Abstract
Repairing damaged DNA is essential for an organism’s survival. UV damage endonuclease (UVDE) is a DNA-repair enzyme that can recognize and incise different types of damaged DNA. We present the structure of Sulfolobus acidocaldarius UVDE on its own and in a pre-catalytic complex with UV-damaged DNA containing a 6-4 photoproduct showing a novel ‘dual dinucleotide flip’ mechanism for recognition of damaged dipyrimidines: the two purines opposite to the damaged pyrimidine bases are flipped into a dipurine-specific pocket, while the damaged bases are also flipped into another cleft.
INTRODUCTION
For DNA-repair enzymes, it is extremely important to discriminate damaged bases from undamaged DNA. Most DNA-repair enzymes, such as O6-meG-DNA methyl transferase, (6-4)-photolyase and uracil-DNA glycosylase (1–3), recognize damage by flipping the damaged base into a selective pocket while the corresponding undamaged base either remains stacked in the DNA helix or is flipped into the solvent. In contrast, damage recognition by T4 endonuclease V is mainly via flipping and specific binding to one of the adenines opposite to the photodimer and leaving the actual damage in the helix (4). Non-sequence-specific recognition of the undamaged bases has also been reported for Rad4 (5) where the flexibility of the DNA is the main contributor to damage recognition while the lesion is flipped into the solvent.
The UV damage endonuclease (UVDE) is a DNA endonuclease that not only recognizes and incises DNA 5′ to cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs) (6), but also non–UV-induced DNA damage such as abasic sites, nicks and gaps (7). The activity on abasic sites, nicks and gaps depends on the presence of neighbouring pyrimidines, suggesting that UVDE’s active site is most optimal for binding distorted pyrimidines (8). The crystal structure of Thermus thermophilus UVDE (9) showed that UVDE has a triosephosphateisomerase (TIM) barrel fold with a large groove with positive charges on the edges proposed to bind DNA and the active site on the bottom of this groove containing three metal ions. Two conserved residues (Gln104 and Tyr105 in TthUVDE) named the ‘probing finger’ were proposed to aid in flipping out the damaged bases from the DNA helix.
To understand how UVDE can recognize damaged DNA, we determined the structure of UVDE from S. acidocaldarius (SacUVDE) on its own and in a pre-catalytic complex with DNA containing a 6-4 photoproduct.
MATERIALS AND METHODS
Cloning
Genomic DNA was isolated from S. acidocaldarius by resuspension of cells in TEN-buffer (20 mM Tris pH 8, 1 mM EDTA, 100 mM NaCl), followed by lysis in TENST-buffer (20 mM Tris pH 8, 1 mM EDTA, 100 mM NaCl, 1.6% sarcosyl, 0.12% Triton) and phenol/ chloroform extraction. The gene for UVDE was amplified using the primers 5′ ATTAATAACATATGAGAGTAGGTTACGTATCCAC 3′ and 5′ TAGGATCCATTAATCCAGTTTGTTTAACTCCTTTAAC 3′. Subsequently, it was cloned into the pETUVDEΔ228 vector (8) using NdeI and BamHI, resulting in the gene for SacUVDE with a N-terminal 10x His-tag and factor Xa cleavage site. Mutants of SacUVDE were created by PCR and cloned into the same vector with NdeI and BamHI. All constructs were verified by sequencing.
Expression and purification
The plasmid with the gene for SacUVDE was transformed to E.coli BL21(DE3)-codon+ and overexpressed for 2 h at 37°C after induction by 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). After harvesting, the pellet was resuspended in Ni buffer A (20 mM Tris pH 7.5, 500 mM NaCl, 50 mM imidazole, 8 mM β-mercaptoethanol and 10% glycerol), and the cells were lysed by sonication. The lysate was spun down at 37 000rpm (100 000g) for 30 min, and the soluble fraction was loaded on a His-trap column (GE healthcare) equilibrated with Ni buffer A. The column was washed with 20 column volumes Ni buffer A, and the protein was then eluted with a 60 column volumes gradient to Ni buffer B (20 mM Tris pH 7.5, 500 mM NaCl, 500 mM imidazole, 8 mM β-mercaptoethanol and 10% glycerol). Fractions containing SacUVDE were dialyzed to 20 mM Tris pH 8 and were then loaded on a HiTrap Q column (GE healthcare) equilibrated with Q buffer A (20 mM Tris pH 8 and 10% glycerol). The column was washed with 10 column volumes Q buffer A and eluted with a 60 column volumes gradient to Q buffer B (20 mM Tris pH 8, 1 M NaCl and 10% glycerol).
For crystallization, the purification protocol was adapted: size exclusion (Superdex 200, GE healthcare) was performed in GF buffer (20 mM HEPES 7.2, 200 mM NaCl, 5 mM DTT) after the Ni purification instead of ion exchange. The protein from either protocol was found to be >95% pure as judged from SDS PAGE. All purification steps were performed at 4°C.
DNA substrates
The sequence of the DNA substrates used are listed in Table 1. The oligos containing CPD or 6-4PP were synthesized as described previously (10). The top strands of the DNA substrates were 5′ radioactively labelled using polynucleotide kinase as reported previously (11).
Table 1.
30-mer DNA substrates used in this study
| No damage | 5′ CTCGTCAGCATCTTCATCATACAGTCAGTG 3′ 3′ GAGCAGTCGTAGAAGTAGTATGTCAGTCAC 5′ |
| CPD and 6-4PP | 5′ CTCGTCAGCATCTTCATCATACAGTCAGTG 3′ 3′ GAGCAGTCGTAGAAGTAGTATGTCAGTCAC 5′ |
| Abasic site | 5′ CTCGTCAGCATCXTCATCATACAGTCAGTG 3′ 3′ GAGCAGTCGTAGAAGTAGTATGTCAGTCAC 5′ |
The positions of the CPD (TT), (6-4)PP (TT) and AP site (X) are indicated in bold.
Incision assays
Labelled DNA substrates (1 nM) were incubated for 15 min at 55°C with the indicated amount of UVDE (in the range 0.05–50 nM) in 20 mM HEPES pH 6.5, 100 mM NaCl and 1 mM MnCl2 in a reaction mix of 20 µl. The reaction was then stopped by adding 3 µl stop mix (0.33 M EDTA, 3.3% SDS), after which 2.4 µl 4 mg/ml glycogen was added and the DNA was precipitated by ethanol. Samples were loaded on a 15% denaturing polyacrylamide gel and visualized by autoradiography. For kinetics incision assays, a mix was prepared of buffer, cofactor, protein (25 nM) and DNA and put at 55°C. At the indicated time points, samples were taken out and the reaction was stopped in these samples.
Crystallization
SacUVDE was concentrated to 3–5 mg/ml with a 3 kDa molecular weight cut off (MWCO) centrifugal filter unit (Millipore). Crystallization trials were performed using the sitting-drop vapour diffusion method and the JCSG+ and PACT (Qiagen) screens. SacUVDE crystals were obtained in 20% PEG3350 with 0.2 M NH4Cl or 0.2 M NaI. The conditions were optimized by a systematic screen around these conditions, and the largest crystals were grown in 14–28% PEG3350 with 0.15–0.3 M NH4Cl.
The damaged strand of the oligo containing 6-4PP for crystallization was synthesized as previously described (10), whereas the undamaged strand was purchased from Eurogentec, Belgium. The sequence of the oligo containing the 6-4PP was 5′ GCGTCCTTGACGACG 3′, with the site of the damage printed in bold, and its complementary strand was 5′ CGTCGTCAAGGACGC 3′. The two strands were hybridized by heating to 80°C for 2 min in 20 mM Tris pH 7 and then allowed to slowly cool down to room temperature. For co-crystallization, protein (at 0.11 mM) and DNA (at 0.21 mM) were incubated on ice for 15 min, after which sitting-drop vapour diffusion experiments were set up in the NucPro screen (Jena Biosciences). Damaged DNA:protein complex crystals appeared after several days in 30% PEG2000-MME, 100 mM acetate buffer pH 4.6, and 200 mM (NH4)2SO4.
Data collection
Crystals were caught with SPINE sample loops and put in cryoprotectant solution (precipitant solution with 10–15% glycerol) and flash-frozen. Data were collected at the European Synchrotron Radiation Facility (Grenoble, France). A total of 180 images were collected with an oscillation angle of 1.0°, with transmission of 13% and exposure time of 0.5 s per frame at 0.9393 Å at 100 K on beamline ID14-4 for the apoprotein crystals. For the DNA:protein complex crystals, 150 images were collected with an 1.0° oscillation angle and an exposure time of 25 s per frame at 0.934 Å at 100 K on beamline ID14-1. The images were processed with iMosflm (12). Scaling and merging were done with SCALA (13) from the CCP4 suite (14). For the apoprotein structure, two data sets (from two different crystals) were merged to yield the final data set used for determining the structure. Data collection statistics are shown in Table 2.
Table 2.
Data collection and refinement statistics
| SacUVDE | SacUVDE with 6-4PP | |
|---|---|---|
| Space group | P1 | C222 |
| Cell dimensions | ||
| a, b, c (Å) | 42.08 × 53.59 × 77.39 | 57.20 × 112.51 × 153.85 |
| α, β, γ (°) | 102.09, 93.02, 111.76 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 46.05–1.50 (1.58–1.50) | 52.83–2.70 (2.85–2.70) |
| Wilson plot B-factor | 17.5 | 64.7 |
| Rpima | 0.061 (0.436) | 0.055 (0.396) |
| <I/σI> | 9.3 (1.9) | 11.1 (1.9) |
| Completeness (%) | 97.3 (94.0) | 99.9 (99.2) |
| Redundancy | 3.4 (2.4) | 5.9 (5.2) |
| Total number of observations | 317 749 | 82 168 |
| Number of unique reflections | 94 666 | 14 041 |
| Refinement | ||
| Resolution (Å) | 46.05–1.50 | 52.89–2.70 |
| No reflections | 89 933 | 13 316 |
| Molecules in ASU | 2 | 1 |
| Rwork/Rfree | 0.177/0.214 | 0.191/0.262 |
| Number of atoms | ||
| Protein | 4706 | 2353 |
| Metal ions | 2 | 0 |
| Water | 579 | 4 |
| DNA | 0 | 609 |
| Other ions | 0 | 10 |
| B-factors | ||
| Protein | 20.37 | 55.28 |
| Metal ions | 50.78 | NA |
| Water | 31.93 | 34.14 |
| DNA | NA | 67.83 |
| Other ions | NA | 60.77 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.026 | 0.011 |
| Bond angles (°) | 2.21 | 1.57 |
| N° TLS bodies | 2 | NA |
| Ramachandran favouredb | 96.87% | 92.8% |
| Ramachandran outliers | 0.00% | 0.69% |
| Rotamer outliers | 1.48% | 2.95% |
Structure solution and refinement
The phase problem for the apoprotein was solved by molecular replacement using the structure of TthUVDE (PDB entry 2j6v) as a search model. The model was automatically rebuilt using Arp/Warp (15) and refined with Refmac (16). Manual fitting was performed using Coot (17). For the DNA:protein complex, the phase problem was solved by molecular replacement using the SacUVDE structure as a search model. Clear difference density was visible for the DNA. The DNA was manually built in with Coot. The model was refined with Refmac and further manual fitting was also performed using Coot. The final R-factor and Rfree for uncomplexed SacUVDE were 0.177 and 0.214, respectively, and 0.191 and 0.262 for SacUVDE in complex with DNA. Refinement statistics are shown in Table 2.
Superpositions were done with the ssm function in Coot. Root-mean-square deviation calculations were done using Theseus (18). Structure-based sequence alignment was performed using the program VAST (19). Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (accession code 3tc3 for the apoprotein and 4gle for the co-crystal structure). All figures were made with CCP4mg (20).
RESULTS AND DISCUSSION
Overall structure of SacUVDE with and without damaged DNA
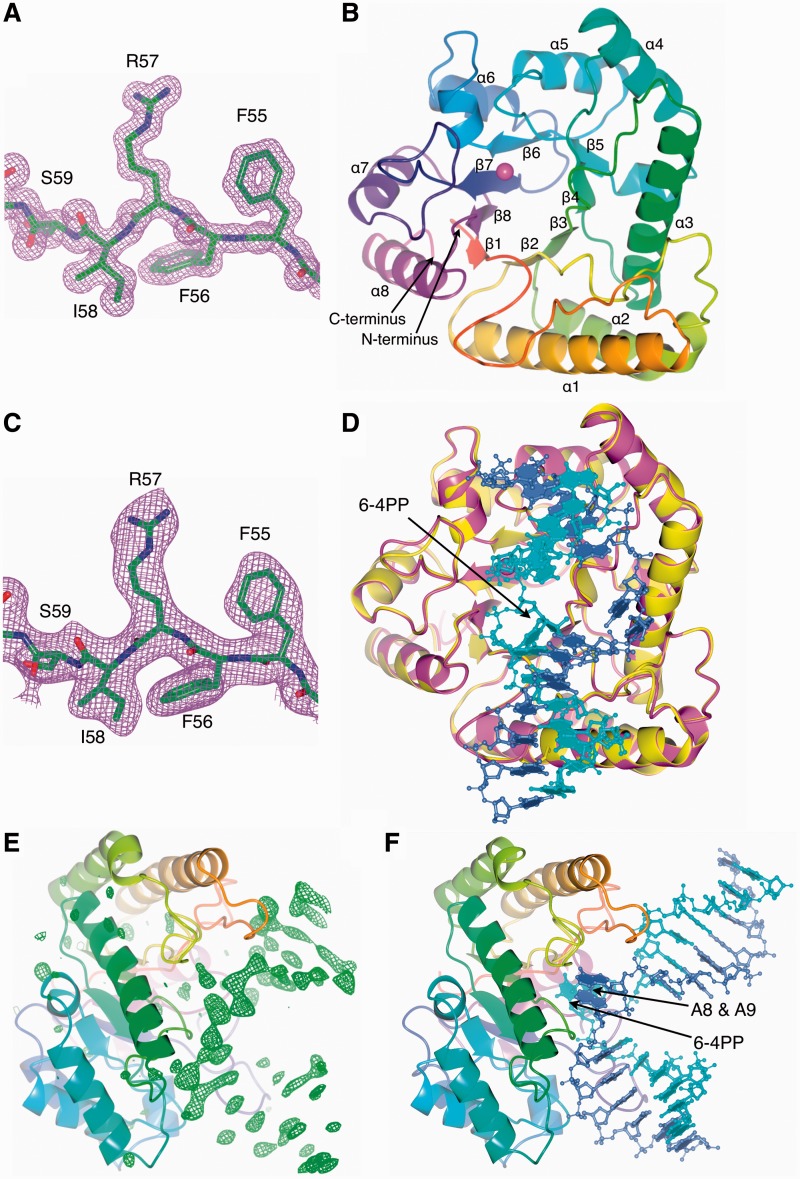
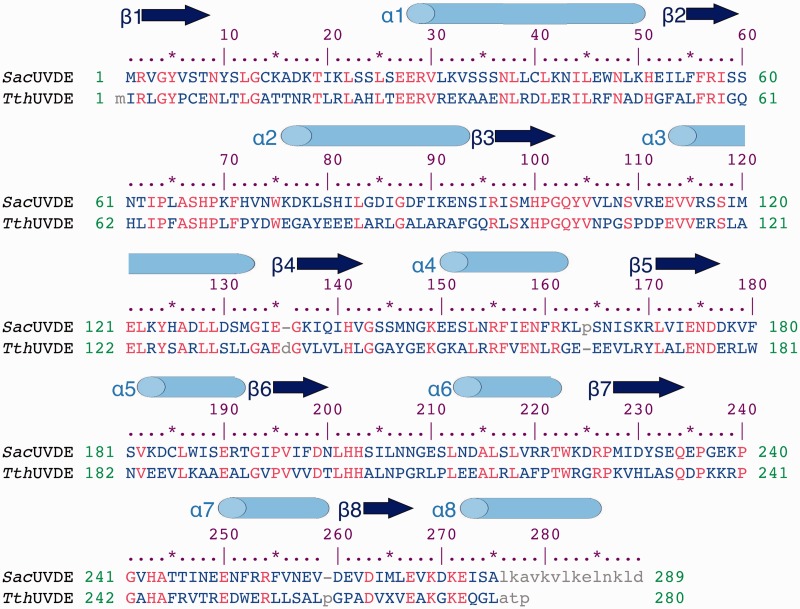
We determined the structure of UVDE from S. acidocaldarius (SacUVDE) on its own to 1.5 Å resolution and in a pre-catalytic complex with DNA containing a 6-4 photoproduct to 2.7 Å (data collection and refinement statistics are shown in Table 2; maps can be seen in Figure 1A and C). SacUVDE has a TIM-barrel fold (Figure 1B), and its backbone structure is similar to the previously determined TthUVDE structure: the root mean square deviation of the Cα atoms is only 0.651 Å (for a sequence alignment, see Figure 2). In contrast to TthUVDE, SacUVDE, like UVDE from most species, has one extra α-helix at its C-terminus and thus has a complete α8β8 TIM-barrel fold. SacUVDE has a disulfide bridge, between cysteines 14 and 40, which is rare amongst UVDEs and probably provides more stability to this protein from a highly thermophilic organism. Hardly any changes are seen in SacUVDE on binding the 6-4 photoproduct UV damaged DNA: the root mean square deviation for the protein in the uncomplexed and the complexed structure of the Cα atoms is only 0.11 Å (Figure 1D). An OMIT map for the DNA is shown in Figure 1E.
Figure 1.
Overall structure of SacUVDE with and without DNA. (A) Representative part of the 2Fo–Fc electron density map of SacUVDE (contoured at 1σ). (B) Overall fold of SacUVDE, which has a TIM-barrel fold. The metal ion is shown in magenta. (C) Representative part of the 2Fo–Fc electron density map of SacUVDE in complex with DNA (contoured at 1σ). (D) Superposition of SacUVDE with (magenta) and without (yellow) DNA (blue and cyan), showing that the two structures are very similar. (E) Omit map of the SacUVDE cocrystal with DNA showing clear positive difference density for the DNA (contoured at 3σ). (F) Overall fold of the SacUVDE-DNA complex, showing the 90° bend in the DNA.
Figure 2.
Structure-based sequence alignment of SacUVDE and TthUVDE. The sequence identity between the two proteins is 38%. Secondary structure elements are indicated with cylinders and arrows for α-helices and β-strands respectively. Identical residues are in red, variable residues in blue and unaligned residues in grey.
Protein–DNA interactions
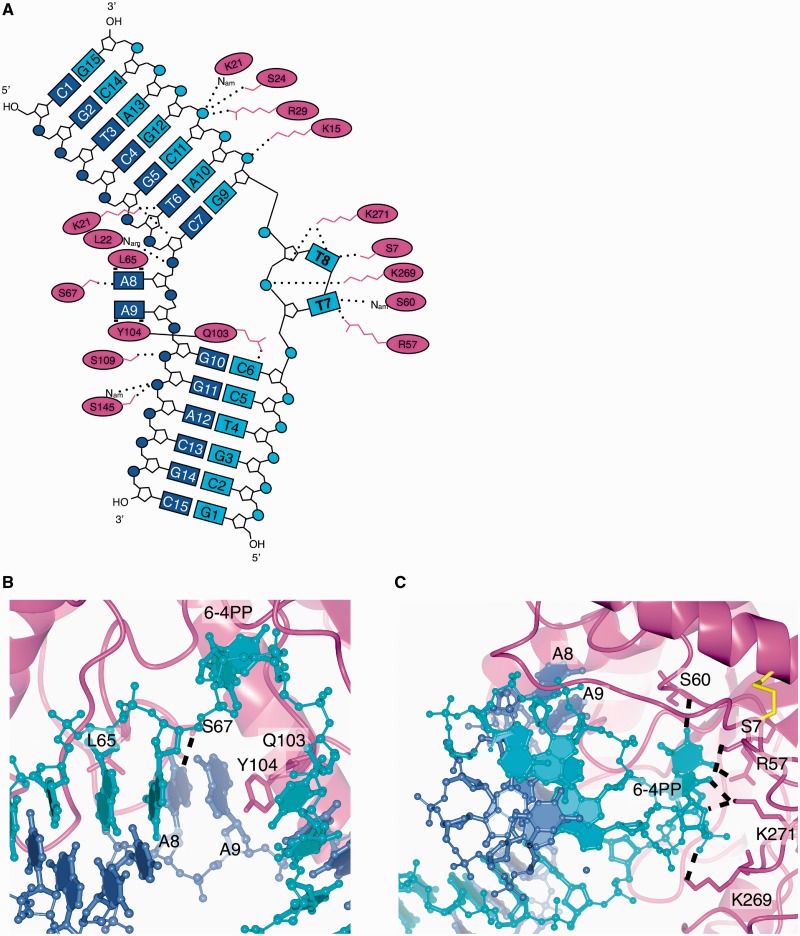
In the DNA:protein complex structure, the DNA is bound in the previously predicted DNA-binding groove and makes a bend of around 90° (Figure 1F), similar to the related TIM-barrel structure of Endonuclease IV with DNA containing an abasic site (21). Numerous interactions of protein side-chains and backbone amides with the DNA phosphates and some with the base of the deoxyribose ring hold the DNA in its binding groove (Figure 3A). The residues responsible for these interactions are partially or fully conserved in UVDE from different organisms, hence the structure provides an explanation for the mode of action of the entire UVDE family.
Figure 3.
UVDE-DNA interactions. (A) Schematic representation of UVDE-DNA interactions with the undamaged DNA strand in blue, the damaged strand in cyan and the protein in magenta. The label Nam points out that it is the nitrogen of the amide bond that is involved in hydrogen bonding. In three dimensions, the damaged bases T7 and T8 and the undamaged bases A8 and A9 are actually below the plane of the figure, since they are flipped out of the helix towards one side of the DNA. The residues Q103 and Y104 are together inserting between the two DNA strands near the damage and the two bases opposite to the damage. (B) Detailed view of the pocket for the undamaged bases. Hydrogen bonds are indicated with dashed lines. (C) Detailed view of the pocket for the damaged bases. Hydrogen bonds are indicated with dashed lines.
A novel ‘dual-flip’ mechanism for recognizing pyrimidine-dimer lesions
Strikingly, the two bases opposite to the damage (nucleotides A8 and A9) are flipped from the DNA helix into an ‘undamaged-bases-binding’ pocket of UVDE (Figure 3B). In this pocket, the ‘probing finger’ residue Tyr104 forms stacking interactions with the base of A9 and thus explains the loss of activity when this residue is mutated (9). Leu65 aids in creating a hydrophobic environment on the other side of this pocket near A8. In other UVDEs, usually a leucine or phenylalanine is found at this position, and in a superposition of TthUVDE with the DNA:protein complex structure, this phenylalanine indeed forms stacking interaction with the base of A8 that likely improves the interaction between the protein and DNA. A hydrogen bond between the conserved Ser67 and the N1 of the A8 base is present that would be lost if a pyrimidine was present in this position owing to its smaller size. The size of the pocket together with these stacking and hydrogen bond interactions provides a customized fit for two purines. The existence of this pocket is an ingenious feature of an enzyme that needs to recognize UV-damaged DNA, as dipyrimidines always have two purines opposite the lesion. It also provides an explanation for the strong preference of UVDE for incising abasic sites flanked by a pyrimidine over those flanked by a purine (8). Moreover, flipping of the bases opposite to the damage into a pocket explains the previously reported fluorescence studies (8) that showed the bases opposite to the damage are flipped from the helix into a partially to not solvent-exposed area and, thus, agreeing with bases flipped into a pocket.
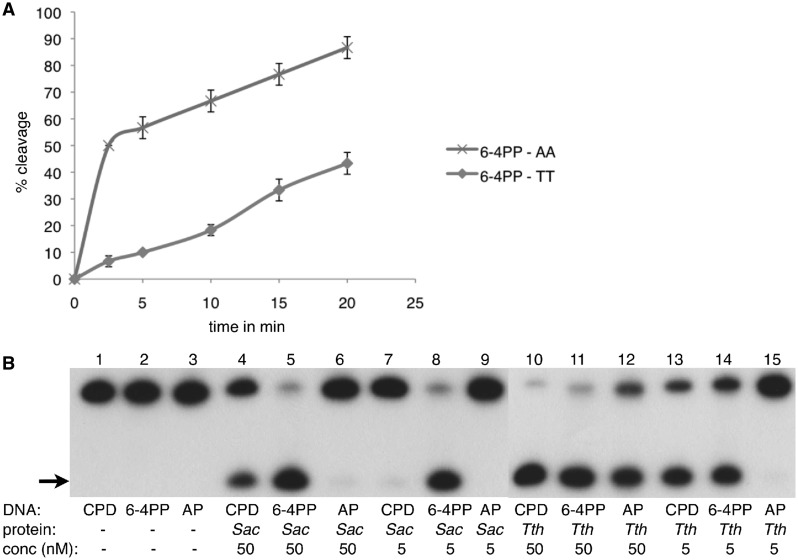
To confirm the importance of two purines opposite the damage for the incision activity of UVDE, we compared its activity on a 6-4PP with two adenines opposite to the damage (the ‘natural’ substrate) to a 6-4PP with two thymines opposite the damage. As can be seen in Figure 4A, the activity of UVDE on 6-4PP with two adenines opposite the damage is considerably higher than with two thymines, demonstrating the relevance of the dipurine stabilization in the custom-fit/non-damaged bases pocket.
Figure 4.
Activity of SacUVDE with a dipurine oppposite to the damage and its activity versus that of TthUVDE. (A) Kinetics incision assay showing a marked preference of SacUVDE for incising 6-4PP with a dipurine opposite the damage (data points shown with x’s) compared with a 6-4PP with a dipyrimidine opposite the damage (data points shown with circles). (B) Incision assay with SacUVDE or TthUVDE (indicated below the lanes) showing that SacUVDE has a preference for incising 6-4PP compared with CPD in contrast to TthUVDE. The incision product is indicated with an arrow. The assay was carried out at 55°C for both proteins.
Not only are the undamaged bases flipped, but the damage itself is also flipped into a protein pocket (Figure 3C). A hydrogen bond by the probing finger residue Gln103 to the base just 5′ to the damage helps in stabilizing this flipped conformation. The damage pocket is lined with several residues making hydrogen bonds to the damage (shown in Figure 3A and C) and is close to the enzyme’s active site. The pocket is tight around the 6-4PP, but the shape does allow a CPD to fit, but purines would be excluded from the pocket owing to their larger size. Other lesions that are larger are also excluded from the pocket, and the pocket probably stabilizes smaller lesions such as abasic sites less, and therefore damaged dipyrimidines are the preferred substrate. To be flipped from the DNA helix into the pocket, the bases lose their interaction in the DNA helix and the DNA backbone must deform substantially. The loss of base-pairing and amount of deformation that an undamaged DNA substrate would have to undergo to enter the pocket is probably unfavourable and might explain the enzyme’s specificity for damaged DNA.
SacUVDE has a preference for 6-4 photoproduct over CPD damaged DNA
Although the structure of the apoprotein shows a high structural similarity to TthUVDE, in vitro assays surprisingly showed that SacUVDE has a high incision activity for the 6-4PP, even higher than that of TthUVDE, but lower for CPD and virtually absent for abasic sites (Figure 4B, lanes 4–9). In contrast, TthUVDE has high incision activity for both CPD and 6-4PP, but lower incision activity for abasic sites (Figure 4B, lanes 10–15). The relative difference in incision was the same over a variety of conditions we tested including a 30-mer or 50-mer DNA substrate, a pH range of 3.5–8.5 and a temperature range of 35–80°C. The optimum activity was found at pH 5.5–6.5 and temperature 55–80°C. The optimal metal co-factor is either manganese or cobalt. The absence of metal ions, calcium, zinc or nickel gives little to no activity and the addition of manganese lowers the activity slightly.
Structural analysis in combination with site-directed mutagenesis studies showed that the different phenotype is not caused by any one specific part of the protein. For example, the deletion by mutagenesis of the extra helix present in SacUVDE, but absent in TthUVDE (Supplementary Figure S1A), led to a poorly soluble protein, hence we concluded that the helix is required for SacUVDE protein solubility. Another difference is the presence of Tyr10 in SacUVDE that could form a hydrogen bond with the DNA damage, whereas TthUVDE has a leucine in this position (Supplementary Figure S1B). Mutation of this tyrosine to an alanine in SacUVDE did not cause any difference in incision activity (Supplementary Figure S2). Furthermore, as mentioned earlier, SacUVDE has a disulfide bond, Cys14 to Cys40 (Supplementary Figure S1C), that is not present in most UVDEs, including TthUVDE and causes a backbone shift up to 2.8 Å of residues 10–18 near the damage-binding pocket. Supplementary Figure S2 (lanes 11–17) shows that SacUVDE C14A has a strongly reduced activity compared with SacUVDE wildtype (lanes 4–10); however, the preference for incision of 6-4PP over CPD still exists. Finally, the SacUVDE probing finger residues Gln103 and Tyr104 that aid in flipping out the damaged bases and the opposite bases from the double DNA helix are situated in a rigid loop with a proline at the beginning and the end (Pro102 and Pro108) in TthUVDE, while SacUVDE only has one proline at the corresponding positions (Pro101 and Leu107) (Supplementary Figure S1D). A more rigid finger loop might be more suited to flip out less distorted DNA substrates and hence a protein with such a rigid finger might have a broader substrate range. Mutation of Leu107 to proline led to a less soluble protein, but the soluble protein fraction was still as active as wildtype (Supplementary Figure S2). The more rigid probing finger is therefore also not the (sole) cause of the difference between SacUVDE and TthUVDE.
We therefore think it is a more global feature of the protein such as the capacity to bend damaged DNA that causes SacUVDE’s preference for incision of 6-4PP. UVDE from S. acidocaldarius might be more rigid to be able to function well at the high temperatures that this organism lives (75–80°C). It should be noted that in solution, 6-4PP is more distorted than CPD and hence might present an easier substrate (22). Nonetheless, SacUVDE’s preference for incising next to 6-4PP may be useful for biochemical assays to distinguish between CPD- and 6-4PP-damaged DNA.
Metal ions in the structure
No metal ions were added during the purification or crystallization of either structure. The uncomplexed SacUVDE structure contains only one of the three metal ions, whereas no metal ion density was found in the complexed SacUVDE structure and thus, the DNA is not incised. The SacUVDE metal coordinating residues are at similar positions to the corresponding residues in TthUVDE, and addition of manganese is needed for activity in incision assays for both proteins. Therefore, it is very likely that active SacUVDE also uses a three-metal ion catalysis. In the DNA:protein complex structure, the scissile P-O bond is still several Angstroms away from the correct position for cleavage, but the positive charge of the metal ions, whose location can be predicted from the metal atoms present in the TthUVDE structure, is likely required to draw the scissile bond inwards to the correct position, allowing incision to take place.
CONCLUSION
We propose the following model for UVDE activity: first, UVDE recognizes a distortion in the DNA and binds to it. The residues of the ‘probing finger’ flip the damaged bases as well as the opposite bases out of the helix into their respective pockets. Optimal binding occurs if the opposite bases are two purines and if the damaged bases fit in the damage pocket. The positive charge of the metal ions then draws in the scissile phosphodiester bond and incision by a hydroxyl ion takes place using three-metal ion-mediated catalysis.
ACCESSION NUMBERS
3tc3 and 4gle.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) [021.002.024 and 700.55.425]. Funding for open access charge: Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities.
REFERENCES
- 1.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 2.Maul MJ, Barends TRM, Glas AF, Cryle MJ, Domratcheva T, Schneider S, Schlichting I, Carell T. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew. Chem. Int. Ed. Engl. 2008;47:10076–10080. doi: 10.1002/anie.200804268. [DOI] [PubMed] [Google Scholar]
- 3.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn M, Tainer JA. Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vassylyev DG, Kashiwagi T, Mikami Y, Arlyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 5.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–576. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 6.Bowman KK, Sidik K, Smith CA, Taylor JS, Doetsch PW, Freyer GA. A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res. 1994;22:3026–3032. doi: 10.1093/nar/22.15.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avery AM, Kaur B, Taylor J, Mello JA, Essigmann JM, Doetsch PW. Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:2256–2264. doi: 10.1093/nar/27.11.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paspaleva K, Moolenaar GF, Goosen N. Damage recognition by UV damage endonuclease from Schizosaccharomyces pombe. DNA Repair. 2009;8:600–611. doi: 10.1016/j.dnarep.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Paspaleva K, Thomassen E, Pannu NS, Iwai S, Moolenaar GF, Goosen N, Abrahams JP. Crystal structure of the DNA repair enzyme ultraviolet damage endonuclease. Structure. 2007;15:1316–1324. doi: 10.1016/j.str.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Iwai S. Chemical synthesis of oligonucleotides containing damaged bases for biological studies. Nucleosides Nucleotides Nucleic Acids. 2006;25 doi: 10.1080/15257770600685826. 561–582. [DOI] [PubMed] [Google Scholar]
- 11.Verhoeven EE, Van Kesteren M, Turner JJ, Van der Maarel GA, Van Boom JH, Moolenaar GF, Goosen N. The C-terminal region of Escherichia coli UvrC contributes to the flexibility of the UvrABC nucleotide excision repair system. Nucleic Acids Res. 2002;30:2492–2500. doi: 10.1093/nar/30.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie AG. Integration of macromolecular diffraction data. Acta Crystallogr. 1999;D55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 13.Evans P. Scaling and assessment of data quality. Acta Crystallogr. 2006;D62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 14.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 16.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macro- molecular crystal structures. Acta Crystallogr. 2011;D67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson KE, Wang Y, Madej T, Bryant SH. Improving protein structure similarity searches using domain boundaries based on conserved sequence information. BMC Struct. Biol. 2009;19:33. doi: 10.1186/1472-6807-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular graphics project. Acta Crystallogr. 2004;D60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 21.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double- nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 22.Kim JK, Choi BS. The solution structure of DNA duplex-decamer containing the (6-4) photoproduct of thymidylyl (3′→5′) thymidine by NMR and relaxation matrix refinement. Eur. J. Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 23.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.