Abstract
Trypanosoma brucei survives in mammals through antigenic variation, which is driven by RAD51-directed homologous recombination of Variant Surface Glycoproteins (VSG) genes, most of which reside in a subtelomeric repository of >1000 silent genes. A key regulator of RAD51 is BRCA2, which in T. brucei contains a dramatic expansion of a motif that mediates interaction with RAD51, termed the BRC repeats. BRCA2 mutants were made in both tsetse fly-derived and mammal-derived T. brucei, and we show that BRCA2 loss has less impact on the health of the former. In addition, we find that genome instability, a hallmark of BRCA2 loss in other organisms, is only seen in mammal-derived T. brucei. By generating cells expressing BRCA2 variants with altered BRC repeat numbers, we show that the BRC repeat expansion is crucial for RAD51 subnuclear dynamics after DNA damage. Finally, we document surprisingly limited co-localization of BRCA2 and RAD51 in the T. brucei nucleus, and we show that BRCA2 mutants display aberrant cell division, revealing a function distinct from BRC-mediated RAD51 interaction. We propose that BRCA2 acts to maintain the huge VSG repository of T. brucei, and this function has necessitated the evolution of extensive RAD51 interaction via the BRC repeats, allowing re-localization of the recombinase to general genome damage when needed.
INTRODUCTION
African trypanosomes, such as Trypanosoma brucei, are protistan parasites that infect mammals in sub-Saharan Africa, where they are responsible for devastating disease in both humans and their domestic animals. Survival of T. brucei in mammals is dependent on antigenic variation, which involves switches in expression of variant surface glycoproteins (VSGs) that form a protective ‘coat’ on the parasite cell surface (1,2). One VSG is expressed at a time, and continual switching to immunologically distinct VSGs allows part of the infecting population to survive successive waves of eradication by host immunity, prolonging the infection and enhancing transmission.
VSG switching involves the activation of silent VSG genes by recombination, which results in copying the silent genes into specialized sites of transcription, termed VSG expression sites (3). VSGs are copied from an enormous (>1000 distinct genes) silent repository, mainly composed of subtelomeric VSG arrays. Most recombination-based VSG switching occurs through gene conversion reactions (2), which can take two forms: activation of functionally intact VSGs (∼5% of the repository) by whole gene conversion or activation of VSG pseudogenes (∼85% of the repository) by segmental gene conversion (2,4). Intact VSG gene conversion involves homologous recombination (HR), a universally conserved process that is critical in all organisms for reversing genotoxic damage and ensuring the completion of DNA replication (5). The key enzyme of eukaryotic HR is Rad51, which forms nucleoprotein filaments on single-stranded (ss) DNA at sites of damage and catalyses the transfer of the broken molecule to homologous sequences in an unbroken DNA molecule, leading to repair. Mutation of RAD51 in T. brucei impairs switching of intact VSGs (6), but the contribution of RAD51 and HR to segmental VSG conversion is unclear. Rad51 HR reactions are mediated by a number of proteins, which either directly influence Rad51 activity or act in upstream or downstream HR reaction steps (5). Trypanosoma brucei HR factors that perform both roles, such as RAD51 paralogues (7,8) and RMI1/TOPO3 (9,10), have been shown to act in VSG switching, reinforcing a close association between general HR and antigenic variation.
BRCA2 has emerged as a key mediator of Rad51 function and is widely conserved, although not ubiquitous, in eukaryotes (11). BRCA2 orthologues vary considerably in size, from >3000 amino acids in mammals to proteins only 30 and 10% that size in Ustilago maydis (Brh2) (12) and Caenorhabditis elegans (CeBRC-2) (13), respectively. BRCA2 sequence conservation is limited out with two domains, termed the BRC repeats and the DSS1-DNA binding domain (DBD). BRC repeats mediate interaction with Rad51 during HR (14) and are a key conserved functional element, as each BRCA2 orthologue seems to retain at least one (11). In vertebrates, BRCA2 also binds Rad51 via an unrelated C-terminal sequence; binding here is regulated by cell cycle-dependent phosphorylation (15) and is specific for Rad51–DNA filaments (16,17). Non-BRC repeat binding of Rad51 may be a conserved feature of BRCA2, as it is also seen in U. maydis (18) and C. elegans (13), but whether it provides a functional link to cell cycle progression is unclear. Indeed, the detailed role of BRCA2 in promoting HR is still being unravelled. Initial models suggested that the BRC repeats of vertebrate BRCA2 bind Rad51 as monomers and counter RAD51 nucleoprotein filament formation on ssDNA. Dephosphorylation of the C-terminal Rad51 binding site then allows this region to promote the formation of Rad51 nucleoprotein filaments, which are disassembled via the BRC repeats to terminate HR when the C-terminus is rephosphorylated as the cells proceed to mitosis (19). However, other work has shown that the BRC repeats of mammalian BRCA2 inhibit Rad51 binding to double-stranded (ds) DNA, promote binding to ssDNA and can support strand exchange in the absence of the C-terminal Rad51 binding site (20–23). Indeed, yet further studies have suggested that vertebrate BRCA2–Rad51 interaction via BRCA2’s C-terminus acts in an HR-independent role to stabilize DNA replication forks whose progression has stalled (24–26). Such issues have been little addressed in other eukaryotes.
Trypanosoma brucei and related kinetoplastid parasites encode BRCA2 orthologues, in which strong homology is seen in the C-terminus around the DBD, but with little obvious sequence conservation with human BRCA2 in the C-terminal Rad51 binding region (Supplementary Figures S1–S3). Upstream of the DBD, kinetoplastid BRCA2 proteins are significantly shorter than human BRCA2, and here, sequence homology seems to be limited to the BRC repeats (Supplementary Figure S1). However, the number of BRC repeats in T. brucei BRCA2 is highly unusual (Figure 1) (27): up to 15 repeats have been described, nearly double the number found in mammalian BRCA2 (eight BRC repeats) (11), and far greater than in BRCA2 of Arabidopsis thaliana (four BRC motifs) (28) or in C. elegans (13) and U. maydis (12), each of which has only a single BRC repeat. The BRC repeat number in T. brucei BRCA2 seems to be a recent evolutionary expansion, as BRCA2 proteins from Trypanosoma cruzi and Leishmania major, related intracellular kinetoplastid parasites, have two non-identical BRC repeats (Supplementary Figure S1). The arrangement of the BRC repeats in T. brucei BRCA2 is also unusual: most of the repeats are identical in sequence and all form a tandem array (27). This contrasts with BRCA2 in humans (Supplementary Figure S1), plants (11) and at least some Drosophila species (29), where the BRC repeats are variable in sequence and dispersed throughout the polypeptide.
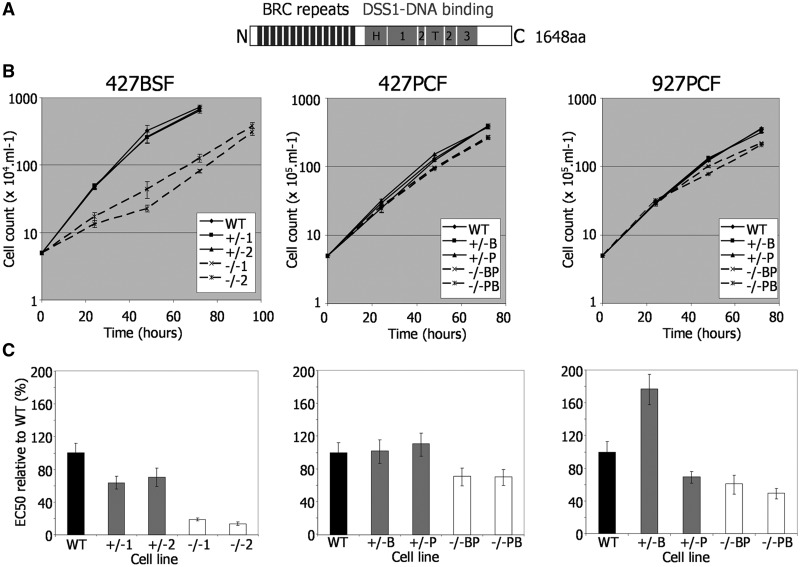
Figure 1.
Growth and repair efficiency of T. brucei BRCA2 mutants. (A) Locations of conserved domains are shown diagrammatically within the T. brucei BRCA2 polypeptide: BRC repeats (black boxes), and sub-domains of the DSS1-DNA binding region (grey boxes: H, helical domain; T, tower domain; 1, 2, 3, OB domains). (B) Growth is shown of WT, BRCA2 heterozygous (+/−) and brca2 homozygous (−/−) mutants cells in T. brucei Lister427 bloodstream form (427BSF) cells and in PCF cells from Lister427 (427PCF) and TREU927 (927PCF) strains. Cell densities were measured in vitro at 24 h intervals; bars indicate standard errors from three experiments. (C) EC50 values of the same T. brucei BSF and PCF strains are shown, comparing WT, BRCA2+/− and brca2−/− cells exposed to phleomycin. EC50 values are the mean from three experimental repetitions expressed as a percentage of WT; bars indicate standard error.
Little work has examined why BRCA2 in some organisms have evolved multiple BRC repeats, whereas in others a single BRC repeat is sufficient for function. In mammals, the BRC repeats fall into two categories (21): isolated BRC repeats in one category display relatively high affinity Rad51 binding, reduce the adenosine triphosphatase activity of the recombinase and stimulate strand exchange in vitro; in contrast, BRC repeats in the second category have low affinity for Rad51 and do not affect the recombinase’s adenosine triphosphatase or strand exchange activities. Thus, it is modelled that BRCA2’s role in Rad51 HR is through a two-step process, directed by the differing BRC–Rad51 interactions, aiding the formation of Rad51 nucleoprotein filaments on ssDNA (21). How this occurs in the context of the complete protein and during in vivo repair remains to be examined. In T. brucei, we tested previously whether the BRC repeat expansion is an adaptation for HR during VSG switching. To our surprise, mammal-derived bloodstream form (BSF) cells modified to express BRCA2 variants with only a single BRC repeat, whereas impaired in general HR and in re-localization of RAD51 to subnuclear foci after damage, were competent for switching of intact VSGs (27). Here, we more comprehensively dissect the need for a BRC repeat array and show that BRC repeat number is a critical determinant of RAD51 subnuclear dynamics. We also analyse BRCA2 and RAD51 localization in T. brucei and show that, contrary to other eukaryotes, these factors rarely co-localize in subnuclear repair foci. In addition, we show that chromosome instability, described previously in BSF T. brucei BRCA2 mutants, is not seen in procyclic form (tsetse midgut-derived; PCF) parasites, where the phenotypes of BRCA2 loss are less severe. Finally, we document replication-associated functions of BRCA2 that localize to the C-terminus of the protein. We propose that these data are explained by BRCA2 acting in VSG array stability in a specific process linked to antigenic variation, which has necessitated the evolution of extensive RAD51 interaction, via expanded BRC repeats, to allow redistribution of RAD51 to general damage when needed.
MATERIALS AND METHODS
Trypanosoma brucei strains, growth and mutation of BRCA2 and RAD51
Trypanosoma brucei BSF cells (strain Lister427 MITat1.2) were grown at 37°C in HMI-9 medium. PCF cells (strain TREU927 or Lister427) were grown in SDM-79 medium at 27°C. BSF BRCA2 mutants were generated as described previously (27); PCF mutants used the same constructs and were selected with 5 µg ml−1 blasticidin and 0.5 µg ml−1 puromycin. Cell growth was analysed in vitro by diluting the BSF cells to 5 × 104 ml−1 and measuring cell density (using a haemocytometer, Bright-line, Sigma) at 24 h time intervals; PCF cells were diluted to 5 × 105 ml−1, and cell density was measured using a Z2 Coulter particle count and size analyser (Beckman). To generate RAD51 mutants, sequences derived from the start and end of the RAD51 ORF, covering equivalent coding sequence to that contained in previous RAD51 disruption constructs (6), were generated by high-fidelity polymerase chain reaction (PCR)-amplification and cloned into pBluescript. The same blasticidin-resistance cassette used for BRCA2 deletion was then cloned between the RAD51 sequences, generating ΔRAD51::BSD. This was transformed into RAD51+/− cells, in which one RAD51 allele had been disrupted by puromycin resistance insertion (6), transformants were selected with 2.5 µg ml−1 blasticidin and 0.5 µg ml−1 puromycin.
Generation of BRCA2 variants
ORFs encoding full-length BRCA2 and the BRCA2 variants were cloned, after high-fidelity PCR-amplification, into EcoRV-digested pRM482, which targets genes to the tubulin array and is identical to pRM481 (8), except that the phleomycin resistance ORF is replaced with G418 resistance (neomycin phosphotransferase). Full-length T. brucei BRCA2 was PCR-amplified with Tbfor (5′-CCCGATATCATGAGCCACAAAAAAGGAAGACAA; EcoRV site underlined) and Tbrev (5′-CCCGATATCCTATTCTCGCATAAGATCAGCGAC). Trypanosoma vivax BRCA2: Tvivfor2 (5′-CCCCGATATCATGAAGCAGCGGCAAGTAGGTGAA) and Tvivrev3 (5′-CCCCGATATCCTACACTGAACTCTCCTCCTGCAT). The BRCrep coding sequence: BRCA-TRUNC 5′ (5′-CCCCTCGCGAATGTACGGGACTGAAAATGGCCAAGAG; NruI) and BRCA-TRUNC 3′ (5′-CCCCTCGCGACTACGGCTTTCTTGCTAGCTTGGATG). BRCA2 Cterm coding sequence: BRCA-NO-BRC (5′-CCCTCGCGAATGTCGGGAGCAGGTGCCTCCTTGTCG) and Tbrev2. The BRC–replication protein A (RPA) fusion was generated by PCR-amplifying the T. brucei BRCA2 BRC repeat region and RPA ORF separately and then cloning them together into pRM482. The BRCrep coding sequence was amplified with BRCA–RPA5′ (5′-CCCCGATATCATGTACGGGACTGAAAATGGCCAAGAG; EcoRV) and BRCA–RPA3′ (5′-CGGCTTTCTTGCTAGCTTGGATG); RPA was amplified with RPA 5′ (5′-CAGCAGCCATCACAACAACAG) and RPA 3′ (CCCCGATATCTTACAAGTAGGCATTAATGC; EcoRV). Trypanosoma brucei BRCA2 with 1 BRC repeat was generated by PCR-amplifying the coding sequence immediately upstream of the BRC repeats and a region encompassing the most C-terminal coding repeat and all downstream sequence separately, then cloning the two fragments together into pRM482. The upstream fragment was PCR-amplified with Tbfor and BRCVAR5′3 (5′-CTCTTGGCCATTTTCAGTCCC); the downstream with BRCVAR3′5 (5′-AGCACTGCGGTACAAGGAAATTCC) and BRCVAR3′3 (5′-CCCTCGCGACTATTCTCGCATAAGATCAGCGAC; NruI). Trypanosoma brucei BRCA2 with 4, 7 and 10 BRC repeats were synthesized and PCR-amplified with Tbfor and Tbrev. To C-terminally tag BRCA2 with a 12Myc epitope, a 491-bp region of the 3′-end of the ORF was PCR-amplified with primers 32 (5′-CCCAAGCTTGAAGTGGAAAGTTTGTAGTGTCC; HindIII) and 33 (5′-CCCTCTAGATTCTCGCATAAGATCAGCG; XbaI) and cloned into the vector pNATX12myc (30); after digestion with SphI, this was transformed into BRCA2+/− cells (::PUR) and transformants selected with 10 µg ml−1 blasticidin.
Microscopy
The 5 × 106 BSF cells or 5 × 105 PCF cells were harvested by centrifugation at room temperature for 10 min at 735g and washed once in phosphate buffered saline (PBS). The cell pellet was resuspended in 50 µl PBS and spread thinly on a silane prepared slide (Sigma) and allowed to air dry. Slides were soaked in ice cold methanol for 15 min and washed in cold PBS twice. Slides were blocked by immersion in cold 5% fetal calf serum (FCS) in PBS for 1 h. Blocking solution was removed, and the slides were transferred to a dark chamber before adding rabbit polyclonal anti-TbRAD51 antibody diluted 1:1000 in 2% of FCS/PBS for 1 h. The slides were washed twice in PBS for 5 min and then returned to the humid chamber. Alexa Fluor conjugated secondary antibodies were added, diluted in 2% of FCS/PBS and incubated for 1 h: Alexa 594-conjugated goat-derived anti-rabbit IgG (1:7000 dilution) was used to detect RAD51, and Alexa 488-conjugated mouse-derived anti-myc IgG (1:7000 dilution) was used to detect BRCA2myc. Finally, the slides were washed twice in PBS for 5 min, allowed to air dry, then mounted in Vectashield with DAPI (4,6-diamidino-2-phenylindole) (Vector Laboratories). Fluorescence microscopic analysis was performed using a Deltavision confocal microscope, and flattened projections of the resulting image slices were obtained using softWoRx explorer (Applied Precision).
Assaying DNA damage sensitivity
Sensitivity of the cell lines to methyl methanesulphonate (MMS) and phleomycin was assessed by the metabolic reduction of Alamar blue (31). Here, cells in mid-logarithmic growth were plated at a density of 2 × 105 cells ml−1 for BSF cells and 5 × 105 cells ml−1 for PCF cells in a 96-well dish (200 µl/well) in medium containing doubling dilution concentrations of drug. After 48 h growth, 20 µl of Alamar blue (from a 0.125 mg ml−1 stock of resazurin, Sigma) was added to each well, and the plates were incubated in the dark for 24 h, when the fluorescence in each well was determined fluorometrically (PerkinElmer LS55) at 530 nm excitation wavelength and 590 nm emission wavelength.
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) was carried out essentially as described in (27). In brief, agarose plugs containing genomic DNA from 1 × 108 cells were prepared and separations of megabase-chromosomes conducted using 1.2% of agarose (Seakem LE) in a CHEF-DR III apparatus (BioRad). Gels were electrophoresed in 2 l of electrophoresis buffer (1× TB1/10E: 90 mM of Tris, 90 mM of boric acid, 2 mM of ethylenediaminetetraacetic acid). Before electrophoresis, the agarose genomic DNA plugs were prepared by three rounds of dialysis in electrophoresis buffer. Gels were electrophoresed at 15°C with an included angle of 120° and 2.5 V cm−1 for 144 h with an initial switch time of 1400 s and final switch time of 700 s.
Aqueous fractionation
Aqueous fractionation was performed essentially as described in (32). The 5 × 108 PCF cells were centrifuged at 1620g and washed twice in 5 ml ice cold fractionation buffer A [FBA; 150 mM of sucrose, 20 mM of KCl, 3 mM of MgCl2, 20 mM of HEPES–KOH (pH 7.9), 1 mM of DTT and 1× complete ethylenediaminetetraacetic acid-free protease inhibitor cocktail (Roche)]. Cells were re-suspended in 1 ml of FBA with 0.2% Nonidet P-40. The cell suspension was passed through a 26-gauge syringe needle three times. The suspension was centrifuged at 20 000g for 10 min at 4°C. The supernatant was separated and centrifuged again at 20 000g at 4°C; this supernatant contains the soluble cytoplasmic proteins. The cell pellet was re-suspended in 500 μl of FBA and passed through a 26-guage syringe needle 15 times. The suspension was centrifuged at 20 000g at 4°C for 10 min. The supernatant was removed and the pellet re-suspended in 500 μl of FBA; this suspension contains the soluble nuclear proteins. The proteins were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) on NuPAGE Novex Bis–Tris 10% mini gels (Invitrogen), western blots prepared using a Mini Trans-Blot Cell (Bio-Rad) and Hybond ECL nitrocellulose (Amersham) and the blots probed with anti-RAD51 (1:500 dilution), anti-OPB1 (1:1000) and anti-NOG1 (1:5000) antiserum. Bound antiserum was detected by horseradish peroxidase conjugated secondary antiserum and the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
RESULTS
BRCA2 mutation causes more pronounced phenotypes in bloodstream form than in procyclic form T. brucei
To examine the functions of BRCA2 in T. brucei, we used reverse genetics to delete the entire BRCA2 ORF in BSF and PCF T. brucei cells of strain Lister427, as well as in PCF TREU927 cells where ∼800 VSGs have been positionally annotated (4). In each case, two independent mutants were derived by two rounds of transformation, generating first BRCA2 heterozygous (+/−) mutants and then homozygous (−/−) null mutants (Figure 1); the mutations were confirmed by Southern analysis (Supplementary Figure S4). Comparing the growth of the −/− mutants with wild-type (WT) cells and +/− predecessors showed that the impact of BRCA2 loss was not equivalent in the two life cycle stages (Figure 1B). In BSF cells, the brca2−/− mutants, although viable, showed significantly impaired growth, with population doubling times (pdts; ∼16 h) nearly 2-fold increased relative to WT and +/− cells (∼8.5 h). In contrast, PCF brca2−/− mutants displayed a more modest increase in pdts (∼15.5 and ∼13 h for TREU927 and Lister427−/− cells, respectively, relative to ∼11 h for WT and +/− cells). In each case, re-expression of BRCA2 (−/−/+ cells, see below) in the brca2−/− cells restored growth to WT rates, demonstrating that this phenotype is a direct consequence of BRCA2 loss rather than secondary effects.
To examine the contribution of BRCA2 to T. brucei DNA repair, the sensitivity of the mutants to induced DNA damage was examined by comparing growth in the presence of either phleomycin (BLE; Figure 1C), a glycopeptide antibiotic that generates ssDNA and dsDNA breaks (33) or MMS (Supplementary Figure S5), an SN2 alkylating agent. To quantify this, the EC50 (concentration that resulted in death of 50% of cells) of the WT, +/− and −/− cell lines in each life cycle stage and in each strain was determined by measuring the metabolic capacity of the cells using Alamar blue as an indicator (31). Here again, the effect of BRCA2 loss was more marked in BSF than in PCF cells. Lister427 BSF brca2−/− mutants were 5–10-fold more sensitive to BLE than WT cells, whereas PCF brca2−/− cells displayed at most 2-fold greater sensitivity. BSF brca2−/− mutants displayed less pronounced increased sensitivity to MMS (∼3 fold), but this was again greater than seen for the PCF homozygous mutants of either strain. Indeed, the brca2−/− mutants in Lister 427 PCF cells showed only a limited increase in MMS sensitivity relative to WT (Supplementary Figure S5).
Trypanosoma brucei BRCA2 BRC repeat number is an important determinant of RAD51 subnuclear redistribution after DNA damage
To ask why T. brucei BRCA2 requires an expanded array of BRC repeats, we expressed a number of BRCA2 variants in the brca2−/− cell lines (Figure 2). In each case, the variants were expressed by integration of the ORFs into the tubulin array, the same strategy used previously to express BRCA2 variants with a single BRC repeat (27) (see later in the text). Here, we constructed and expressed BRCA2 variants with increasing BRC repeat number (Figure 2A). We refer to these as 1BRC, 4BRC, 7BRC and 10BRC; 1BRC retains only the C-terminal, sequence degenerate BRC repeat, whereas the others have additionally 3, 6 or 9 upstream conserved BRC repeats, respectively. Integration of each BRCA2 variant, as well as the larger full-length BRCA2 allele from the Lister427 genome (12 BRC repeats), was analysed by Southern analysis (Supplementary Figure S6) and confirmed for all except 7BRC in Lister427 PCF cells, where two BRCA2 gene copies were apparent. Expression of mRNA for each variant was analysed by quantitative reverse transcriptase-PCR (as we could not detect BRCA2 protein with an anti-peptide antiserum): BRCA2 mRNA was present in each variant expresser at ∼50% of the quantity seen in WT cells, consistent with single gene integrations; the exception to this was 7BRC in PCF Lister 427, which showed approximate WT levels of mRNA (data not shown).
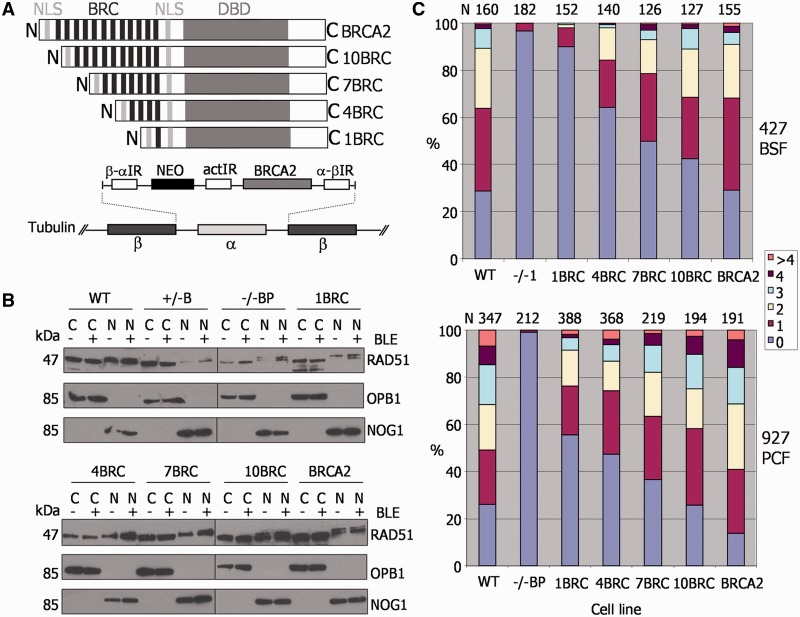
Figure 2.
The BRC repeat number of T. brucei BRCA2 affects the efficiency of RAD51 subnuclear foci formation. (A) A diagrammatic illustration of the strategy for expression of BRCA2 variants with differing numbers of BRC repeats. BRCA2 variant ORFs were targeted to the tubulin array for expression in brca2−/− cells, using flanking β-α and α-β tubulin intergenic regions (IR) for homologous integration. Transformants were selected by G418 resistance using a neomycin phosphotransferase ORF (NEO), which was upstream of the BRCA2 variant ORF; actin IR sequence between the NEO and BRCA2 ORFs was for mRNA processing. The BRCA2 variants differed only in the number of BRC repeats they encode (black bars): 1, 4, 7, 10 or 12, indicated by 1BRC, 4BRC, 7BRC, 10BRC or BRCA2, respectively. Other conserved domains are indicated: DBD and two putative NLS. (B) Aqueous fractionation was performed to generate protein fractions enriched in soluble cytoplasmic (C) and nuclear proteins (N) from WT TREU927, BRCA2+/−, brca2−/− and BRCA2 BRC variant expresser cells; ‘hyphen’ indicates fractions prepared without phleomycin (BLE) treatment, and ‘plus’ indicates fractions prepared after phleomycin treatment (1 μg ml−1 for 18 h). Fractions were separated by SDS–PAGE and western blotted before being sequentially probed with anti-RAD51, anti-OPB1 and anti-NOG1 antiserum; protein sizes are indicated (kDa). (C) For PCF TREU927 and BSF Lister427 T. brucei, WT, brca2−/− mutants and BRC variant expressers were treated with 1 μg ml−1 phleomycin for 18 h, and the number of cells with a specific number of subnuclear RAD51 foci (0, 1, 2, 3, 4, >4) counted; graphs represent the numbers of foci-containing cells as a percentage of the total number of cells counted (N).
We first considered the possibility that the increased BRC repeat number may be because BRCA2 acts as a carrier for transit of RAD51 into the nucleus, as has been described in human cells (34) and Leishmania (35). If so, greater BRC numbers may reflect the need for abundant nuclear RAD51 in T. brucei. To test this, aqueous fractionation was carried out on T. brucei whole cell extracts (32) from PCF WT TREU927 and BRCA2 mutant cells, and from the BRC variant expressers. In all cases, nuclear and cytoplasmic extracts were examined before and after BLE treatment (1 μg ml−1 for 18 h) by probing western blots of the separated proteins sequentially with anti-RAD51, anti-OPB1 (cytoplasmic) (36) and anti-NOG1 (nuclear) (37) antiserum (Figure 2B). In all the cells, including the brca2−/− mutants, RAD51 was detected in both the nuclear and the cytoplasmic fractions before and after BLE-induction of DNA damage. There was also no evidence that the number of BRC repeats affected the relative amount of RAD51 in the nucleus and cytoplasm. We conclude that BRCA2 is not a critical nuclear transport factor for RAD51 in T. brucei, despite the absence of detectable nuclear localization signals in the polypeptide sequence of the recombinase (data not shown).
Relocalization of HR factors, including Rad51, to microscopically detectable foci after DNA damage is a conserved response in eukaryotes and bacteria, and such foci represent the concentration of factors into putative repair centres (38,39). The brca2−/− mutants in BSF Lister427 T. brucei display a marked deficiency in their ability to form RAD51 foci after DNA damage (27). To date, RAD51 foci have not been described in PCF cells. PCF TREU927 BRCA2 mutant cells were, therefore, treated with BLE (1 μg ml−1 for 18 h) and RAD51 visualized with anti-RAD51 antiserum by immunofluorescence. In the absence of DNA damage, nuclear RAD51 foci were seen in only ∼2% of cells (data not shown), whereas ∼75% of WT and BRCA2+/− cells had detectable foci (1 to >4 foci/nucleus) after BLE treatment (Supplementary Figure S7A), a quantitatively similar response to that of BSF T. brucei (7,8). The brca2−/− cells seemed to have lost the ability to form RAD51 foci (Figure 2C and Supplementary Figure S7), suggesting a key role for BRCA2 in this process, as seen in BSF cells. The ability of BRCA2 BRC variants to support RAD51 foci formation was next examined in both the BSF and PCF cells in the same way (Figure 2C; examples of foci in Supplementary Figure S8). In the absence of DNA damage, RAD51 foci were again rarely seen, irrespective of the BRC repeat number (data not shown). However, after BLE treatment, the number of cells with detectable RAD51 foci increased linearly as the BRC repeat number increased. As described previously (27), BSF cells expressing the 1BRC variant were markedly impaired in the formation of RAD51 foci, which were seen in <10% of these cells. As the number of BRC repeats in the expressed BRCA2 variants increased, the number of cells in the population that displayed RAD51 foci increased, as did the number of foci seen in individual cells. The same trend was also seen in PCF cells, although in this life cycle stage, each BRC variant seemed better at supporting the formation of RAD51 foci than the same variant in BSF cells (e.g. ∼45% of cells had detectable foci when expressing 1BRC). To ensure that these findings are not because of differences in the levels of RAD51 in the BRC variants, western analysis was performed in all cells before and after BLE treatment (Supplementary Figure S9). RAD51 levels seemed equivalent in all the BRC variants (and in the brca2−/− cells) in both life cycle stages, and in none of the cells did we detect an increase in RAD51 expression after BLE-induced DNA damage. These data indicate that the BRC repeat number of T. brucei BRCA2 is a key determinant of the efficiency with which RAD51 is relocalized to foci after BLE damage. Moreover, they may suggest that the need for an expanded array of BRC repeats in BRCA2 is more pronounced in BSF cells, as each BRC repeat variant was less effective at moving RAD51 to BLE damage in these cells relative to PCF cells.
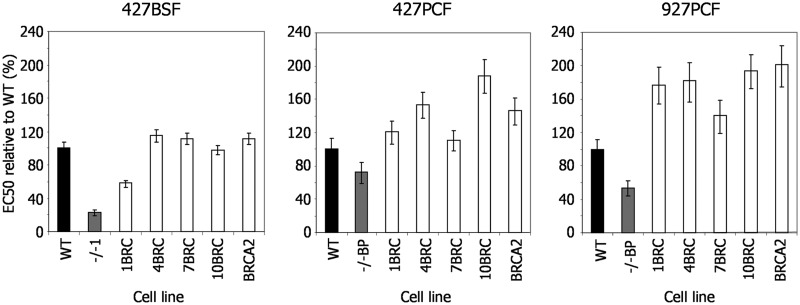
Given the above relationship between RAD51 foci formation efficiency and BRCA2 BRC repeat number, we next asked whether the efficiency of DNA damage repair is also dependent on BRC number. Growth curves showed that the increased pdts of the brca2−/− cells were reverted to WT rates by expression of any BRC variant (Supplementary Figure S10), even in the BSF cells that show a more pronounced growth impediment. Next, we measured the EC50 values of the BSF and PCF BRC variant expressers in BLE and MMS and compared these with WT and brca2−/− mutants (Figure 3 and Supplementary Figure S10). BSF cells expressing the 1BRC variant displayed significantly decreased survival in both BLE and MMS relative to WT cells, suggesting repair deficiency, although their EC50 values were slightly higher than the brca2−/− cells, suggesting some ability to complement the mutation. Nonetheless, this is consistent with the finding that this variant, in this life cycle stage, is largely deficient in RAD51 foci formation (Figure 2C) and HR (27). In contrast, in PCF cells of either strain, there was no evidence for such repair deficiency of the 1BRC variant, which showed EC50 values of at least that of the WT cells. Expression of BRCA2 with 4, 7 or 10 BRC repeats, in either BSF or PCF cells, restored survival of the brca2−/− mutants to at least WT levels, with each BRC variant displayed EC50 values essentially indistinguishable from the cells expressing full-length BRCA2. Thus, although reducing the number of BRC repeats in T. brucei BRCA2 affects the capacity of the protein to mediate relocalization of RAD51 to foci after BLE treatment, this is not reflected in the ability of such variants to support the repair of BLE-induced damage. Indeed, only when the BRC number is reduced to a single unit of the repeat array, is an effect seen on repair efficiency, and even then only in parasites that reside in the mammal.
Figure 3.
Repair efficiency of T. brucei cells expressing BRCA2 variants with altered BRC repeat number. EC50 values after exposure to phleomycin are shown of WT cells, brca2−/− mutants and brca2−/− mutants expressing either BRCA2 variants with altered numbers of BRC repeats (1BRC, 4BRC, 7BRC and 10BRC) or full-length BRCA2 (see Figure 2 for details); values are the mean from three experiments expressed as a percentage of WT, and vertical lines indicate standard error. This analysis was conducted T. brucei Lister427 bloodstream form (427BSF) cells, and in procyclic form cells from Lister427 (427PCF) and TREU927 (927PCF) strains.
Trypanosoma brucei genome instability in BRCA2 mutants is detectable only in mammalian stage parasites
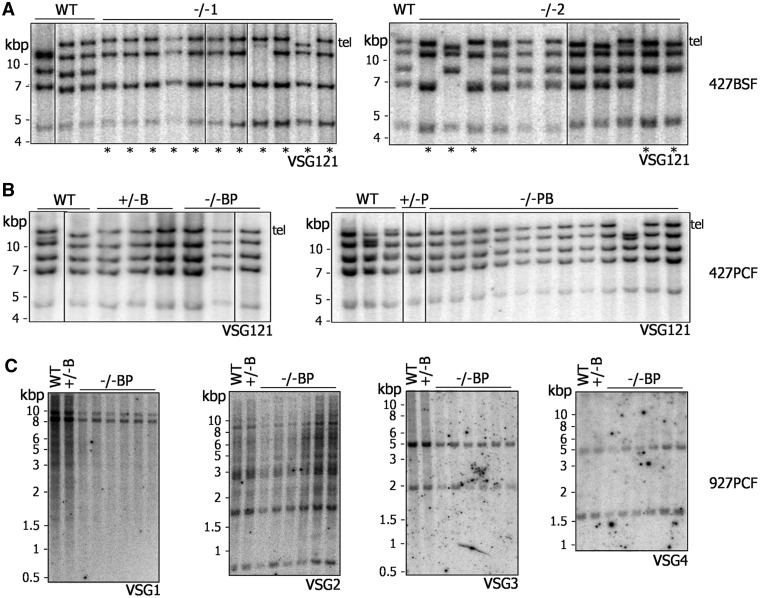
After prolonged growth (∼290 generations), brca2−/− mutants in BSF Lister427 T. brucei display genomic instability, detectable by visible reductions in the size of many megabase-chromosomes, at least some of which arises because of VSG loss (27). Southern analysis of clones of BSF Lister427 brca2−/− mutants after only ∼150 generations revealed loss of one or more copies of the multicopy VSG121 gene (Figure 4A) to the same extent as seen after 2-fold longer growth (27), suggesting instability arises more quickly than previously thought. To date, the VSG repertoire in the T. brucei genome has been positionally annotated only in strain TREU927 (4). To allow a finer examination of the loss of VSGs and genome instability after BRCA2 mutation, PCF TREU927 brca2−/− mutants, as well as WT and +/− mutants, were grown for ∼380 generations, cloned and the genome analysed. Our assumption was that the instability observed previously reflects a general role for BRCA2 in genome maintenance during growth. To our surprise, on using PFGE (Supplementary Figure S11A), we found little evidence for the extensive changes in size of the megabase-chromosomes in the brca2−/− mutants seen previously in BSF cells (27). Southern blots were performed on the clones (Figure 4C), probing with four annotated VSGs (gene IDs: Tb09.244.0140, Tb08.27P2.610, Tb0827P2.530 and Tb09.244.1310). Each of these genes belong to subtelomeric array VSG ‘families’, within which we could predict at least four VSGs that showed 56–100% sequence identity. For each VSG, there was no evidence for either gene loss or rearrangements that led to changes in restriction digest patterns. Thus, even after more extensive growth, brca2−/− mutants in PCF TREU927 cells do not display equivalent levels of genome instability to that seen in BSF Lister427 brca2−/− mutants.
Figure 4.
Genome instability after BRCA2 mutation is seen only in bloodstream form T. brucei. Southern blots are shown of genomic DNA from Lister427 BSF (A), Lister427 PCF (B) and TREU927 PCF (C) T. brucei, comparing WT, BRCA2+/− and brca2−/− clones that had been grown in vitro for ∼150, ∼380 and ∼230 generations, respectively. Lister427 genomic DNA was digested with XmnI, and each blot was probed with VSG121, which is found as a telomeric (Tel) and four putative subtelomeric copies; lanes with clones in which at least one VSG121 copy has been lost are indicated by asterisk. DNA from TREU927 PCF cells was digested with HindIII, and the blots probed with DNA fragments recognizing one of four distinct VSG families, designated VSG1, VSG2, VSG3 or VSG4. Size markers (kb) are indicated.
A potential explanation for the aforementioned dichotomy between BSF and PCF T. brucei could be differences in the relative size or composition of the VSG repertoire in the TREU927 and Lister427 strains. The genome size of TREU927 is among the smallest of the T. brucei strains examined (40), and microarray data suggest that the larger size of the Lister427 genome, as well as chromosome size polymorphisms between the strains, is accounted for by greater numbers of VSGs in the subtelomeres of the megabase-chromosomes (41). Assuming that the main cause of instability in brca2−/− cells is VSG-associated rearrangements, then the putatively smaller VSG repertoire of TREU927 may mean that rearrangements are less frequent or less detectable. Alternatively, the VSG composition of strain Lister427 may contribute to instability: if greater numbers of high sequence identity families of VSGs exist in Lister427 relative to TREU927, then they may be more prone to HR reactions. To test these possibilities, genome stability was examined in clones of PCF Lister427 brca2−/− mutants grown in culture for ∼230 generations. Southern analysis of VSG121 showed none of the characteristic loss of copies seen in the BSF mutants (Figure 4B) (27). PFGE (Supplementary Figure S11B) confirmed this greater stability across the PCF genome: a predominant uniformity in the size of each chromosome in the mutant clones was observed, with only one exception (visible reduction in size of a single chromosome of ∼2.3 Mb in one clone). Taken together, the above data suggest, remarkably, that genome instability after loss of BRCA2 in T. brucei, rather than being common to PCF and BSF cells, is either a BSF-specific process or is more prevalent in that life cycle stage.
Trypanosoma brucei BRCA2 and RAD51 display only partial subnuclear co-localization, even after induced DNA damage
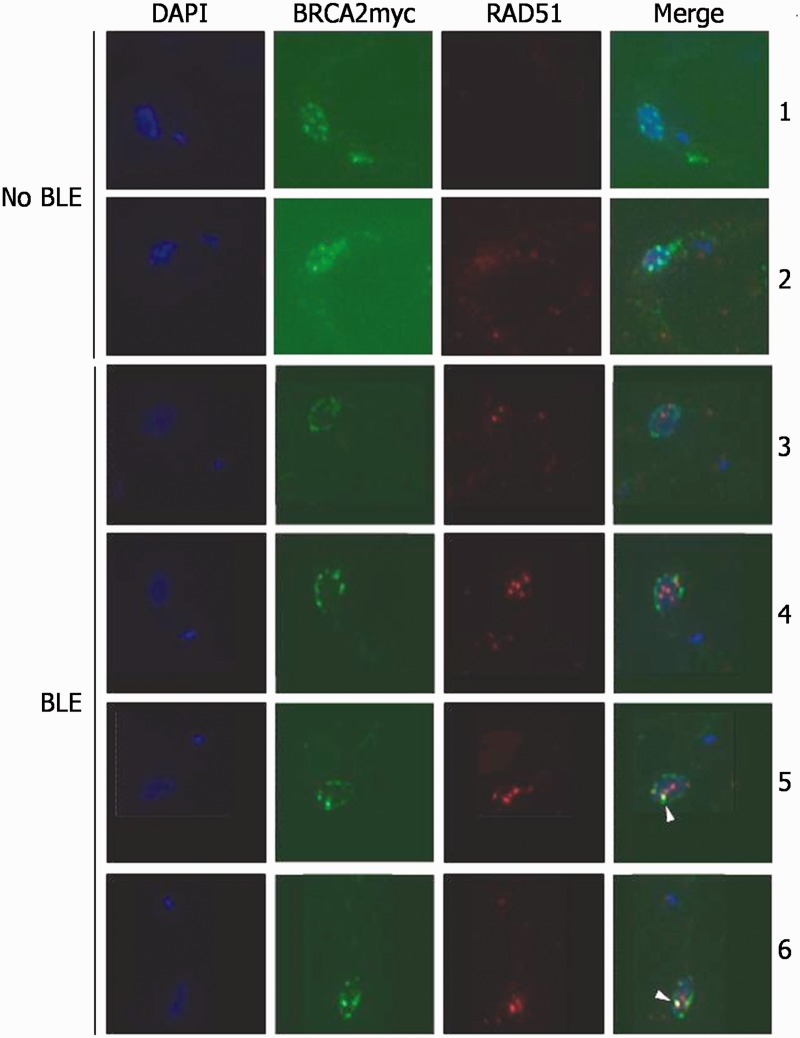
Although T. brucei BRCA2 influences the formation or maintenance of RAD51 nuclear foci after damage, it is not clear whether the two proteins (as well as other repair factors) interact in these putative repair structures in the parasite, as has been described in other eukaryotes (42–44). We, therefore, performed co-immunofluoresence microscopy to localize BRCA2 and RAD51. To do this, we transformed PCF TREU927 BRCA2+/− cells with a construct that modifies the remaining BRCA2 ORF, such that it expresses C-terminally myc-tagged BRCA2 (BRCA2myc). Southern blotting confirmed that the only BRCA2 allele in this strain was myc-tagged, and assaying MMS sensitivity showed that such epitope addition did not impede BRCA2 function (Supplementary Figure S12). The proteins were then visualized (BRCA2 with anti-myc antibody, and RAD51 with polyclonal anti-RAD51 antiserum) in cells grown in the absence of induced DNA damage, and after 5 (Figure 5) or 24 h (data not shown) growth in BLE. Unlike RAD51, for which no signal is seen by co-immunofluoresence in the absence of induced damage, BRCA2myc was readily detected in the nucleus before BLE treatment. In most cells (67%), BRCA2myc was found in multiple foci-like punctate structures (Images 1 and 2, Figure 5), positioned either around the nuclear periphery or throughout the nucleus. In other cells, BRCA2myc localization was less focal, either because staining was visible throughout the nucleus or because no clear subcellular staining was apparent (27%), or because BRCA2myc was seen in peripheral nuclear ‘rings’(6%). After BLE treatment, as has been described previously, RAD51 formed clear subnuclear foci, and the numbers of cells which showed such foci increased between 5 and 24 h (40 and 62% of cells, respectively). The relative localization of BRCA2myc and RAD51 in these conditions can be broadly classified into two distinct types. First, BRCA2myc localization in rings, where the signal was generally continuous around the edge of the DAPI-stained DNA, increased to ∼20% of cells after BLE treatment (either 5 or 24 h). In most of these cells (59%), no RAD51 foci were observed, but when RAD51 foci were seen, the signal was found within the BRCA2myc ring (image 3, Figure 5), and thus the two signals did not overlap. Second, although BLE treatment caused no clear change in the number of cells with punctate BRCA2myc localization, whether spread throughout the nucleus or peripheral, more of these cells displayed RAD51 foci. Despite this, co-localizaton of the two proteins was remarkably rare. In ∼50% of the cells, there was no overlap between the BRCA2myc and RAD51 signals (image 4, Figure 5). Moreover, in the remaining 50% of cells where BRCA2myc and RAD51 co-localization was observed, in no cases was there complete co-localization of all ‘foci’ and, instead, it seemed that co-localization was essentially always between a single BRCA2myc ‘focus’ (that often made up part of a punctate ring) and a single RAD51 focus, frequently out of many (images 5 and 6, Figure 5). These data reveal that T. brucei BRCA2 displays DNA damage-independent localization to discrete subnuclear domains, including focal-like structures, which is not observed for RAD51. In addition, and in contrast to other eukaryotes, DNA damage in T. brucei does not result in extensive co-localization of the two repair factors in putative repair foci; instead, although RAD51 forms such foci in these conditions and they can be abundant in the nucleus, co-localization of BRCA2 and RAD51 is mainly limited to a single detectable focus. The same, limited co-localization is seen in T. brucei BSF cells (data not shown).
Figure 5.
Immunolocalization of BRCA2 and RAD51 in T. brucei. Representative images are shown of BRCA2 and RAD51 localization in individual PCF TREU927 T. brucei grown in the absence of phleomycin (no BLE) or after exposure to phleomycin (2 μg ml−1) for 5 h (BLE). BRCA2 is an endogenously expressed Myc-tagged variant (BRCA2myc) and was detected with anti-Myc Alexa Fluor 488-conjugated antiserum (1:7000 dilution). RAD51 was detected with polyclonal anti-RAD51 antiserum (1:1000 dilution) and secondary hybridization with Alexa Fluor 594 conjugated anti-rabbit antiserum (1:7000 dilution). Each cell is shown after staining with DAPI and as a merge of the BRCA2myc and RAD51 images (Merge). White arrows indicate co-localization between BRCA2myc and RAD51 signals.
Trypanosoma brucei BRCA2 mutants have cell division defects, not found in other DNA repair mutants, that require the C-terminus of the protein
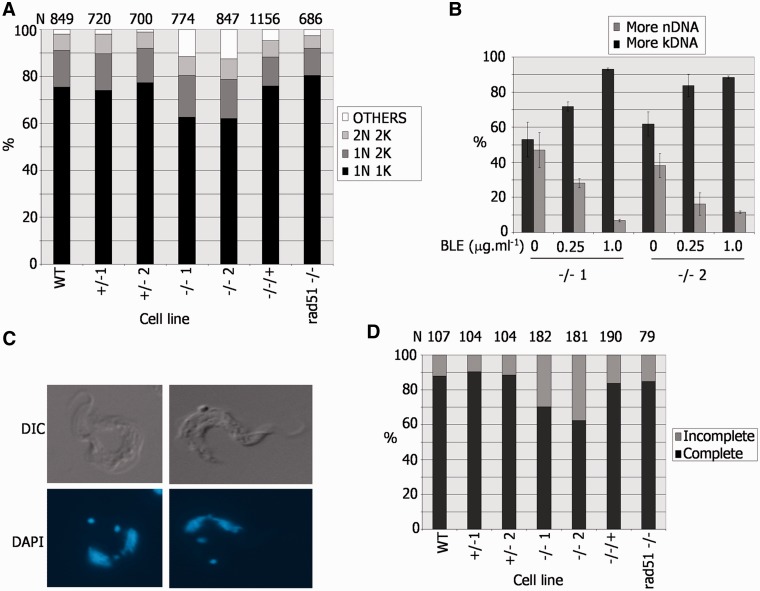
To ask whether we could identify a basis for the pronounced growth impairment of the BSF T. brucei brca2−/− mutants, the cell cycle stage of individual BSF cells was characterized by DAPI staining for DNA content (45), which revealed that the brca2−/– population displayed abnormalities compared with WT, +/− and −/−/+ cells (Figure 6A). The −/− mutants showed an ∼12% reduction in the proportion of cells that contain one nucleus and one kinetoplast (1N1K), which are in the G1 or S phases of the cell cycle (45). This was not associated with a concomitant accumulation of 1N2K or 2N2K cells, showing that the mutants do not arrest in either the G2 or M phases, respectively. Instead, an increase in aberrant cell types (‘others’) was found. Detailed examination showed that this population consisted of roughly equal number of cells with raised kDNA and nDNA content (Figure 6B); raised kDNA content was primarily in the form of cells with 0N1K, 0N2K or 1N3K DNA configurations, whereas those with raised nDNA content were primarily 1N0K, 2N1K or 2N0K cells (data not shown). A number of findings argue that this phenotype is not a straightforward result of the DNA repair deficiency of the brca2−/− cells. First, the same phenotype is not found in T. brucei mutants of other DNA repair/recombination factors that result in growth impairment: comparable analysis of rad51−/− mutants is shown in Figure 6A, and previous work has examined mutants of MRE11 (46) and the RAD51 paralogues, RAD51-3 or RAD51-5 (8). Second, although it is possible that BRCA2 has a more important role in DNA damage repair than these other factors, meaning that only brca2−/− cells manifest a severe enough phenotype to visualize, this is not reflected in obviously greater sensitivity to induced DNA damage compared with other key repair/recombination factors (data not shown). Third, if the phenotype was a consequence of unrepaired DNA damage because of a DNA repair deficiency, then it might be expected that induction of such damage would amplify this phenotype. In fact, the cell cycle pattern observed after BLE treatment of brca2−/− cells was distinct: an accumulation of 1N2K cells, not seen in undamaged cells, was observed (data not shown); in addition, the DNA content of the ‘other’ cells in these conditions displayed a pronounced accumulation of cells with raised kDNA (or lowered nDNA) content (Figure 6B), primarily because of the generation of 0N1K and 1N3K cells (data not shown).
Figure 6.
A DNA replication or segregation defect in T. brucei bloodstream form BRCA2 mutants. (A) Nucleus (N) and kinetoplast (K) configurations of individual cells in a population were analysed by DAPI staining and microscopic examination. The percentage of cells with standard (2N2K, 1N2K and 1N1K) or aberrant (other) N–K configurations is shown in WT Lister427 BSF T. brucei, in BRCA2+/− or brca2−/− mutants, in brca2−/− cells in which full-length BRCA2 was re-expressed (−/−/+), and in rad51−/− mutants; the number of cells counted (N) is shown above the graph. (B). Analysis of aberrant N–K configurations in two independent brca2−/− mutants (1 or 2), detailing whether kDNA or nDNA content was raised, before and after growth for 18 h in phleomycin (BLE). (C) Examples of 2N2K brca2−/− cells undergoing nuclear segregation. (D). Analysis of 2N2K cells from the populations described in A, detailing whether the nuclei were in the process of (incomplete) or had completed (complete), segregation; N is indicated.
Examining 2N2K cells, which are undergoing mitosis and are about to enter cytokinesis, suggested an explanation for the generation of aberrant cells after BRCA2 loss. In WT, BRCA2+/− and BRCA2−/−/+ parasites, the majority of 2N2K cells had clearly separated nuclei, with only ∼10–15% still visibly connected (Figure 6C and D). In contrast, in a larger proportion (30–40%) of brca2−/− 2N2K cells, the two nuclei were visibly connected, and, therefore, were still being segregated. Again, this aberrant phenotype was not seen in rad51−/− mutants (Figure 6D) or in rad51-3−/− or rad51-5−/− mutants (data not shown), suggesting it is not merely a result of impaired growth. As the numbers of 2N2K cells in the brca2−/− mutants were not greater than in WT, BRCA2+/− or BRCA2−/−/+ cells (Figure 6A), this indicates that in the absence of BRCA2 cytokinesis proceeds while nuclear segregation is still occurring, resulting in daughter cells that inherit both nuclei or none.
To dissect the role that BRCA2 plays in this function, we expressed a number of BRCA2 variants in BSF cells, again by integration into the tubulin array in brca2−/− mutants (Figure 7A), which have been described before (27). In brief, full-length BRCA2 from both T. brucei and T. vivax was examined. The latter protein possesses a single BRC repeat, and the two proteins share 26% sequence identity over their whole length, increasing to 42% C-terminal of the BRC repeats. We also examined the 1BRC variant, which retains only the C-terminal repeat (Figure 4). Next, two truncated versions of T. brucei BRCA2 were examined: one encompassed only the BRC repeat region plus the C-terminal predicted nuclear localization signal (NLS); the other encompassed all sequence C-terminal to the BRC repeats, including the same NLS. Finally, we examined a BRCA2 variant in which the T. brucei BRC repeat region plus NLS was translationally fused to the T. brucei RPA 50 kDa subunit, which is homologous to the 70 kDa RPA-1 protein in other eukaryotes. Previously, we have shown that this fusion, which is structurally equivalent to similar fusions examined in U. maydis (47) and mammalian cells (23), is functional in supporting the relocalization of RAD51 to subnuclear foci and in VSG switching, although it is inefficient in HR (27). Integration of each variant was confirmed by Southern analysis, and the expression of each mRNA was shown to be equivalent by northern blotting (27).
Figure 7.
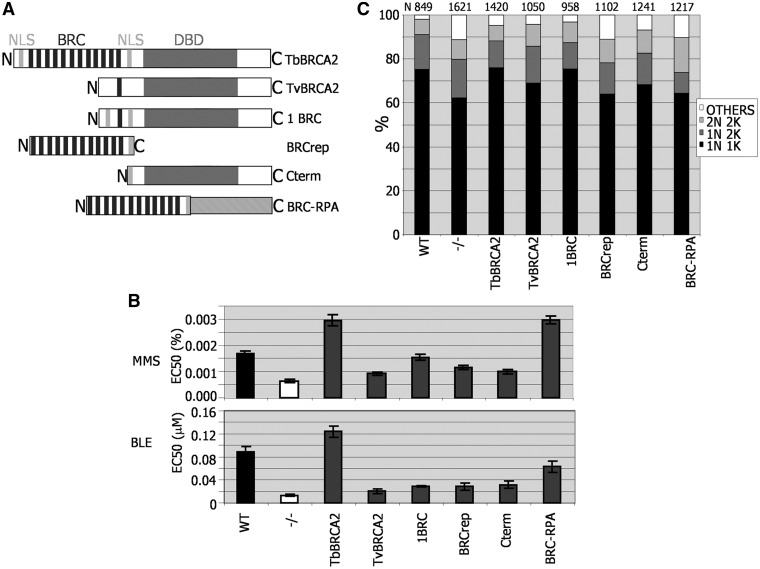
Assessing T. brucei BRCA2 variant function in DNA repair and replication. (A) Diagrammatic representation of BRCA2 variants expressed in Lister427 BSF brca2−/− mutants, using the same strategy as described in Figure 3. The full-length T. brucei BRCA2 used here (TbBRCA2) contains 12 BRC repeats, a conserved DBD and two putative NLS. BRCA2 from T. vivax (TvBRCA2) has a single BRC repeat and a conserved DBD, but no NLSs have been predicted. 1BRC is identical to full-length T. brucei BRCA2, but the BRC array is reduced to a single repeat (Figure 3). BRCrep is a polypetide fragment of T. brucei BRCA2 encompassing only the BRC repeats and 33 downstream amino acids, including a bipartite NLS. Cterm is a 747 amino acid residue N-terminal truncation of T. brucei BRCA2. BRC–RPA is a fusion of the BRCrep polypeptide to the 50 kDa T. brucei replication protein A subunit. (B) The concentration of MMS or phleomycin (BLE) that caused 50% growth inhibition (EC50) of WT cells is compared with brca2−/− mutants, and with −/− cells expressing the BRCA2 variant polypeptides detailed in A. Values are the means from three experiments; bars indicate standard error. (C) The relative proportions of cells with standard (2N2K, 1N2K and 1N1K) or aberrant (other) nuclear (N) and kinetoplast (K) configurations are compared in populations of WT cells, brca2−/− mutants and −/− cells expressing the BRCA2 variant polypeptides detailed in A. N–K configurations were analysed by DAPI staining and microscopic examination; the number of cells counted (N) is indicated.
To examine the ability of the BRCA2 variants to function in DNA repair, the sensitivity of each cell type to MMS and BLE was compared (Figure 7B). As described earlier in the text, ectopic expression of full-length T. brucei BRCA2 by this approach not only complemented the sensitivity of brca2−/− cells to each agent but also (in particular for MMS) resulted in greater resistance than WT cells. The BRC–RPA fusion protein functioned as efficiently as full-length BRCA2 in response to MMS, and it provided partial complementation of BLE sensitivity, indicating that repair can be catalysed in the absence of BRCA2 C-terminal sequence. This is consistent with the ability of this fusion to support RAD51 foci formation after BLE treatment in T. brucei (27) and, more broadly, with the functioning of a BRC–RPA fusion in response to ultraviolet irradiation in U. maydis (47) and in response to mitomycin C or ionising radiation damage in mammals (23). Neither of the BRCA2 variants with only one BRC repeat supported fully efficient repair. Expressing the T. brucei 1BRC variant, as described earlier in the text, partly alleviated the brca2−/− mutant’s sensitivity to BLE and MMS but to a lesser extant that the BRC–RPA fusion. Cells expressing T. vivax BRCA2 were profoundly sensitive to both DNA damaging agents, to an extent comparable with the BRC repeat polypeptide and the polypeptide C-terminal to the repeats.
To test the ability of the BRCA2 variants to provide for efficient cell division, we analysed the cells’ DNA content by DAPI staining, asking whether the different cells displayed the abnormalities noted in the brca2−/− mutants (Figure 7C). The BRC–RPA expresser cells, which were significantly more proficient in DNA repair than the brca2−/− cells, continued to display an increased number of aberrant cells, indicating that the brca2−/− replication defect is not complemented by this protein. In contrast, the cells expressing either of the BRCA2 variants with 1BRC repeat, which were deficient in DNA repair, no longer displayed an increase in ‘others’. Remarkably, the cells that expressed the C-terminus of BRCA2, missing all BRC repeats, showed partial complementation, as the number of ‘others’ was significantly fewer than seen in the brca2−/− mutants and in the BRC–RPA-expressing or BRC repeat polypeptide-expressing cells. Taken together, these findings demonstrate that the replication or cell division deficiency seen in brca2−/− mutants is a consequence of a function that can be separated from the BRC repeats and resides, at least partially, in the C-terminal portion of T. brucei BRCA2.
Reducing RAD51 expression levels in T. brucei leads to DNA repair and recombination defects
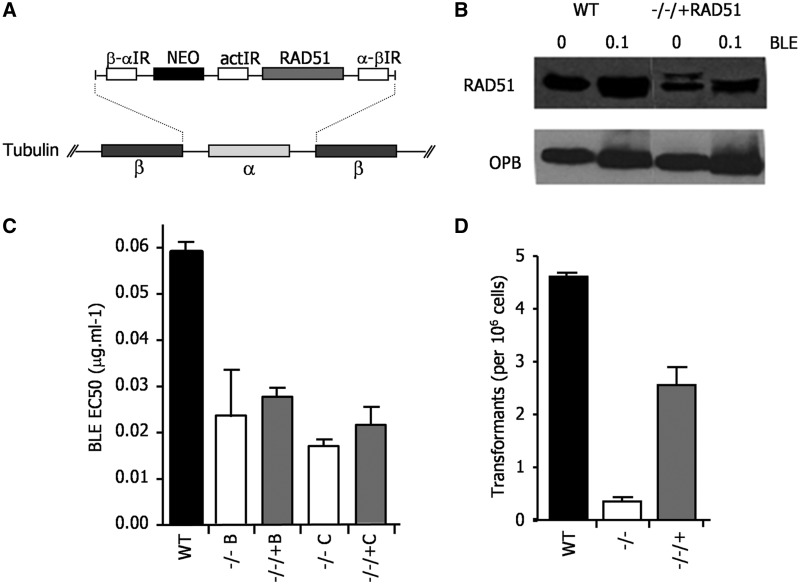
In the above analysis, although we detect visible relocalization of T. brucei RAD51 to subnuclear foci after BLE-induced damage, an increase in the expression of the recombinase is not seen (Supplementary Figures S7 and S9). Moreover, induction of a T. brucei locus-specific DNA DSB also does not result in RAD51 upregulation (48). Such lack of RAD51 expression upregulation is surprising, as this conserved response to DNA damage is seen in closely related kinetoplastid parasites: increased levels of RAD51 mRNA and protein are seen after BLE treatment of Leishmania (35,49) and ionising radiation of T. cruzi (50) (our unpublished data). Given this, we decided to ask whether the abundance of RAD51 is important in T. brucei. To do this, we re-expressed RAD51 in BSF Lister427 rad51−/− mutants using the tubulin integration strategy adopted for BRCA2 (Figure 8A). To ensure reproducibility, two independent rad51−/− mutant clones (Figure 8C) were generated, in each case by two rounds of transformation that were performed in parallel from the same WT cells, each leading to ORF disruption (data not shown); in each mutant, the RAD51 ORF was then re-introduced (−/−/+RAD51). Western blots showed that this expression strategy resulted in reduced levels of RAD51 protein relative to WT cells and that, like in WT, these levels were not altered by BLE treatment (Figure 8B). BSF rad51−/− cells are impaired in in vitro growth and re-expression of RAD51, even at these reduced levels, complemented this deficiency (data not shown). However, each of the RAD51 re-expresser cells were deficient in repair of BLE-induced damage, displaying EC50 values that were close to those of the rad51−/− mutants and ∼2–3-fold lower than WT (Figure 8C). Using a transformation assay described previously (27), it was revealed that limiting the amount of RAD51 also affected the efficiency of HR: the re-expressers were only capable of integrating DNA at ∼60% of the frequency of WT, indicating impairment, although not as severe as in rad51−/− cells (Figure 8D). These data indicate that the levels of RAD51 expressed from the tubulin array are insufficient to support DNA repair and HR at efficiencies seen in WT cells, indicating that, unlike BRCA2, RAD51 abundance seems limiting for these processes.
Figure 8.
Expression of RAD51 from tubulin in T. brucei lowers the cellular levels of the protein and leads to repair and recombination deficiency. (A) RAD51 was re-expressed in Lister427 BSF rad51−/− cells using the same strategy for ORF integration into tubulin used for BRCA2 (see Figure 3). (B) Western blots are shown of whole cell extracts from WT cells and from a RAD51 re-expresser (−/−/+) cells, each grown in the absence or presence of phleomycin (BLE; 1µg ml−1 for 18 h). RAD51 was detected using polyclonal anti-RAD51 antiserum, and the blot was then stripped and re-probed with anti-OPB antiserum to compare loading. (C) The concentration of BLE that caused 50% growth inhibition (EC50) of WT cells is compared with two independently generated rad51−/− mutants (B and C), and with each −/− cell re-expressing RAD51 from tubulin (−/−/+); values are the means from three experiments and bars indicate standard error. (D) To assay recombination, the number of antibiotic resistant clones recovered when the construct tub-HYG-tub (27) was transformed into WT cells, rad51−/− mutants and RAD51 re-expresser (−/−/+) cells was determined; values shown are the means from three experiments (expressed as number of transformants per 106 cells put on selection), and bars indicate standard error.
DISCUSSION
The initial impetus for this study was to understand why BRCA2 in T. brucei, a diverged eukaryote that exploits HR for antigenic variation, has evolved a striking expansion in the number of BRC repeat motifs that mediate interaction with RAD51, the key eukaryotic HR catalytic factor. In examining this, we have revealed a number of further aspects of BRCA2 function. We propose that these findings can be explained by a model (Figure 9) in which the BRC repeat expansion has evolved to allow BRCA2 to operate in dual functions in T. brucei: a putative repair-independent role in the maintenance of the VSG repository, and HR repair of generalized genome damage. Later in the text we outline these findings and discuss this proposal.
Figure 9.
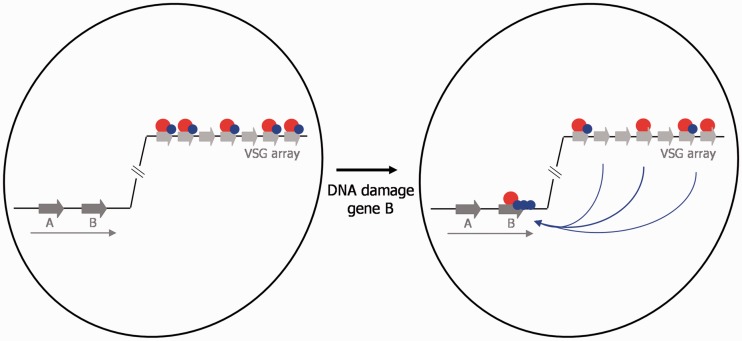
A model for the dual function of BRCA2 in T. brucei genome stability. The genome of T. brucei is represented by a single chromosome (black line inside the nucleus, which is shown as black circle) in which silent VSGs (light grey arrows) in the subtelomere are shown, as well as two genes (A and B; dark grey arrows) that represent the heavily transcribed chromosome core (transcription represented by a long arrow). BRCA2 (red circle) is shown to predominantly localize to the VSGs in undamaged cells, where is acts to ensure stable transmission of these genes and recruits RAD51 (blue circle). After DNA damage in the core (gene B in this example) repair is mediated by BRCA2 acting to relocalize RAD51 to the site of damage, where RAD51 becomes concentrated in microscopically detectable subnuclear foci.
BRC repeats are a critical motif of BRCA2, conserved in all homologues (11), but their detailed function is still unclear. One important question in considering this function is why some organisms, such as mammals, plants and T. brucei, encode BRCA2 homologues with multiple BRC repeats, whereas others, such as C. elegans and U. maydis, function with single-BRC repeat homologues. Taking advantage of the unusual arrangement of the T. brucei BRC repeats as an array, we have been able to generate T. brucei cells expressing BRCA2 variants in which the BRC repeat copy number is systematically reduced. This revealed that BRC repeat number in T. brucei is a critical determinant of the efficiency with which RAD51 localizes to visible subnuclear foci after induced DNA damage. This is not because BRCA2 acts to transit RAD51 to the T. brucei cell nucleus, which seems to be a functional divergence from human and Leishmania BRCA2 (34,35). Instead, T. brucei BRCA2’s role via the BRC repeats is in the subnuclear redistribution of RAD51. Despite this, reducing the BRC repeat number has a remarkably limited effect on the efficiency of DNA damage repair. BRCA2 with a single-BRC repeat is repair-deficient in BSF T. brucei cells, whereas variants with four or more repeats function as efficiently as BRCA2 with a full complement of 12 BRC repeats. In PCF cells, even one BRC repeat can support fully efficient DNA damage repair. This provides a more complete picture of the functional significance of the BRCA2 BRC repeat expansion that we reported previously (27), and indicates that this adaptation is not because of T. brucei-specific aspects of general HR repair.
Gross chromosomal rearrangements (GCRs) are a hallmark phenotype of BRCA2 mutation or loss: these are manifest as chromosome loss, breakage and translocation (51) in mammals, and similar genome instability is seen in U. maydis (12) and during meiosis in A. thaliana (52) and C. elegans (53). In T. brucei, loss of BRCA2 also leads to genome instability (27). However, we now show that this is limited to mammal-derived BSF cells, with PCF cells derived from the tsetse fly midgut showing no evidence for substantial chromosome alterations. Such a life cycle stage- or cell type-specific role for BRCA2 in genome stability is unprecedented to our knowledge. It seems likely that this is because the BSF instability is not because of genome-wide GCRs, but is limited to the VSG repository. Intermediate and mini-chromosomes, which do not harbour silent VSG arrays, show no instability in brca2−/− BSF cells (27). Furthermore, in an attempt to track rearrangements in the megabase-chromosomes, we probed Southern blots of BSF brca2−/− mutant clones for ingi retrotransposons, which are found in non-coding regions throughout the T. brucei genome, including within the subtelomeric VSGs arrays (54). No evidence for rearrangements was seen (Supplementary Figure S13), indicating that genome instability may largely be limited to loss of VSGs, which is consistent with the predominant reduction in chromosome size in the mutants (27) and may reflect the plasticity of the VSG repertoire between and within T. brucei strains (41).
Examination of the DNA content of individual BSF brca2−/− cells indicates that T. brucei BRCA2 provides genome maintenance functions that can be separated from DNA repair. Unique among mutants of T. brucei DNA repair factors examined to date, brca2−/− mutants display an accumulation of cells with abnormal ratios of nuclear (N) and kinetoplast (K) DNA. These do not arise because of changes in the number of cells with 1N2K and 2N2K composition, indicating that this phenotype is a consequence of impairment in nuclear DNA replication or segregation (separation of the nucleus or nuclear genome during mitosis), and does not affect the kinetoplast. A number of lines of evidence indicate that DNA replication/segregation and DNA damage repair are separate functions of BRCA2. First, BRCA2 variants with only one BRC repeat are impaired in repair of DNA damage in BSF cells, but function normally in DNA replication/segregation. Second, a fusion of BRC–RPA, as has been seen in other organisms (23,47), is capable of functioning in T. brucei DNA repair and in localizing RAD51 to subnuclear foci (27), but cannot complement the replication/segregation defect of brca2−/− cells. This suggests that the C-terminus of BRCA2 (downstream of the BRC repeats) is needed for this role, and, indeed, we find that this C-terminal region, when expressed in isolation, is able to partially complement the replication/segregation deficiency. These data seem consistent with findings from vertebrate BRCA2 studies. Schlacher et al. (25) have shown that the C-terminus of mammalian BRCA2 acts to stabilize Rad51 filaments that form at stalled replication forks, protecting them from MRE11 degradation; indeed, a BRC–RPA fusion was impaired in this process, consistent with the lack of functionality we observed for an equivalent T. brucei fusion in replication/segregation. It is likely that such a role is related to the demonstration that Xenopus Rad51 binds ssDNA gaps that can form behind replication forks, again protecting them from MRE11 degradation (26). Our work may then reveal that this BRCA2 function is widely conserved in eukaryotes, with some caveats. We do not yet know whether T. brucei BRCA2’s replication/segregation function underlies the genome instability we observe; therefore, it is unclear whether this is BSF-specific or is a general role whose importance becomes more pronounced in this life cycle stage. In addition, we do not yet know whether T. brucei RAD51 acts in this function (see later in the text), although it is notable that although the C-terminus of T. brucei BRCA2 is highly diverged from vertebrates, it does interact with the recombinase (our unpublished data) and that it has been suggested to bind CDC45 (55), perhaps providing a direct link with the replication machinery. Finally, we cannot exclude the possibility that T. brucei BRCA2’s role is due to a function in mitosis or cytokinesis, activities that some work in vertebrate cells has linked BRCA2 with (24,56–59), although we find no evidence for specific localization of BRCA2 in mitotic (2N2K) cells (data not shown).
The function of BRCA2 in DNA damage repair has been closely linked with Rad51 through cytological studies of repair foci induced by exogenous DNA damage. In the absence of induced damage, mammalian BRCA2 is nuclear but shows little subnuclear localization (60), whereas after damage, BRCA2 relocalizes to discrete foci that co-localize extensively with Rad51 foci (61). The same behaviour is seen for Drosophila BRCA2 and Rad51 (44). Although co-localization of Rad51 and Brh2/BRCA2 has not been described in U. maydis, each factor localizes to detectable subnuclear foci at similar times after DNA damage (62). The subnuclear dynamics of T. brucei BRCA2 and RAD51 differ from these findings in several important ways. First, T. brucei BRCA2 in the absence of induced damage displays substantial nuclear localization, predominantly in punctate, focal-like structures around the nuclear periphery or throughout the nucleus, as well as in peripheral ‘rings’. This extensive nuclear localization contrasts with RAD51, which is detectable in subnuclear foci rarely (∼2% of cells) in these conditions. This dichotomy is not because of detection of BRCA2 via a Myc epitope, as RAD51 tagged in the same way displays the same behaviour as endogenous RAD51 detected by polyclonal antiserum (our unpublished data). Although it is formally possible that the tagging strategy altered BRCA2 protein levels, for instance, by changing mRNA abundance because of altering the 3′-UTR, we have not seen any indication of this. Second, T. brucei BRCA2 and RAD51 rarely co-localize in multiple foci after BLE-induced damage. Although both proteins can be seen in multiple foci in a single cell, we rarely detect co-localization beyond a single focus. These findings are compatible with BRCA2 in the parasite acting both in DNA repair and in an additional role. Co-localization could represent T. brucei BRCA2 establishing RAD51 repair foci and then moving away, leaving the recombinase foci in place. The subnuclear localization of T. brucei BRCA2 that is distinct from RAD51 may be because of a pronounced genome maintenance role; RAD51 may be recruited to this function without forming cytologically detectable foci or it may not act.
Taken together, we suggest that above data are consistent with a model (Figure 9) in which T. brucei BRCA2, like that of mammals, acts through the C-terminus to ensure correct transmission of nuclear DNA. In T. brucei, this has additional importance in maintenance and transmission of the VSG-containing subtelomeres, which can be peripheral in the nucleus (63) and would explain the persistent localization we see of BRCA2. Moreover, this role has selected for an expanded array of BRC repeats, which are needed to allow BRCA2 to be re-directed from this function to act in general repair when needed. This would match the BRCA2 variant data that we describe. The T. brucei BRCA2 variant in which the normal C-terminus has been replaced, like the same fusion in mammals, is free to mediate repair via the BRC repeats but does not act in replication/segregation (25). Furthermore, in T. brucei, a BRCA2 variant with a single BRC repeat linked to the normal C-terminus is repair-deficient in BSF cells because it acts preferentially in replication/segregation, to the detriment of repair. We suggest that T. brucei BRCA2’s C-terminus, like that of mammalian BRCA2, acts in this role through targeting RAD51. The importance of this role has selected for T. brucei RAD51 expression becoming non-responsive to damage (and thus distinct from T. cruzi and Leishmania) (49,50), leading to an abundant pool of the recombinase. This would explain why BRCA2 variants with increasing BRC repeat number display greater numbers of RAD51 foci: when DNA damage is present in excess (e.g. through treatment with BLE), these variants are increasingly capable of relocating the abundant RAD51 from the replication/segregation role to sites of damage. In addition, if RAD51 is preferentially targeted to the transmission role, then reducing the expression level of RAD51 would lead to the observed repair deficiency. The problems associated with maintenance or transmission of the subtelomeric VSG arrays are, as yet, unknown. One possibility is that there is frequent and unseen recombination among the VSGs, leading to gaps that must be protected to avoid them leading to gene loss, as seen in brca2−/− mutants. Whether this is associated with replication is unknown, although we see no evidence that the BRCA2 localization alters in S-phase cells (data not shown). This process may be common to both PCF and BSF cells, as the punctate BRCA2 localization is seen in the former. However, we suggest that the frequency of the putative gaps increases in BSF cells, either because the VSG arrays undergo a change to make them more ‘fragile’ or because the VSG expression sites begin to ‘sample’ the VSG arrays during antigenic variation, placing a further burden on BRCA2 and RAD51. This would explain why VSG instability in brca2−/− mutants is seen specifically in BSF cells, why PCF brca2−/− mutants have less severe repair defects and why BRC variants with lowered BRC repeat number are more capable of redirecting RAD51 to induced damage in PCF cells than in BSF cells.
The model for BRCA2 function we propose is supported by a recent comparison of VSG composition in T. brucei, T. congolense and T. vivax (64). Each of these Trypanosoma species contains a repository of VSGs that allows immune evasion by antigenic variation, but we have previously reported that this is not reflected in a common BRCA2 architecture: T. congolense BRCA2 possesses three highly sequence-related arrayed BRC repeats, whereas T. vivax BRCA2 has a single BRC repeat (27). These differences in BRCA2 organization can now be explained, as differences in VSG recombination frequency and mechanism are found between the species (64). In T. brucei and T. congelense, recombination between VSGs is more frequent than in T. vivax. Furthermore, in T. brucei recombination seems to be able to occur relatively freely throughout the VSG repository, leading to a greater number of VSG pseudogenes, whereas in T. congolense, the VSGs fall into more clearly defined clades, with fewer pseudogenes, indicative of recombination that is more restricted. Our work shows that this ongoing VSG diversification, which is accelerated in T. brucei, has had an impact on the structure of a core element of the HR machinery, emphasizing how antigenic variation and HR intersect.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–13.
FUNDING
The Wellcome Trust [083223, 089172] and from the Medical Research Council. The Wellcome Trust Centre for Molecular Parasitology is supported by core funding from the Wellcome Trust [085349]. Funding for open access charge: the Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to David Horn, Sam Alsford, Lucy Glover and many colleagues in the University of Glasgow for their comments, and to Marilyn Parsons and Jeremy Mottram for the gift of anti-NOG1 and anti-OPB antiserum.
REFERENCES
- 1.Vink C, Rudenko G, Seifert HS. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol. Rev. 2011 doi: 10.1111/j.1574-6976.2011.00321.x. Dec 25 (doi: 10.1111/J.1574-6976.2011.00321.x;epup ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009;11:1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 3.Becker M, Aitcheson N, Byles E, Wickstead B, Louis E, Rudenko G. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 2004;14:2319–2329. doi: 10.1101/gr.2955304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobson R, Stockdale C, Lapsley C, Wilkes J, McCulloch R. Interactions among Trypanosoma brucei RAD51 paralogues in DNA repair and antigenic variation. Mol. Microbiol. 2011;81:434–456. doi: 10.1111/j.1365-2958.2011.07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proudfoot C, McCulloch R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res. 2005;33:6906–6919. doi: 10.1093/nar/gki996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HS, Cross GA. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000992. doi: 10.1371/journal.ppat.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HS, Cross GA. Identification of Trypanosoma brucei RMI1/BLAP75 homologue and its roles in antigenic variation. PLoS One. 2011;6:e25313. doi: 10.1371/journal.pone.0025313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amst.) 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 12.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 13.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc. Natl Acad. Sci. USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 15.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 16.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 17.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Q, Kojic M, Cao Z, Lisby M, Mazloum NA, Holloman WK. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol. Cell Biol. 2007;27:2512–2526. doi: 10.1128/MCB.01907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lord CJ, Ashworth A. RAD51, BRCA2 and DNA repair: a partial resolution. Nat. Struct. Mol. Biol. 2007;14:461–462. doi: 10.1038/nsmb0607-461. [DOI] [PubMed] [Google Scholar]
- 20.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl Acad. Sci. USA. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl Acad. Sci. USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, Han M, Zdzienicka MZ, Stark JM, Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl Acad. Sci. USA. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayoub N, Rajendra E, Su X, Jeyasekharan AD, Mahen R, Venkitaraman AR. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr. Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol. Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dray E, Siaud N, Dubois E, Doutriaux MP. Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 2006;140:1059–1069. doi: 10.1104/pp.105.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett SM, Noor MA. Molecular evolution of a Drosophila homolog of human BRCA2. Genetica. 2009;137:213–219. doi: 10.1007/s10709-009-9376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alsford S, Horn D. Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 2008;161:76–79. doi: 10.1016/j.molbiopara.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68:139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
- 32.Zeiner GM, Sturm NR, Campbell DA. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell. 2003;2:222–230. doi: 10.1128/EC.2.2.222-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giloni L, Takeshita M, Johnson F, Iden C, Grollman AP. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J. Biol. Chem. 1981;256:8608–8615. [PubMed] [Google Scholar]
- 34.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 35.Genois MM, Mukherjee A, Ubeda JM, Buisson R, Paquet E, Roy G, Plourde M, Coulombe Y, Ouellette M, Masson JY. Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2012;40:6570–6584. doi: 10.1093/nar/gks306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munday JC, McLuskey K, Brown E, Coombs GH, Mottram JC. Oligopeptidase B deficient mutants of Leishmania major. Mol. Biochem. Parasitol. 2011;175:49–57. doi: 10.1016/j.molbiopara.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JH, Jensen BC, Kifer CT, Parsons M. A novel nucleolar G-protein conserved in eukaryotes. J. Cell Sci. 2001;114:173–185. doi: 10.1242/jcs.114.1.173. [DOI] [PubMed] [Google Scholar]
- 38.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair. 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, Centore R, Boonsombat R, Sandler SJ. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol. Microbiol. 2005;57:1074–1085. doi: 10.1111/j.1365-2958.2005.04755.x. [DOI] [PubMed] [Google Scholar]
- 40.Melville SE, Leech V, Gerrard CS, Tait A, Blackwell JM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- 41.Callejas S, Leech V, Reitter C, Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 2006;16:1109–1118. doi: 10.1101/gr.5147406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, Jenkins NA, Lalande M, Alt FW. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl Acad. Sci. USA. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 44.Brough R, Wei D, Leulier S, Lord CJ, Rong YS, Ashworth A. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair. 2008;7:10–19. doi: 10.1016/j.dnarep.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 45.McKean PG. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr. Opin. Microbiol. 2003;6:600–607. doi: 10.1016/j.mib.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Robinson NP, McCulloch R, Conway C, Browitt A, Barry JD. Inactivation of Mre11 does not affect VSG gene duplication mediated by homologous recombination in Trypanosoma brucei. J. Biol. Chem. 2002;277:26185–26193. doi: 10.1074/jbc.M203205200. [DOI] [PubMed] [Google Scholar]
- 47.Kojic M, Zhou Q, Lisby M, Holloman WK. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol. Cell Biol. 2005;25:2547–2557. doi: 10.1128/MCB.25.7.2547-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glover L, McCulloch R, Horn D. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 2008;36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKean PG, Keen JK, Smith DF, Benson FE. Identification and characterisation of a RAD51 gene from Leishmania major. Mol. Biochem. Parasitol. 2001;115:209–216. doi: 10.1016/s0166-6851(01)00288-2. [DOI] [PubMed] [Google Scholar]
- 50.Regis-da-Silva CG, Freitas JM, Passos-Silva DG, Furtado C, Augusto-Pinto L, Pereira MT, DaRocha WD, Franco GR, Macedo AM, Hoffmann JS, et al. Characterization of the Trypanosoma cruzi Rad51 gene and its role in recombination events associated with the parasite resistance to ionizing radiation. Mol. Biochem. Parasitol. 2006;149:191–200. doi: 10.1016/j.molbiopara.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 51.Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 52.Dumont M, Massot S, Doutriaux MP, Gratias A. Characterization of Brca2-deficient plants excludes the role of NHEJ and SSA in the meiotic chromosomal defect phenotype. PLoS One. 2011;6:e26696. doi: 10.1371/journal.pone.0026696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko E, Lee J, Lee H. Essential role of brc-2 in chromosome integrity of germ cells in C. elegans. Mol. Cells. 2008;26:590–594. [PubMed] [Google Scholar]
- 54.Bringaud F, Ghedin E, El Sayed NM, Papadopoulou B. Role of transposable elements in trypanosomatids. Microbes. Infect. 2008;10:575–581. doi: 10.1016/j.micinf.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Oyola SO, Bringaud F, Melville SE. A kinetoplastid BRCA2 interacts with DNA replication protein CDC45. Int. J. Parasitol. 2008;39:59–69. doi: 10.1016/j.ijpara.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Lee H, Trainer AH, Friedman LS, Thistlethwaite FC, Evans MJ, Ponder BA, Venkitaraman AR. Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol. Cell. 1999;4:1–10. doi: 10.1016/s1097-2765(00)80182-3. [DOI] [PubMed] [Google Scholar]
- 57.Futamura M, Arakawa H, Matsuda K, Katagiri T, Saji S, Miki Y, Nakamura Y. Potential role of BRCA2 in a mitotic checkpoint after phosphorylation by hBUBR1. Cancer Res. 2000;60:1531–1535. [PubMed] [Google Scholar]
- 58.Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA, Yen TJ, Shiekhattar R. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 59.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 60.Jeyasekharan AD, Ayoub N, Mahen R, Ries J, Esposito A, Rajendra E, Hattori H, Kulkarni RP, Venkitaraman AR. DNA damage regulates the mobility of Brca2 within the nucleoplasm of living cells. Proc. Natl Acad. Sci. USA. 2010;107:21937–21942. doi: 10.1073/pnas.1009577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kojic M, Zhou Q, Lisby M, Holloman WK. Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis. Mol. Cell Biol. 2006;26:678–688. doi: 10.1128/MCB.26.2.678-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung HM, Shea C, Fields S, Taub RN, Van der Ploeg LH, Tse DB. Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 1990;9:2611–2619. doi: 10.1002/j.1460-2075.1990.tb07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson AP, Berry A, Aslett M, Allison HC, Burton P, Vavrova-Anderson J, Brown R, Browne H, Corton N, Hauser H, et al. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc. Natl Acad. Sci. USA. 2012;109:3416–3421. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.