Abstract
Recognition of the 3′-splice site is a key step in pre-mRNA splicing and accomplished by a dynamic complex comprising splicing factor 1 (SF1) and the U2 snRNP auxiliary factor 65-kDa subunit (U2AF65). Both proteins mediate protein–protein and protein–RNA interactions for cooperative RNA-binding during spliceosome assembly. Here, we report the solution structure of a novel helix-hairpin domain in the N-terminal region of SF1 (SF1NTD). The nuclear magnetic resonance- and small-angle X-ray scattering-derived structure of a complex of the SF1NTD with the C-terminal U2AF homology motif domain of U2AF65 (U2AF65UHM) reveals that, in addition to the known U2AF65UHM–SF1 interaction, the helix-hairpin domain forms a secondary, hydrophobic interface with U2AF65UHM, which locks the orientation of the two subunits. Mutational analysis shows that the helix hairpin is essential for cooperative formation of the ternary SF1–U2AF65–RNA complex. We further show that tandem serine phosphorylation of a conserved Ser80-Pro81-Ser82-Pro83 motif rigidifies a long unstructured linker in the SF1 helix hairpin. Phosphorylation does not significantly alter the overall conformations of SF1, SF1–U2AF65 or the SF1–U2AF65–RNA complexes, but slightly enhances RNA binding. Our results indicate that the helix-hairpin domain of SF1 is required for cooperative 3′-splice site recognition presumably by stabilizing a unique quaternary arrangement of the SF1–U2AF65–RNA complex.
INTRODUCTION
Removal of non-coding sequences (introns) from pre-mRNAs is a key step in mammalian gene expression performed by the spliceosome, a large and dynamic ribonucleoprotein particle. Alternative splicing is essential to generate different proteins from a given primary transcript by differential inclusion or exclusion of coding sequences (exons) (1). Rather weakly conserved RNA sequence motifs located at the 5′- and 3′-ends of an intron are first recognized by splicing factors (SFs) in the early complex E during the assembly of the spliceosome, which is then converted into complexes A, B and finally into complex C where splicing catalysis occurs (2). Given the importance of splicing for gene expression, numerous diseases have been linked to aberrant splicing and mutations within the consensus sequences at the 5′- and 3′-ends of introns interfere with spliceosome assembly (3). Recently, frequent missense mutations in genes of SFs that mediate 3′-splice site recognition, such as SF1, and the U2 snRNP auxiliary factor (U2AF) large (U2AF65) and small subunits (U2AF35) have been linked to tumourigenesis (4).
The regulation of splicing by extracellular signals and signal transduction cascades is poorly understood. With respect to 3′-splice site recognition, phosphorylation of conserved serine residues (Ser20, Ser80 and Ser82) in SF1 has been implicated in regulating the stability of the ternary SF1–U2AF65–RNA complex in vitro (5,6). However, both the structural and functional roles of SF1 phosphorylation remain elusive.
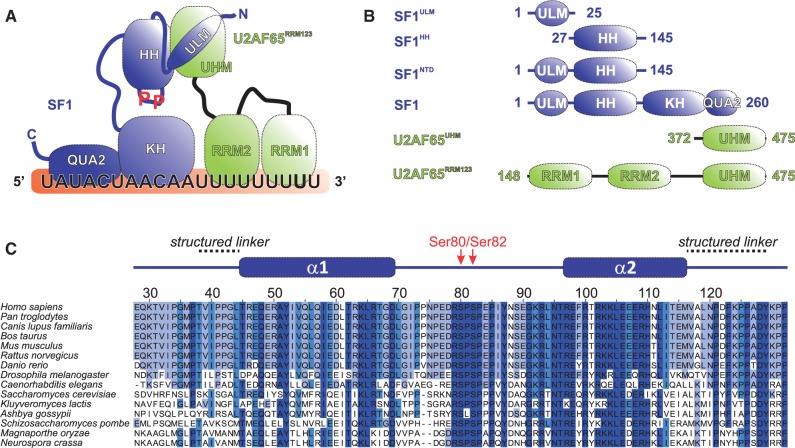
Early recognition of the intron/exon junctions is a key regulatory step in splicing. In the initial complex E, the U1 snRNP binds to a short stretch of 6 nt at the 5′-splice site, whereas the 3′-splice site is defined by the binding of SF1 to the branch point sequence (BPS), of U2AF65 to the poly-pyrimidine tract (Py tract), and of U2AF35 to the AG dinucleotide defining the 3′-splice site, with the additional involvement of proof-reading factors (7,8) (Figure 1A). The structure of the U1 snRNP involved in 5′-splice site recognition has been reported recently (9). Structural information about 3′-splice site recognition is so far limited to binary protein–protein and protein–RNA complexes (10–14), involving SF1, U2AF and RNA. However, given the dynamic nature of protein–RNA interactions that mediate 3′-splice site recognition (12), its complete structural analysis should include solution approaches to detect and characterize functionally relevant conformational dynamics (15). Details of intron RNA recognition are available based on 3D structures of the RNA-binding regions of SF1 (comprising the KH and QUA2 domains, SF1KHQUA2) bound to BPS RNA (10), and of the tandem U2AF65 RRM1–RRM2 domains (U2AF65RRM1,2) with Py tract RNA (12). Structural details of a protein–protein interaction between U2AF65 and SF1 have been reported (11). These involve the non-canonical U2AF65 RRM3, a founding member of the U2AF homology motif domain (U2AF65UHM) (16) and a tryptophan-containing peptide sequence, called UHM ligand motif (17), in SF1 (SF1ULM) (11) (Figure 1B).
Figure 1.
Proteins and domains involved in 3′-splice site recognition. (A) Diagram of the SF1–U2AF65 3′-splice site recognition complex representing domains of SF1 and U2AF65 (HH, helix hairpin; KH, hnRNP K homology; QUA2, quaking homology-2; RRM, RNA recognition motif; ULM, U2AF homology domain (UHM) ligand motif). SF1 and U2AF65 are coloured blue and green, respectively, with the same colour code for the proteins being used in the NMR spectra throughout the article. (B) Diagram of the protein constructs used. (C) Sequence alignment of SF1HH domains. Sequences were taken from the UniProt database (www.uniprot.org) and residue numbers are given for Homo sapiens SF1. The secondary structure of SF1HH is depicted above the alignment. The phosphorylated serine residues (Ser80 and Ser82) are indicated in red. Sequences were aligned with ClustalW (18) and analysed with Jalview 2 (19).
Here, we present structural and biochemical analyses of novel interactions in the SF1–U2AF65 complex in solution by combining nuclear magnetic resonance (NMR) spectroscopy and small-angle X-ray scattering (SAXS). We show that the N-terminus of SF1 (SF1NTD) comprises a helix hairpin fold (SF1HH) providing an additional binding interface to U2AF65UHM. This interface involves mainly hydrophobic interactions between the SF1NTD helical hairpin and U2AF65UHM. SF1NTD is essential for cooperative formation of the ternary SF1–U2AF65–RNA complex. To assess the effects of the recently reported tandem serine phosphorylation of a Ser80–Pro81–Ser82–Pro83 (SPSP) motif in SF1NTD, we performed structural and functional studies of phosphorylated SF1 alone and in complex with U2AF65. These studies show that phosphorylation does not noticeably affect the conformation of SF1 or SF1–U2AF65 and does not modulate cooperative RNA binding, thus suggesting additional roles for SF1 phosphorylation. Our results represent a significant step towards solving the structure of the ternary SF1–U2AF65–RNA complex and thus understanding the molecular basis of 3′-splice site recognition.
MATERIALS AND METHODS
Protein expression and NMR sample preparation
Plasmids for expressing human U2AF65UHM (residues 372–475), U2AF65RRM123 (residues 147–475), as well as human SF1ULM (residues 1–25), SF1HH (residues 27–145), SF1NTD (residues 1–145) and SF1 (residues 1–260) were prepared in a modified pET-M11 vector containing an N-terminal His6 tag followed by a tobacco etch virus (TEV) protease cleavage site. SF1 deletions of helix α1 (Δ35–68), α2 (Δ96–128), the complete helix hairpin (Δ35–128) or the α1–α2 linker (Δ75–90) were generated by polymerase chain reaction (PCR) of SF12-320 in pTRCHis A (20) with reverse and forward primers complementary to sequences upstream and downstream, respectively, of the desired deletion. The primers contained KpnI restriction sites and PCR products were ligated after KpnI digestion. The deleted protein sequences were thus replaced by Gly–Thr. Full-length KIS kinase (comprising the kinase domain and a UHM domain) was cloned by PCR ligation of the DNA encoding the short KIS isoform (residues 1–344) and the KIS UHM (residues 315–419). All plasmids were verified by sequencing.
Expression plasmids were transformed into Escherichia coli BL21(DE3) cells, grown in standard LB medium or in minimal M9T medium supplemented with 2 g/l [U-13C]-glucose and/or 1 g/l [15N]-ammonium chloride as the sole carbon and nitrogen sources. SF1 (residues 1–260) and U2AF65RRM123 were prepared as [U-2H,15N]-labelled proteins for NMR titrations and relaxation measurements or as [U-2H,13C,15N]-deuterated samples with methyl protonation of isoleucine, leucine and valine residues as described (21) for chemical shift assignments. Protein synthesis was induced by the addition of 0.5 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) at OD600 ∼0.8. After protein expression for 16 h at 25°C, cells were collected by centrifugation, lysed by sonication in the presence of lysozyme and ethylenediaminetetraacetic acid (EDTA)-free ‘complete protease inhibitor’ (Roche Applied Science), then re-suspended in binding buffer consisting of 50 mM Tris (pH 8.0), 500 mM NaCl, 5% (v/v) glycerol and 5 mM imidazole. The sample was loaded onto Ni-NTA chromatography resin (Qiagen) and washed with 20 column volumes of binding buffer followed by five column volumes of the same buffer but with 25 mM imidazole, and then the sample was eluted with the buffer containing 250 mM imidazole. The His6-tag was cleaved by incubating samples with 0.1 mg TEV proteinase/mg protein sample and 2 mM DTT at 4C for 12 h. The TEV protease, the histidine-tag and uncleaved protein were removed by a second passage of the sample through Ni-NTA resin. The eluate was further purified by gelfiltration on a Superdex 75 column (GE) using sodium phosphate (pH 6.5), 50 mM NaCl, 1 mM Dithiothreitol (DTT) as buffer. NMR samples were concentrated from 100 to 600 µM in NMR buffer consisting of 20 mM sodium phosphate (pH 6.5), 50 mM NaCl, 0.1% sodium azide, 1 mM DTT and 1 mM EDTA.
KIS kinase was expressed in standard LB medium following the same protocol. The protein was purified at 4°C without cleavage and removal of the His6-tag. Aliquots of the recombinant KIS kinase were stored at −80°C in buffer consisting of 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 8.0), 15 mM MgCl2, 25% (v/v) glycerol, 2 mM DTT and 1 mM EDTA.
His6-tagged SF12-320 and deletion mutants for gel shift experiments were expressed in E. coli and purified as described (20). Proteins were dialyzed against 20 mM Hepes–KOH pH 7.9, 100 mM KCl, 20% (v/v) glycerol, 0.2 mM EDTA and 0.5 mM DTT.
A 20-mer RNA containing the BPS and the Py tract (5′-UAUACUAACAAUUUUUUUUU-3′) was purchased from BioSpring GmbH, dissolved in H2O to a final concentration of 10 mM and added to the protein samples before the NMR and SAXS measurements.
In vitro phosphorylation
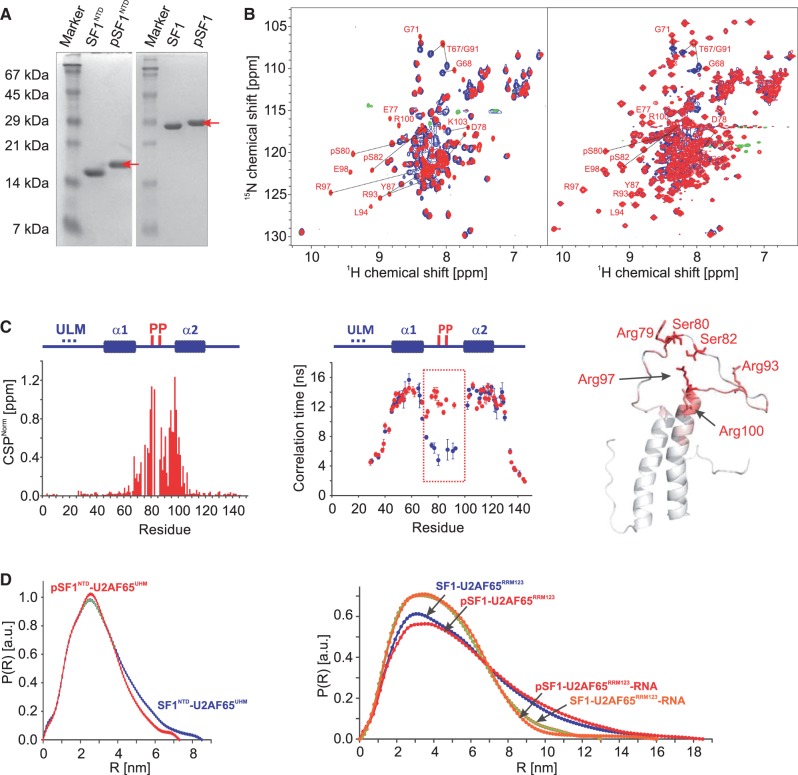
Phosphorylation of SF1NTD and SF1 for NMR analysis was performed in 10 ml kinase buffer containing 50 mM MES (pH 8.0), 15 mM MgCl2, 25% (v/v) glycerol, 5 mM DTT, 1 mM EDTA, 0.5 ml of 100 mM adenosine triphosphate (ATP) stock solution, 1 ml 10× phos-stop (Roche), 40 µg of KIS kinase and 2 mg of substrate; 100 mM ATP stock solution was prepared by dissolving ATP powder (Sigma) in kinase buffer and adjusting the pH to 8.0. Aliquots were stored at −20°C. The reaction typically required 24–48 h at 30°C to fully phosphorylate both Ser80 and Ser82. The phosphorylation products were purified with a MonoQ ion exchange column (GE). The reaction products were buffer exchanged to MonoQ buffer consisting of 20 mM Tris–HCl (pH 7.2) and loaded onto the column. The NaCl concentration in the elution buffer was gradually increased over 40 column volumes from 0 to 1 M, and 1-ml fractions were collected. The phosphorylation products were separated depending on the phosphorylation state. The double phosphorylation of Ser80 and Ser82 in phosphorylated SF1NTD (pSF1NTD) and SF1 (pSF1) was confirmed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), mass spectrometry (Supplementary Figure S1) and, for isotope labelled samples, by NMR spectroscopy (Figure 5).
Figure 5.
NMR and SAXS analysis of the effect of tandem serine phosphorylation of SF1. (A) SDS–PAGE analysis of phosphorylation of SF1NTD and SF1. The phosphorylated protein (red arrow) migrates slower on the gel than the non-phosphorylated. (B) Superposition of 1H,15N HSQC NMR spectra of non-phosphorylated (black) and phosphorylated SF1NTD and SF1 (red). Selected residues, which are shifted upon phosphorylation are annotated. (C) CSPs (red) and residue-specific local correlation times τc are shown for SF1NTD (blue) and pSF1NTD (red). The secondary structures and the phosphorylation sites are depicted above the diagram. The tandem phosphorylated linker region that rigidifies upon phosphorylation is highlighted by a box. A ribbon representation of SF1HH colour coded with the phosphorylation-induced CSPs is shown. (D) SAXS data showing comparisons of radial density distributions of non-phosphorylated and phosphorylated SF1NTD and SF1, in complex with U2AF65UHM, U2AF65RRM123 and with U2AF65RRM123–RNA, respectively.
NMR spectroscopy
All samples were measured at 298 K in NMR buffer with 10% 2H2O added for the lock. NMR spectra were recorded on AVIII 500, AVIII 600, AVIII 750, AVIII 800 or AVI 900 Bruker NMR spectrometers, equipped with cryogenic triple resonance gradient probes (with the exception of the AVIII 750 MHz). Data were processed in NMRPipe/Draw (22) and analysed in Sparky 3 (T.D. Goddard and D.G. Kneller, University of California). Protein backbone assignments were obtained from HNCACB and HNCA spectra, and by comparison of 1H,15N-HSQC and -TROSY spectra with previously reported data (11,23). Amino acid side chain resonance assignments were obtained from HCCH-TOCSY, 15N- and 13C-edited NOESY-HSQC/HMQC experiments (24). Intermolecular NOEs between the U2AF65UHM and SF1NTD were identified for well-resolved peaks in the 3D 13C- and 15N-edited NOESY-HSQC experiments. HN-N, N-C′ and HN-C′ residual dipolar couplings for SF1HH were recorded using HNCO-based NMR experiments (24). Alignment media consisted of 15 mg/ml Pf1 phage (Profos AG, Regensburg, Germany) (25). 15N R1, R1ρ relaxation rates and {1H}-15N heteronuclear NOEs of the SF1NTD and U2AF65UHM complexes were recorded at 750 MHz, of SF1NTD at 600 MHz and of SF1 at 800 MHz proton Larmor frequency at 298 K as described (26). Local correlation times were derived from the 15N R2/R1 ratio (26).
Structure calculations
The structure of SF1HH was determined using standard approaches. Automated NOESY cross-peak assignments and structure calculations with torsion angle dynamics were performed with CYANA 3.0 (27). NOE-derived distance restraints derived from CYANA together with φ and ψ backbone dihedral angle restraints derived from TALOS+ (28) based on secondary chemical shifts and residual dipolar couplings were used during the structure calculation and for final water refinement (29). Structures were validated with iCing (http://nmr.cmbi.ru.nl/icing/). Molecular images were generated with PyMol (Schrödinger).
The structure of the SF1NTD–U2AF65UHM complex was calculated using our previously reported protocol (30). The SF1HH structure determined here and the structure of the U2AF65UHM/SF1ULM complex (11) were used as input structures for semi-rigid calculation of the complex structure. The input structures were maintained using non-crystallographic symmetry restraints with a modified version of Aria1.2/CNS (30). Distance restraints derived from intermolecular NOEs and backbone torsion angle restraints derived from TALOS+ (28) were employed during molecular dynamics and simulated annealing. The final structures were refined in a shell of water molecules (29).
Small-angle X-ray scattering
SAXS data for solutions of SF1NTD, pSF1NTD, SF1, pSF1, U2AF65UHM, U2AF65RRM123, SF1NTD–U2AF65UHM, pSF1NTD–U2AF65UHM, SF1–U2AF65RRM123, pSF1–U2AF65RRM123, SF1–U2AF65RRM123–RNA and pSF1–U2AF65RRM123–RNA were recorded at the X33 beamline of the European Molecular Biology Laboratory at Deutsches Elektronen Synchrotron (DESY, Hamburg) using a MAR345 image plate detector. The scattering patterns were measured with a 2-min exposure time (eight frames, each 15 s) for several solute concentrations in the range from 1 to 10 mg/ml. Radiation damage was excluded based on a comparison of individual frames of the 2-min exposures, where no changes were detected. Using the sample-detector distance of 2.7 m, a range of momentum transfer of 0.01 < s < 0.6 Å−1 was covered (s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength).
All SAXS data were analyzed with the package ATSAS. The data were processed using standard procedures and extrapolated to infinite dilution with the program PRIMUS (31). The forward scattering, I(0), and the radius of gyration, Rg, were evaluated using the Guinier approximation. The values of I(0) and Rg as well as the maximum dimension, Dmax, and the inter-atomic distance distribution functions, (P(R)), were computed with the program GNOM (32). The scattering from the high-resolution models was computed with the program CRYSOL (33). The masses of the solutes were evaluated by comparison of the forward scattering intensity with that of a bovine serum albumin reference solution (molecular mass 66 kDa). For back calculation of SAXS data from the NMR ensemble of the SF1NTD–U2AF65UHM complex, the flexible regions (SF1NTD: 1–12, 26–38, 70–95, 128–145) were randomized with CORAL (34).
Isothermal titration calorimetry
Calorimetric titrations were performed using an iTC200 microcalorimeter (MicroCal) at 25°C. The buffer used for the protein and ligand samples was 20 mM sodium phosphate (pH 6.5) and 50 mM NaCl. The 200 -µl sample cell was filled with a 5 or 10 µM solution of protein and the 40 -µl injection syringe with a 50 or 100 µM of the titrating ligand. Each titration consisted of a preliminary 0.2 -µl injection followed by 20 subsequent 2 -µl injections. The heat of the injections was corrected for the heat of dilution of every ligand into the buffer. At least two replicas were performed for each experiment. Binding thermodynamic models corresponding to bimolecular complex formation were fitted using routines provided by the manufacturer.
Electrophoretic mobility shift assays
The 3′-splice site RNA (GGUCAUACUAACCCUGUCCCUUUUUUUUCCACAG|C; | denotes the 3′-splice site) is derived from the 3′-splice site of AdML intron 1 by replacing the original BPS with a consensus BPS (underlined). The RNA was synthesized with the T7-MEGA shortscript kit (Ambion) in the presence of [α-32P] UTP and gel purified. RNA binding was performed for 15 min at room temperature in 10 -µl reactions containing U2AF65RRM123, SF1 proteins and 50 pmol [α-32P] UTP–RNA. Assays with SF12-320 and deletion mutants were performed in the presence of 12% (v/v) glycerol, 12 mM Hepes–KOH (pH 7.9), 4 mM potassium–phosphate (pH 6.5), 60 mM KCl, 10 mM NaCl, 1.8 mM MgCl2, 0.5 mM DTT, 0.12 mM EDTA, 5 µg tRNA and 8U RiboShield™ ribonuclease inhibitor (Dundee Cell Products). Assays with SF1 and pSF1 contained final concentrations of 8 mM potassium–phosphate (pH 6.5), 20 mM NaCl, 0.5 mM DTT, 5 µg tRNA and 8U RiboShield™ ribonuclease inhibitor. Reaction products were resolved in native 5% polyacrylamide gels (acrylamide:bisacryamide = 80:1) in 0.5× Tris/Borate/EDTA (TBE) at 4°C for 3 h at 150 V.
RESULTS
The N-terminal region of SF1 adopts a novel helix-hairpin structure
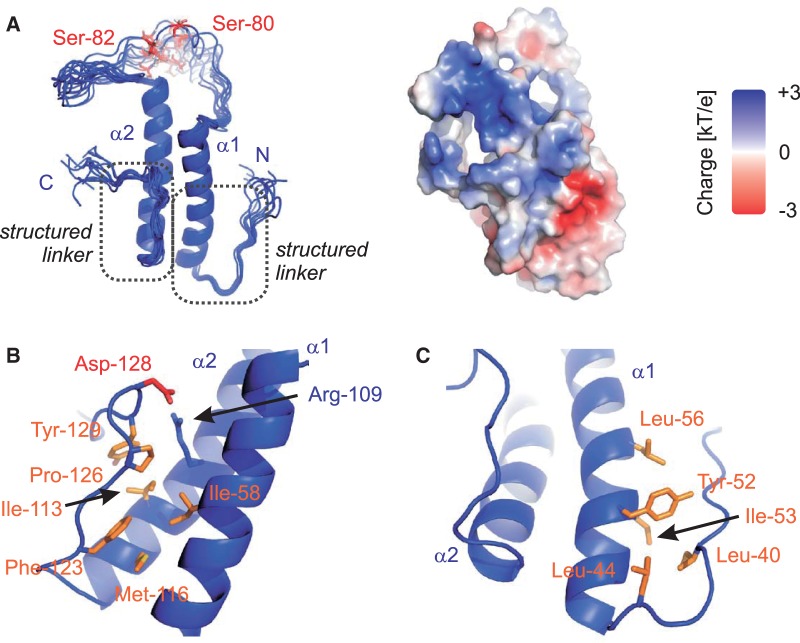
The 3D structures of the SF1ULM and SF1KHQUA2 regions bound to U2AF65 and intron RNA, respectively, have been reported previously (11,23). However, the region C-terminal of the SF1ULM up to the KH–QUA2 domain of SF1 has not been studied. A multiple sequence alignment of SF1 shows that this region, which also harbours the tandem serine motif that is phosphorylated by KIS kinase (5), is highly conserved (Figure 1C). We therefore cloned and expressed recombinant proteins comprising residues 1–145 (SF1NTD) and 27–145 (SF1HH) for NMR analysis (Figure 1B). NMR data demonstrate that SF1NTD comprises a structured domain consisting of two α-helical regions that are interrupted by a long disordered linker (residues 69–95) (Supplementary Figures S2 and S3). As the free ULM region is intrinsically disordered (11), we determined the 3D structure of the region comprising residues 27–145 (SF1HH) based on distance, dihedral angle and residual dipolar coupling restraints (Figure 2, Supplementary Table S1). The structure was further validated by comparison of measured and back-calculated relaxation rate enhancements upon addition of the paramagnetic co-solvent Gd(DTPA-BMA) (Supplementary Figure S3) (35). SF1HH comprises a helix-hairpin motif with two α-helices in an anti-parallel arrangement that are connected by a flexible linker (Figure 2), which contains the SPSP tandem phosphorylation motif. Hydrophobic residues within the two helices are spaced every three to four residues, i.e. Ala51, Val54, Ile58, Leu61, and Leu65 in α1 contact Leu105, Leu112, Met116 and Leu119 in α2, respectively, and thereby stabilize the arrangement of the two helices. Arg109 in helix α2 is engaged in a potential salt bridge with Asp128 C-terminal of helix α2 (Figure 2B). The C-terminal region adopts an extended conformation and packs against the helix hairpin by hydrophobic interactions of Phe123, Pro126 and Tyr129 with residues in both α-helices (Ile58, Ile113 and Met116) (Figure 2B). In addition, residues N-terminal of α1 (Ile40 and Leu44) form hydrophobic contacts with residues located within α1 (Tyr52, Ile53 and Leu56; Figure 2C). The additional interactions involving regions preceding the N- and C-terminal ends of the helix hairpin are confirmed by 15N relaxation data (Supplementary Figure S2A), which show that these extensions are rigid and thus stably interact with the helix hairpin. Thus, SF1HH comprises a helix-hairpin fold, which exposes the SPSP phosphorylation motif in a flexible linker.
Figure 2.
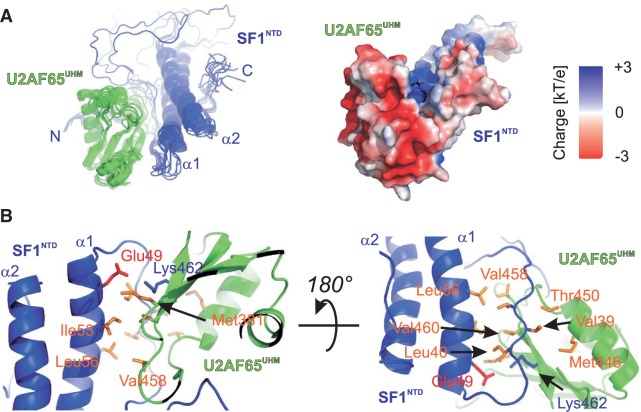
Structure of a novel helix hairpin in the N-terminal domain of SF1. (A) Ribbon representation of the ensemble of the 10 lowest energy structures and surface representation coloured according to electrostatic surface potential at 3 kT/e− for positive (blue) or negative (red) charge potential using the program APBS (36). Close-up view of the N-terminal (B) and C-terminal (C) structured extensions. Side chains of key residues mediating the interactions between the α-helices and the N/C termini are shown as sticks.
SF1NTD provides a secondary interface with U2AF65UHM
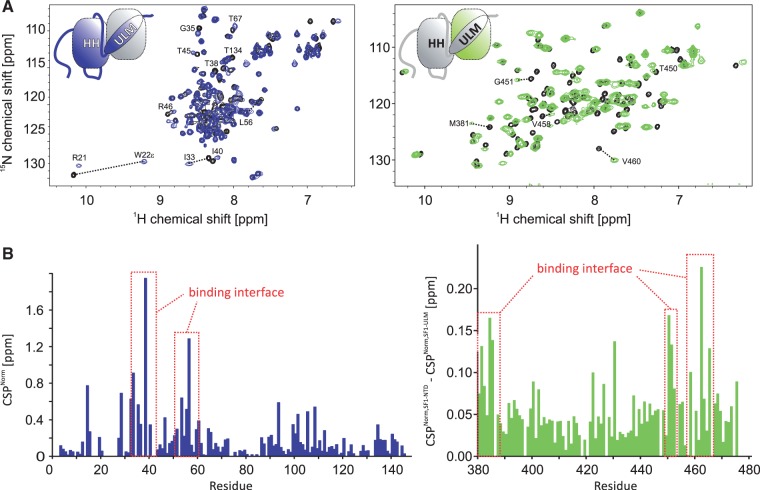
To determine whether the helix hairpin contributes to the SF1–U2AF65 interaction, we determined binding affinities of SF1 and U2AF65 fragments using isothermal titration calorimetry (Table 1). As reported previously (11), U2AF65UHM binds SF1ULM with Kd = 127 ± 48 nM. Inclusion of the helix hairpin in SF1 (SF1NTD) shows a slightly increased affinity (Kd = 84 ± 24 nM), indicating that this region contributes to the protein–protein interaction. No further increase of binding affinity is detected when studying the SF1–U2AF65 complex using almost full-length U2AF65 (U2AF65RRM123) and a region comprising all structural domains in SF1 (residues 1–260) (Kd = 114 ± 24 nM). We therefore conclude that SF1NTD and U2AF65UHM harbour all relevant binding sites for the U2AF65–SF1 interaction and represent a minimal complex. To identify the binding interface between SF1NTD and U2AF65UHM, we performed NMR chemical shift titrations of the isotope-labelled recombinant proteins with the unlabelled binding partner. The NMR signals of both SF1NTD and U2AF65UHM are shifted upon addition of unlabelled U2AF65UHM or SF1NTD, respectively (Figure 3A). Some of the NMR signals in the interface exhibit line broadening, which is characteristic for medium- to high-affinity complexes with micromolar dissociation constants, and thus consistent with the ITC data. We observed large chemical shift perturbations (CSPs) of SF1NTD and residues in U2AF65UHM including numerous residues, which are not in contact with SF1ULM in the previously reported NMR structure (Figure 3B). In NMR relaxation measurements of the complex, both subunits show similar 15N R1 and R1ρ relaxation rates (Supplementary Figure S2C) and tumbling correlation times (τc, Supplementary Figure S2D), thus indicating that they tumble as a single entity. The average τc of 14 ns is in good agreement with the correlation time expected for the molecular mass of the complex (29 kDa; τccalculated = 17 ns) (37).
Table 1.
Binding affinities determined from isothermal titration calorimetry
| Sample | Dissociation constant [Kd] |
|---|---|
| SF1ULM–U2AF65UHM | 127 ± 48 nM |
| SF1HH–U2AF65UHM | n.d. |
| SF1NTD–U2AF65UHM | 84 ± 23 nM |
| SF1–U2AF65RRM123 | 114 ± 23 nM |
| pSF1–U2AF65RRM123 | 96 ± 32 nM |
Figure 3.
NMR analysis of the SF1NTD–U2AF65UHM interaction. (A) Superposition of 1H,15N HSQC NMR spectra of labelled SF1NTD and U2AF65UHM free (black) and when bound to unlabelled U2AF65UHM (blue) or SF1NTD (green), respectively. Selected residues, which are shifted upon formation of the SF1NTD–U2AF65UHM complex are annotated. (B) CSPs of amides linked to U2AF65UHM (blue) and SF1NTD (green) binding. To selectively analyse the contributions of the secondary binding interface, the CSPs obtained in a titration of labelled SF1ULM with unlabelled U2AF65UHM were subtracted from the SF1NTD CSPs. Residues with strong CSPs that are therefore located in the secondary binding interface are annotated.
To determine the 3D structure of the SF1NTD–U2AF65UHM complex, we employed inter- and intra-molecular distance restraints derived from 15N- and 13C-edited NOESY spectra. To unambiguously identify intermolecular NOEs, a set of isotope-edited and -filtered NOESY spectra was recorded (24,38) and specifically isotope-labelled protein complexes were used where one of the subunits (either SF1NTD or U2AF65UHM) was deuterated and methyl-protonated. Several NOEs were detected for 1H,13C-labelled methyl groups in the binding interface (SF1NTD V39, I40, I53, L56 and U2AF65UHM V458, V460; Supplementary Figure S4). For structure calculation of the SF1NTD–U2AF65UHM complex, we used a protocol described recently (30). Shortly, semi-rigid body modelling was performed using the previously determined structures of SF1ULM–U2AF65UHM and SF1HH as input. Comparison of secondary chemical shifts between the free and bound proteins and overall similarity of NMR spectra confirm that both binding partners do not undergo substantial structural changes upon complex formation compared with the input structure. Structures were calculated based on inter-molecular NOE distances and dihedral angle restraints derived from TALOS+ (28). The final ensemble of structures was refined in a shell of water molecules (29) and validated by comparing experimental and back-calculated SAXS data of the complex (Supplementary Table S1).
The final ensemble of structures of the SF1NTD–U2AF65UHM complex is shown in Figure 4A and Supplementary Figure S4. SF1NTD and U2AF65UHM form an additional hydrophobic interface involving U2AF65UHM Met381, Val458, Val460 and SF1NTD Val39, Ile40, Ile53, Leu56, which is further stabilized by a potential salt bridge between SF1NTD Glu49 and U2AF65UHM Lys462 (Figure 4B). This secondary interface buries an additional solvent-exposed surface of approximately 500 Å2 [determined with PDBePISA (39)]. Notably, the additional interface in SF1HH is remote from the ULM; it is located on the opposite side of the SF1ULM-binding site of U2AF65UHM and therefore does not interfere with binding of SF1ULM. The additional hydrophobic interface moderately strengthens the SF1–U2AF65 interaction, with ITC-derived dissociation constants of U2AF65UHM for SF1ULM and SF1NTD of Kd = 127 and 85 nM, respectively. Consistent with this moderate contribution to the overall affinity, the interaction of the helix hairpin (SF1HH) alone with U2AF65UHM is weak and not detectable by ITC (Kd >> 100 µM) (Table 1 and Supplementary Figure S5).
Figure 4.
Structure of the SF1NTD and U2AF65UHM complex. (A) Ribbon representation of the ensemble of the 10 lowest energy structures and surface representation coloured according to electrostatic surface potential at 3 kT/e for positive (blue) or negative (red) charge potential using the program APBS (36). (B) Close-up view of SF1NTD–U2AF65UHM complex interface. Side chains of key residues mediating the interactions between the α-helices and the N/C-termini are shown as sticks.
Tandem serine phosphorylation structures and rigidifies the SF1HH linker
Phosphorylation of two serine residues within the linker connecting the two α-helices in SF1NTD has been reported recently to enhance formation of the ternary SF1–U2AF65–RNA complex (5). To study potential structural effects linked to this observation, we prepared phosphorylated SF1NTD (pSF1NTD) and SF1 (pSF1) by in vitro phosphorylation with recombinant KIS kinase (Figure 5A). An overlay of NMR spectra comparing non-phosphorylated and phosphorylated proteins (Figure 5B) reveals large chemical shift changes linked to Ser80/Ser82 tandem phosphorylation. NMR chemical shifts were re-assigned using a set of standard triple resonance NMR experiments (24). Most CPSs are observed in close proximity to the phosphorylation sites at Ser80 and Ser82 and at the N-terminal end of helix α2 (Figure 5C). Strongly affected residues include many positively charged residues in helix α2 such as Arg97 and Arg100. 15N R1 and R1ρ relaxation rates and local tumbling correlation times indicate that the linker connecting helices α1 and α2 in SF1HH becomes rigid upon phosphorylation (Figure 5C). Taken together these observations suggest that the side chains of Arg93 (linker), Arg97 and Arg100 (α2) mediate salt bridges with the two phosphate groups in the SF1HH linker and thereby strongly reduce the conformational flexibility of this linker.
Phosphorylation of SF1 does not significantly alter the overall conformation of SF1–U2AF65
We next tested the structural impact of phosphorylation on SF1 as well as on the SF1NTD–U2AF65UHM and SF1–U2AF65RRM123 complexes using NMR and SAXS. CSPs induced by phosphorylation are very similar for SF1NTD and SF1 (cf. Figure 5C and Supplementary Figure S6) and are mainly localized within the SF1 helix-hairpin domain. This indicates that phosphorylation does not induce strong intra-molecular contacts between the NTD and the KH–QUA2 regions of SF1. A comparison of local mobility along the protein backbone derived from 15N NMR relaxation data of phosphorylated SF1 with the non-phosphorylated protein (Supplementary Figure S6A) reveals that the overall backbone dynamics of the protein does not significantly change upon phosphorylation. SAXS data further corroborate the NMR results. The radii of gyration (Rg) of SF1 and pSF1 are comparable (32.5 ± 0.3 versus 29.1 ± 0.3 Å) and only minor differences are seen in the pairwise distance distribution functions (P(R), Supplementary Figure S6B). Although these changes might indicate compaction of a minor fraction of ‘open’ species in the non-phosphorylated protein, the NMR relaxation data and the absence of phosphorylation-induced CSPs in SF1KHQUA2 indicate that phosphorylation does not significantly affect the conformation of SF1.
We next studied the impact of phosphorylation on the overall structure of the SF1NTD–U2AF65UHM, SF1–U2AF65RRM123 and the ternary SF1–U2AF65RRM123–RNA complexes using SAXS analysis. Only minor differences are observed for the pairwise distribution functions of the two protein complexes (Figure 5D) and the derived radii of gyration (Table 2). In contrast, and similar to a recent report (40) binding of a 20-mer RNA containing the BPS and the Py tract regions induces large overall changes for the structure and/or dynamics of the SF1–U2AF65RRM123 complex and leads to formation of a compact SF1–U2AF65RRM123–RNA arrangement with Rg = 39.4 ± 0.4 and 34.2 ± 0.1 Å in the absence and presence of the RNA ligand, respectively (Table 2). Notably, tandem serine phosphorylation of SF1 introduces only minor differences to the overall arrangement of the SF1–U2AF65 complexes (with or without RNA) (Figure 5D). This indicates that SF1 Ser80/Ser82 phosphorylation is mainly limited to minor conformational changes within SF1 while RNA binding leads to a large change in the overall structure and/or dynamics of the U2AF65–SF1 complex.
Table 2.
SAXS data and analysis
| Sample | Rg [nm] | Dmax [nm] |
|---|---|---|
| SF1NTD | 2.24 ± 0.11 | 7.8 |
| pSF1NTD | 2.18 ± 0.11 | 7.6 |
| SF1 | 3.25 ± 0.16 | 10.8 |
| pSF1 | 2.91 ± 0.09 | 10.2 |
| SF1NTD–U2AF65UHM | 2.48 ± 0.03 | 8.7 |
| pSF1NTD–U2AF65UHM | 2.32 ± 0.02 | 7.6 |
| SF1–U2AF65RRM123 | 3.94 ± 0.04 | 14.0 |
| pSF1–U2AF65RRM123 | 4.18 ± 0.04 | 14.0 |
| SF1–U2AF65RRM123–RNA | 3.42 ± 0.01 | 11.0 |
| pSF1–U2AF65RRM123–RNA | 3.36 ± 0.01 | 11.0 |
Role of SF1HH and phosphorylation for cooperative RNA binding
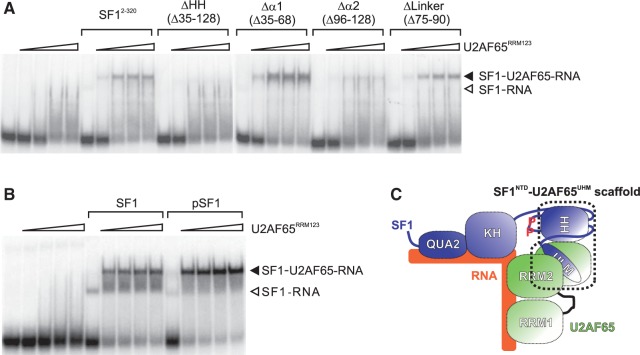
The role of the helix-hairpin structure and tandem serine phosphorylation of SF1 for cooperative binding of SF1 and U2AF65 to the pre-mRNA was tested in electrophoretic mobility shift assays (EMSAs). Increasing concentrations of U2AF65RRM123 added to a 3′-splice site RNA result in a smear of U2AF65–RNA complexes (Figure 6A). His6-tagged SF12-320 also binds the RNA, although weakly. In the presence of both proteins, a ternary SF1–U2AF65–RNA complex forms at lower U2AF65 concentrations, consistent with cooperative binding. The ternary complex is barely evident in the presence of SF1-ΔHH and is strongly reduced in the presence of SF1-Δα2. Deletion of helix α1 slightly increases ternary complex formation and deletion of the linker does not have any effect. Thus, in agreement with the data shown above, the helix-hairpin domain in the N-terminus of SF1 is necessary for cooperative RNA binding, with helix α2 playing a more important role than helix α1. In addition, phosphorylated SF1 shows a slightly higher efficiency of ternary complex formation with U2AF65RRM123 than non-phosphorylated SF1 (Figure 6B), consistent with the results of Manceau et al. (5).
Figure 6.
Cooperative binding of U2AF65RRM123 and SF1 to a 3′-splice site RNA. RNA was incubated with buffer or U2AF65RRM123 (0.2, 0.5, 1 and 2 µM; indicated by triangles) in the absence or presence of 1.5 µM His6-tagged SF12-320 or internal deletion mutants (A) or with 6.6 µM SF1 or pSF1 (B). Reaction products were separated by native PAGE and visualized by autoradiography. The migration of SF1–U2AF65–RNA complexes (closed arrowhead) and SF1–RNA complexes (open arrowhead) is indicated. (C) Role of SF1HH and the SF1NTD–U2AF65UHM interaction in the formation of the 3′-splice site recognition complex. SF1HH may establish an optimal orientation of the SF1 (KH–QUA2) and U2AF65RRM1,2 RNA-binding subunits in the complex and thereby support cooperative RNA binding. Tandem phosphorylation of SF1 might contribute to the formation of the ternary complex by stabilizing a yet unknown interface.
DISCUSSION
Here, we show that the N-terminal region of SF1 (SF1NTD) comprises a helix-hairpin fold. RNA-binding assays show that SF1HH is essential for formation of the ternary SF1–U2AF65–RNA complex, as deletion of the helix hairpin abolishes cooperative RNA binding. Interestingly, deletion of either of the two α-helices does not abrogate formation of the ternary complex, although complex formation is reduced in the absence of helix α2 (Figure 6A). This suggests that (i) SF1HH mainly acts as spacer between the U2AF65UHM-bound SF1ULM and the SF1KHQUA2 region and may thus provide an optimal orientation of the proteins within the ternary complex (Figure 6C). Although deletion of both α-helices strongly shortens the distance between the ULM and KH–QUA2 regions in SF1, and therefore may not allow proper arrangement of these regions needed for cooperative RNA binding, the presence of one of the two helices is able to rescue this effect. (ii) The observation that deletion of helix α2 did not affect formation of the SF1–U2AF65–RNA complex may indicate that residues located in helix α2 are involved in stabilization of the ternary complex. Future structural studies of the ternary complex should clarify this point.
Our NMR and structural studies reveal that U2AF65UHM forms a secondary interface with SF1NTD in addition to the previously reported interaction with SF1ULM. The additional interface locks the relative orientation of U2AF65UHM and SF1NTD and is thus likely to be important for the specific quaternary arrangement of the proteins in the SF1–U2AF65 complex. In addition to providing a proper geometry of SF1 and U2AF65 for RNA binding, the secondary interface involving the SF1HH and U2AF65UHM may reduce the entropic costs linked to SF1–U2AF65–RNA complex formation by providing a prearranged protein–protein scaffold for RNA binding. Thus, although the secondary interface has a rather small contribution to the overall affinity between SF1 and U2AF65 (Table 1), it is an essential feature for formation of the ternary complex. A specific arrangement of SF1 and U2AF enforced by the helix hairpin of SF1 may be required to accommodate variations in the distance between the BPS and Py tract regions of introns by providing a prearranged scaffold of the SF1 KH–QUA2 and U2AF65RRM1,2 regions. Furthermore, the hydrophobic surface on U2AF65UHM might mediate additional protein–protein interactions with additional SFs that could modulate complex assembly and splicing regulation.
Our data show that tandem serine phosphorylation of the conserved SPSP site within the linker connecting SF1NTD helices α1 and α2 has little effect on the conformation and overall structure of SF1 alone, or the SF1NTD–U2AF65UHM and the SF1–U2AF65RRM123 complexes. NMR- and ITC-binding data also suggest that phosphorylation does not alter the protein–protein-binding affinities and interactions (Supplementary Figure S7 and Table 1) and thus further corroborate these results. Nevertheless, the EMSA data indicate that SF1 phosphorylation enhances cooperative assembly of the SF1–U2AF65–RNA complex (Figure 6B) consistent with a previous report (5). As SAXS data do not indicate large conformational rearrangements linked to phosphorylation of SF1 alone or bound to U2AF65 (Figure 5D, Supplementary Figure S6B), the contribution to RNA binding must be small. The slight improvement in cooperative assembly could reflect either additional direct contacts with U2AF65 involving the serine-phosphorylated SF1HH linker or contribute to the overall stability (and rigidity) of a prearranged SF1–U2AF65 complex, by reducing entropy loss linked to RNA binding. Structural studies to analyze these effects in the ternary complex are currently on-going in our laboratory.
Tandem serine phosphorylation of SF1 may have additional roles beyond a function in cooperative assembly of the constitutive splicing complex. Previous studies in mammalian and yeast systems have established that SF1 mainly affects the kinetics of spliceosome assembly, as genetic or biochemical depletion of SF1 does not abolish splicing (41,42). In Saccharomyces cerevisiae, it has been shown that SF1 is involved in removing introns with suboptimal splice sites and in nuclear pre-mRNA retention (43,44), whereas knockdown of human SF1 in HeLa cells did not result in a general splicing phenotype (45). However, Corioni et al. (46) have recently shown that SF1 is not a constitutive SF, but is required for the splicing of certain introns and affects alternative splice site choice. In this respect, SF1 phosphorylation may mediate interactions with other factors that could regulate alternative splicing by modulating 3′-splice site recognition. For example, it has been suggested that the phosphorylated SPSP motif of SF1 is recognized by the tandem WW domains of FBP11 (47) and the interaction of the SF1 and FBP11 orthologues of S. cerevisiae has been implicated in mediating intron definition by connecting the 5′- and 3′-splice sites (48). However, we have not detected any interaction of tandem WW domains with phosphorylated SF1 (data not shown). Nevertheless, the substantial conformational changes in the SF1NTD linker associated with tandem serine phosphorylation by KIS kinase suggest that interactions with so far unknown factors are modulated which may contribute to regulate alternative splicing by SF1.
STRUCTURE COORDINATES AND NMR DATA
The coordinates for the SF1HH structure and the structure of the U2AF65UHM/SF1NTD complex are deposited in the PDB with accession numbers 2 m09 and 2 m0g, respectively. Chemical shifts are deposited in the BMRB, accession codes: 18802 and 18808.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–7, Supplementary Methods and Supplementary References [36,49,50].
FUNDING
The Deutsche Forschungsgemeinschaft [SFB1035, project A03 to M.S., Emmy Noether Grant MA 5703/1-1 to T.M.]; European Commission [NMI3 project to M.S.]; Swiss National Science Foundation (to A.K.); Bavarian Ministry of Sciences, Research and the Arts [Bavarian Molecular Biosystems Research Network to T.M.]; Austrian Academy of Sciences [APART-fellowship to T.M.]; a fellowship from the China Scholarship Council (CSC) and the Helmholtz Association (to Y.Z.) and an EMBO Longterm Postdoctoral fellowship (to H.K.). Funding for open access charge: Grants/internal budget.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Masaki Mishima (Tokyo Metropolitan University, Japan) Cameron Mackereth (IECB Bordeaux, France) and Ana Messias (Munich) for initial NMR assignments, Maria Macias (IRB Barcelona) for FBP expression plasmids. They thank Bertrand Séraphin (IGBMC, Strasbourg, France) and Juan Valcárcel (CRG, Barcelona, Spain) for helpful discussion. DESY (Hamburg) and ESRF (Grenoble) are acknowledged for SAXS measurements, the Bavarian NMR Centre (BNMRZ, Garching, Germany) for NMR measurement time.
REFERENCES
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 5.Manceau V, Swenson M, Le Caer JP, Sobel A, Kielkopf CL, Maucuer A. Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. FEBS J. 2006;273:577–587. doi: 10.1111/j.1742-4658.2005.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Krämer A, Robinson PJ. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 1999;18:4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol. Cell. 2012;45:314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 8.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 9.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Krämer A, Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 11.Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF(65) and SF1/mBBP. Mol. Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 12.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 13.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 14.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackereth CD, Sattler M. Dynamics in multi-domain protein recognition of RNA. Curr. Opin. Struct. Biol. 2012;22:287–296. doi: 10.1016/j.sbi.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rain JC, Rafi Z, Rhani Z, Legrain P, Krämer A. Conservation of functional domains involved in RNA binding and protein–protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Liu ZH, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 24.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Mag. Res. Sp. 1999;34:93–158. [Google Scholar]
- 25.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 26.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Formankay JD, Kay LE. Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studied by N-15 NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 27.Guntert P. Automated structure determination from NMR spectra. Eur. Biophys. J. Biophy. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 30.Simon B, Madl T, Mackereth CD, Nilges M, Sattler M. An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew. Chem. Int. Ed. 2010;49:1967–1970. doi: 10.1002/anie.200906147. [DOI] [PubMed] [Google Scholar]
- 31.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 32.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 33.Svergun D, Barberato C, Koch MHJ. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 34.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madl T, Bermel W, Zangger K. Use of relaxation enhancements in a paramagnetic environment for the structure determination of proteins using NMR spectroscopy. Angew. Chem. Int. Ed. 2009;48:8259–8262. doi: 10.1002/anie.200902561. [DOI] [PubMed] [Google Scholar]
- 36.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daragan VA, Mayo KH. Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog. NMR Spectrosc. 1997;31:63–105. [Google Scholar]
- 38.Otting G, Wuthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Gupta A, Jenkins JL, Kielkopf CL. RNA induces conformational changes in the SF1/U2AF65 splicing factor complex. J. Mol. Biol. 2011;405:1128–1138. doi: 10.1016/j.jmb.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guth S, Valcarcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 43.Rutz B, Séraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 45.Tanackovic G, Kramer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corioni M, Antih N, Tanackovic G, Zavolan M, Kramer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2011;39:1868–1879. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 49.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.