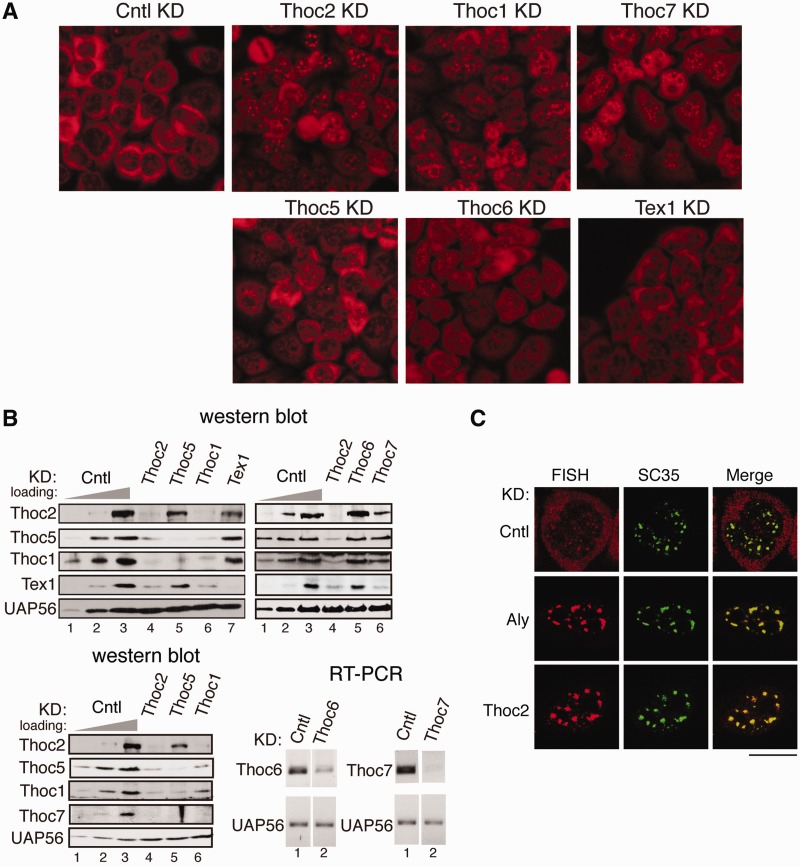
Efficient mRNA export requires THO
The yeast THO has been reported to be required for bulk mRNA export, whereas in Drosophila THO is not required for the export of total mRNA (3,7). In humans, studies on different THO subunits have reached different conclusions on the role of THO in mRNA export (28–30). Our finding that THO is required for stable binding of Aly and UAP56 with spliced mRNAs raises the possibility that THO plays key roles in general mRNA export pathway. To investigate this possibility, we knocked down all THO subunits and examined the effects on the nucleocytoplasmic distribution of polyA+ RNA. Significantly, Thoc2, Thoc1 and Thoc7 knockdown led to severe polyA+ RNA export block, whereas Thoc5, Thoc6 and Tex1 knockdown only had mild effects (Figure 6A and Supplementary Figure S5). We next investigated whether these various effects resulted from different knockdown and co-knockdown efficiencies of THO subunits. As shown in Figure 6B, all THO subunits were efficiently knocked down by the corresponding shRNA. Significantly, Thoc2, Thoc1 and Thoc7 knockdowns, which resulted in severe export block, all led to significant decrease in levels of Thoc2, Thoc7 and Tex1, suggesting that one of these THO proteins might be the determinant factor for mRNA export function of THO. The fact that Tex1 knockdown barely affects mRNA export excludes the Tex1 possibility. Similarly, the possibility for Thoc7 was ruled out by the data showing that although Thoc5 knockdown led to apparent co-knockdown of Thoc7, it only had mild effect on mRNA export (Figure 6A). Thus, these results suggest Thoc2 might be the determinant factor for THO functioning in mRNA export. Together our data indicate that THO as a complex is required for the export of polyA+ RNA and Thoc2 might be the determinant factor.
Figure 6.
THO is required for efficient mRNA export. (A) FISH for polyA+ RNA were performed for control or THO subunits knockdown cells. DAPI staining indicated nuclei. Scale bar, 10 µm. (B) Western analyses of whole-cell lysates of control and THO subunits knockdown cells with indicated antibodies. UAP56 served as a loading control. To determine the knockdown efficiencies for Thoc6 and Thoc7, total RNA was extracted from control, Thoc6 and Thoc7 knockdown cells followed by RT–PCR analysis. UAP56 served as a loading control. (C) Confocal microscope images of FISH for polyA+ RNA, SC35 IF and merged images are shown. Scale bar, 10 µm.
We next examined whether THO is required for the export of reporter spliced mRNA. To do this, we microinjected the CMVSmad DNA construct into the nuclei of control or Thoc2 knockdown cells, followed by FISH to detect Smad mRNA at 0.5, 1.5 and 2.5 h after microinjection. This export time course revealed that compared with control knockdown, in Thoc2 knockdown cells, Smad mRNA had a slow export kinetic, indicating that THO is required for efficient export of spliced mRNA (Supplementary Figure S5).
In a recent study, both UAP56 and Aly have been implicated in mRNA release from the nuclear speckle domains (31). We next determined whether THO is involved in this process. Aly knockdown was used as a positive control. In the negative control knockdown, the polyA+ RNA was mostly accumulated in the cytoplasm (Figure 6C). In Aly knockdown cells, nuclear-retained polyA+ RNA was co-localized with SC35, which is a standard speckle marker (Figure 6C) (32). Significantly, in Thoc2 knockdown cells, the nuclear-retained polyA+ RNA was also exclusively co-localized with SC35 (Figure 6C), indicating that THO also plays a key role in releasing mRNA from nuclear speckle domains. Collectively, we conclude that THO plays critical roles in mRNA export.
DISCUSSION
TREX is a highly conserved protein complex that was thought to contain Aly, UAP56, THO and Tex1. In this study, we found that Tex1 is a subunit of the human THO. The three key TREX components Aly, UAP56 and THO interact with one another in an interdependent manner. Both Aly and THO function in mediating CBC–TREX interaction and are required for stable binding of TREX components with the spliced mRNA. Consistent with these roles of THO, it is required for efficient export of both polyA+ RNA and spliced reporter mRNA.
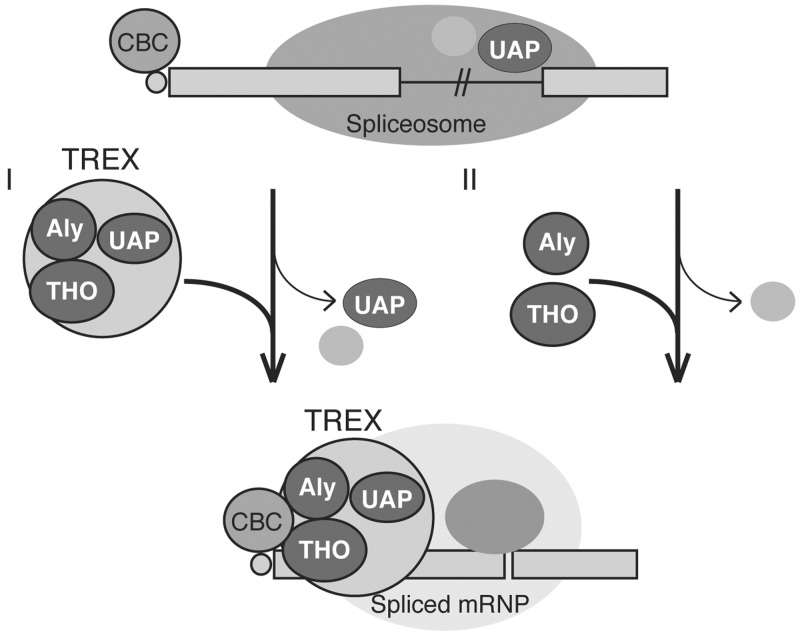
Models for TREX recruitment
Two possible models for recruitment of the human TREX are shown in Figure 7. Pre-mRNA is capped at the 5′-end and associates with the nuclear CBC, which is not available for binding with Aly and THO on pre-mRNAs. During an early stage of splicing, UAP56 is recruited to pre-mRNAs and functions in splicing. During a late step of splicing, some changes occur to CBC and it now can interact with Aly and THO. In one model, upon completion of splicing, UAP56 might be released from the mRNA and subsequently Aly, THO and UAP56 are recruited to the spliced mRNA as a pre-formed particle. In an alternative model, UAP56 associating with the pre-mRNA might be transferred to the spliced mRNA and interacts with Aly and THO recruited by CBC. Aly, THO and UAP56 stabilize one another on the spliced mRNA.
Figure 7.
Models for TREX recruitment. In model (I), UAP56 recruited to the pre-mRNA is released upon completion of splicing. Subsequently, Aly, UAP56 and THO are recruited to the spliced mRNA as a preformed particle. In model (II), UAP56 recruited to the pre-mRNA is transferred to the spliced mRNA, where it forms TREX together with Aly and THO recruited by CBC. See ‘Discussion’ section for details.
In a previous study, all eight TREX components were found to be present in the same fraction in gel filtration of TREX pulled down from RNased nuclear extract, indicating that TREX exists as a preformed particle in the nuclear extract (2). Our data indicating that Aly, UAP56 and THO associate with one another in an interdependent manner further support that TREX is a highly integrated particle in the nuclear extract. Based on these data, it is puzzling how UAP56 associates with the pre-mRNA independent of other TREX components. One possibility is that UAP56 associated with the pre-mRNA is distinct from that binding with the spliced mRNA. Our data showing that even in low salt condition (60 mM), only a small portion of UAP56 co-migrates with THO indicates that in the nuclear extract, most UAP56 exists free from TREX (Supplementary Figure S6). It is possible that this TREX-free UAP56 is recruited to the pre-mRNA and functions in splicing. These are consistent with the former model. However, other possibilities remain. For example, it is known that UAP56 associates with many splicing factors including U2AF65 (5,25). It is possible that on the pre-mRNA, these splicing factors interact with UAP56 and exclude Aly for binding. Upon completion of splicing, these factors are released and UAP56 is transferred to the spliced mRNA where its binding site for Aly is exposed. Thus, recruitment of UAP56 to the pre-mRNA is a prerequisite for its association with the spliced mRNA. This possibility is consistent with the latter model that explains the splicing-dependent TREX recruitment. Nevertheless, in both models, Aly, UAP56 and THO associate with the spliced mRNA as a highly integrated particle and play critical roles in mRNA export as a functional unit.
Formation of the TREX complex
In a recent study, UAP56 has been proposed to be the core component of TREX (5). Here, we found that Aly is required for the interaction between UAP56 and THO, and conversely THO is also needed for stable Aly–UAP56 interaction. These results together indicate that interactions between Aly, UAP56 and THO are interdependent. Our in vitro pull down data show that Aly, but not UAP56, directly interacts with Thoc2 and Thoc5. This bypassed requirement of UAP56 for Aly–Thoc2/Thoc5 interaction in vitro might result from mixing excess amount of recombinant proteins together. In agreement with this possibility, although stable Aly–UAP56 interaction in the nuclear extract requires THO, when excess amount of Aly and UAP56 are mixed together, they strongly interact with each other independent of THO (Figures 2 and 3) (11). Based on our data and previous reports, Aly seems to exist in the center of TREX interacting directly with both UAP56 and THO. The mechanism underlying TREX assembly remains to be determined. One possibility is that some conformational changes might occur to each component when they interact with one another. UAP56 is an ATP-dependent helicase, and a recent study reported that Aly interacts with UAP56 in an ATP-dependent manner (5,12). It is possible that Aly and THO are both required for stimulating the helicase activity of UAP56, which in turn induces conformational changes of Aly and THO, resulting in assembly of TREX. Structural insights of Aly, THO and UAP56 individually and collectively would be required to understand the mechanism for TREX assembly. It would also be interesting to investigate how TREX components interact with one another in yeast, Drosophila and other organisms.
A role for THO in mRNA export
Up to date, the role of mammalian THO in mRNA export has been unclear, and different conclusions have been reached (28–30,33). We found that knockdown of Thoc2, Thoc1 and Thoc7 had dramatic effects on mRNA export, whereas Thoc5, Thoc6 and Tex1 only had minor effects. Our data demonstrate that the extent of mRNA export blockage positively correlates with the knockdown/co-knockdown efficiency of Thoc2. Thus, it seems that Thoc2 is the determinant factor for the role of THO in mRNA export. In a previous study, the unaffected bulk mRNA export by Thoc5 knockdown was also reported (28). In the same report, it was shown that the level of Thoc1 was not affected in Thoc5 knockdown cells. This negative result might result from the relatively low knockdown efficiency of Thoc5. Consistent with this possibility, our result as well as a more recent study indicate that Thoc1 requires Thoc5 for its stability (8). Our data indicating that Thoc2 is the determinant THO protein for its role in mRNA export are consistent with our finding that Thoc2 directly interacts with Aly. It is possible that although both Thoc2 and Thoc5 directly interact with Aly, Thoc2 might play a dominant role in mediating Aly–THO interaction. The yeast Thoc2 was recently shown to be the THO protein making direct contact with the mRNA (9). It is possible that this property of THO is conserved in humans and thus Thoc2 is critical for mRNA export. Thoc5 has been reported to be an alternative mRNA export adaptor and in a most recent study, Thoc5 together with Aly was shown to induce the conformational change of TAP required for mRNA export (34). The direct interaction between Aly and Thoc5 might provide the binding scaffold for TAP.
Similar to Aly and UAP56 knockdown, in THO knockdown, the nuclear-retained polyA+ RNA was also accumulated in nuclear speckle domains, indicating that THO is also involved in release of mRNA from these domains. Our biochemical results demonstrating that THO is required for stable association of Aly and UAP56 with the spliced mRNA indicate that THO functions in mRNA export by ensuring efficient association of Aly and UAP56 with the spliced mRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–6, Supplementary Methods and Supplementary Reference [35].
FUNDING
Ministry of Science and Technology 973 project [2011CB811304 and 2013CB910402]; National Natural Science Foundation of China [30970583]; Shanghai Pujiang Program [09PJ1411300] and Chinese Academy of Sciences [2008OHTP03]. Funding for open access charge: Ministry of Science and Technology 973 project [2011CB811304]; National Natural Science Foundation of China [30970583].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Haixin Lei and Cheng Lab members for useful discussion.
REFERENCES
- 1.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki T, Fujiwara N, Yukinaga H, Ebisuya M, Shiki T, Kurihara T, Kioka N, Kambe T, Nagao M, Nishida E, et al. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol. Biol. Cell. 2010;21:2953–2965. doi: 10.1091/mbc.E09-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010;24:2043–2053. doi: 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- 8.Mancini A, Niemann-Seyde SC, Pankow R, El Bounkari O, Klebba-Farber S, Koch A, Jaworska E, Spooncer E, Gruber AD, Whetton AD, et al. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol. 2010;8:1. doi: 10.1186/1741-7007-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pena A, Gewartowski K, Mroczek S, Cuellar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 31:1605–1616. doi: 10.1038/emboj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 11.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 12.Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008;22:1796–1803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol. Cell Biol. 2008;28:601–608. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Custodio N, Carvalho C, Condado I, Antoniou M, Blencowe BJ, Carmo-Fonseca M. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 2004;10:622–633. doi: 10.1261/rna.5258504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 23.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 24.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 25.Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 26.Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl Acad. Sci. USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 28.Katahira J, Inoue H, Hurt E, Yoneda Y. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 2009;28:556–567. doi: 10.1038/emboj.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominguez-Sanchez MS, Saez C, Japon MA, Aguilera A, Luna R. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer. 11:77. doi: 10.1186/1471-2407-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo S, Hakimi MA, Baillat D, Chen X, Farber MJ, Klein-Szanto AJ, Cooch NS, Godwin AK, Shiekhattar R. Linking transcriptional elongation and messenger RNA export to metastatic breast cancers. Cancer Res. 2005;65:3011–3016. doi: 10.1158/0008-5472.CAN-04-3624. [DOI] [PubMed] [Google Scholar]
- 31.Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010;1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 33.Guria A, Tran DD, Ramachandran S, Koch A, El Bounkari O, Dutta P, Hauser H, Tamura T. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA. 2011;17:1048–1056. doi: 10.1261/rna.2607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.