Abstract
The translation of mammalian messenger RNAs (mRNAs) can be driven by either cap-binding proteins 80 and 20 (CBP80/20) or eukaryotic translation initiation factor (eIF)4E. Although CBP80/20-dependent translation (CT) is known to be coupled to an mRNA surveillance mechanism termed nonsense-mediated mRNA decay (NMD), its molecular mechanism and biological role remain obscure. Here, using a yeast two-hybrid screening system, we identify a stem-loop binding protein (SLBP) that binds to a stem-loop structure at the 3′-end of the replication-dependent histone mRNA as a CT initiation factor (CTIF)-interacting protein. SLBP preferentially associates with the CT complex of histone mRNAs, but not with the eIF4E-depedent translation (ET) complex. Several lines of evidence indicate that rapid degradation of histone mRNA on the inhibition of DNA replication largely takes place during CT and not ET, which has been previously unappreciated. Furthermore, the ratio of CBP80/20-bound histone mRNA to eIF4E-bound histone mRNA is larger than the ratio of CBP80/20-bound polyadenylated β-actin or eEF2 mRNA to eIF4E-bound polyadenylated β-actin or eEF2 mRNA, respectively. The collective findings suggest that mRNAs harboring a different 3′-end use a different mechanism of translation initiation, expanding the repertoire of CT as a step for determining the fate of histone mRNAs.
INTRODUCTION
A series of molecular processes is involved in the coordinated regulation of gene expression in mammals (1,2). Typically, the 5′-end of newly synthesized pre-messenger RNAs (pre-mRNAs) is capped. The cap structure of the pre-mRNAs is recognized by nuclear cap-binding protein complex (CBC), which is composed of a heterodimer of cap-binding proteins 80 and 20 (CBP80/20) (1,2). If pre-mRNAs undergo splicing, during which the introns are removed and the neighboring exons are connected, an exon junction complex (EJC) will be deposited 20–24 nucleotides upstream of the exon–exon junctions (1–3). The resulting mRNAs are then exported to the cytoplasm via the nuclear pore complex (NPC) with the 5′-cap bound by CBP80/20. After mRNA export, CBP80/20 is replaced by the cytoplasmic cap-binding protein, eukaryotic translation initiation factor (eIF) 4E.
The cap structure of the mRNAs is bound by either CBP80/20 or eIF4E (1,2,4). These proteins have the ability to recruit the small subunit of ribosome (40S) to initiate translation in the cytoplasm (4–6). The CBP80/20-dependent translation initiation factor (CTIF), which is localized to the cytoplasmic side of the nuclear envelope, binds to CBP80 in the mRNA being exported (5). The CBP80–CTIF complex then recruits eIF3 via the CTIF–eIF3g interaction (7), which, in turn, recruits the 40S ribosome, triggering the first round of translation (5,8,9). This round of translation, which is defined as the translation of CBC-bound mRNA and occurs for the first time during the entire mRNA lifespan, is called the pioneer round of translation (1,2,4). Notably, here and elsewhere, the data imply that CBP80/20-bound mRNA undergoes multiple rounds of the pioneer round of translation (5,9–11). We will hereafter refer to this as CBP80/20-dependent translation (CT) as long as the translation occurs on the CBP80/20-bound mRNAs, regardless of the number of translation initiations (5).
After CT, CBP80/20 is replaced by eIF4E in a translation-independent manner (12), and the eIF4E-bound mRNAs are subject to active translation (6). The eIF4E recruits eIF4GI/II, which, in turn, recruits eIF3 and eventually the 40S ribosome, initiating the bulk of cellular translation in the cytoplasm (6). We hereafter refer to this as eIF4E-dependent translation (ET).
CT differs mechanistically from ET in several aspects. First, CT can occur on mRNAs that harbor EJCs deposited as a result of splicing, whereas ET occurs on mRNAs that harbor undetectable levels of EJCs (4,8,13). Second, nonsense-mediated mRNA decay (NMD), the best-characterized mRNA quality control mechanism (1–3), is tightly coupled to CT (1,2,4). Third, our group recently showed that, whereas ET uses eIF4GI/II, CT preferentially uses a specific factor, CTIF, to recruit eIF3 and the 40S ribosome (5). Fourth, CT can be maintained even when ET is compromised (1,2). However, like ET, the CT can be targeted for microRNA-mediated gene silencing (14,15).
Eukaryotic cells maintain the balance between the rates of DNA replication and histone protein synthesis so as to coordinate DNA replication and proper chromosomal DNA packaging during the cell cycle (16,17). Any disturbances of this balance could be toxic to cell proliferation. One mechanism to prevent toxic effects of excess histone is the rapid degradation of histone mRNA when DNA synthesis is inhibited.
Most eukaryotic mRNAs are polyadenylated. However, replication-dependent histone mRNAs (hereafter called histone mRNAs, if not specified) are not polyadenylated and instead contain an evolutionarily conserved stem-loop structure that consists of 25 or 26 nucleotides (16,17). This structure is essential for rapid degradation of histone mRNA on the inhibition of DNA replication or at the end of S phase. The stem-loop structure is bound by a stem-loop binding protein (SLBP, also called hairpin binding protein), which plays multiple roles in histone mRNA metabolism, including histone pre-mRNA processing, mRNA export, translation and degradation (16,17).
Here, we identify SLBP as a new CTIF-interacting protein. We found that histone mRNA is primarily translated as CBP80/20 bound mRNAs, which has been previously unappreciated. As a consequence, these mRNAs are the primary substrate for histone mRNA degradation. Our results reveal a new biological role for CT in the control of histone gene expression.
MATERIALS AND METHODS
Plasmid constructions, yeast two-hybrid screening, bimolecular fluorescence complementation assay and polysome fractionation
The details are described in Supplementary Materials and Methods.
Cell culture and transfections
The details are described in Supplementary Materials and Methods.
Reverse transcription-polymerase chain reaction, quantitative real-time reverse transcription-polymerase chain reaction and northern blotting
The details and the oligonucleotides information are provided in Supplementary Materials and Methods.
Immunoprecipitations and western blotting
Immunoprecipitations (IPs) were performed as previously described (5,14,18,19). The details are provided in Supplementary Materials and Methods. The following antibodies were used for western blotting: FLAG (Sigma-Aldrich), Myc (Calbiochem), eIF4E (BD Biosciences or Cell Signaling), eIF3 (20), eIF3b (Santa Cruz Biotechnology), eIF4GI (a gift from S. K. Jang), CTIF (5), CBP80 (7), SLBP (21), SLBP-interacting protein 1 (SLIP1) (22), β-actin (Sigma-Aldrich), glutathione S-transferase (GST; Amersham) and 6xHis (GE Healthcare).
RESULTS
Identification of SLBP as a CTIF-interacting protein
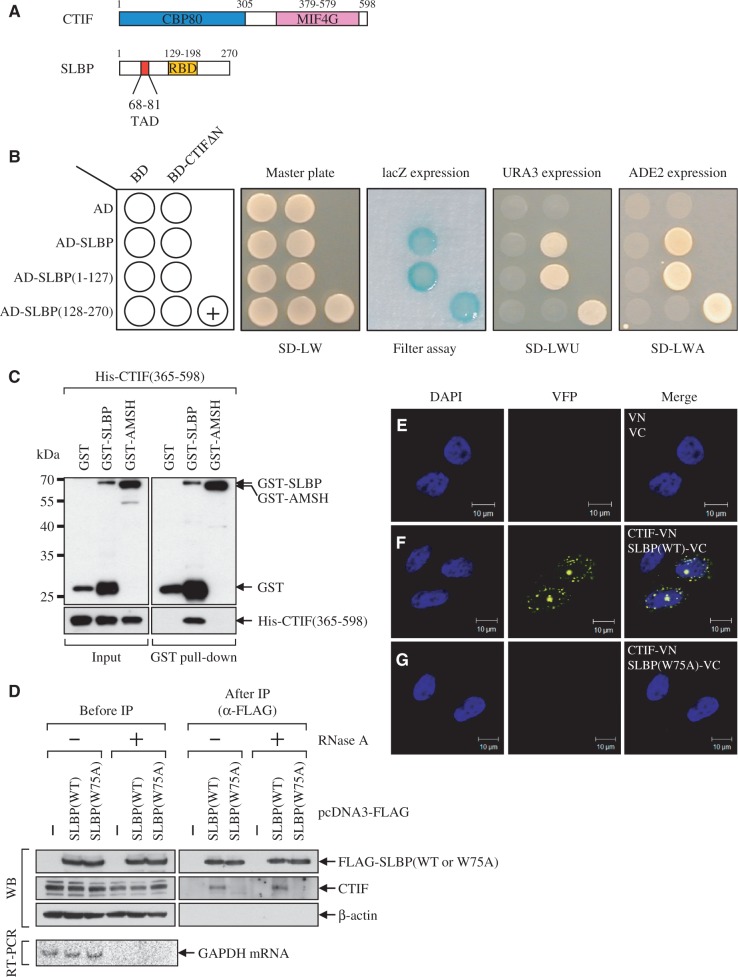
To search for CTIF-interacting proteins, we carried out yeast two-hybrid screening analysis using GAL4 DNA binding domain (BD)-fused human CTIFΔN, which is an N-terminally deleted variant of CTIF, as bait and a GAL4 transcription activation domain (AD)-fused human thymus complementary DNA (cDNA) library. We could not use the full-length and N-terminal half of CTIF (Figure 1A) because of self-activation under our condition (data not shown). Using this approach, we obtained 13 independent positive colonies that contained cDNA fragments encoding SLBP out of 7.9 × 106 transformants. Retransformation of plasmids expressing BD-CTIFΔN and AD-SLBP in yeast strains confirmed a specific interaction between CTIFΔN and SLBP (Figure 1B). The interaction between two proteins produced a blue color on medium containing X-gal (Figure 1B, filter assay), growth on medium lacking uracil (Figure 1B, SD-LWU) and growth on medium lacking adenine (Figure 1B, SD-LWA).
Figure 1.
SLBP interacts with CTIF. (A) Schematic diagram of CTIF and SLBP. CBP80 and MIF4G in CTIF indicate CBP80-interacting domain and a middle domain of eIF4GI, respectively. TAD and RBD in SLBP indicate a TAD- and a RNA-binding domain, respectively. (B) Identification of SLBP as CTIF-interacting protein by yeast two-hybrid screening. Yeast strains were co-transformed with a bait plasmid expressing either BD-CTIFΔN or BD and a prey plasmid expressing AD-SLBP, AD-SLBP(1–127), AD-SLBP(128–270) or AD. Transformed yeast cells were spread on selective medium lacking leucine and tryptophan (SD-LW) to select for co-transformants. Specific interactions between two proteins were tested by monitoring (i) the appearance of visible blue color (filter assay); (ii) the growth on selective medium lacking leucine, tryptophan and uracil (SD-LWU); and (iii) the growth on selective medium lacking leucine, tryptophan and adenine (SD-LWA). BD-polypyrimidine tract-binding protein (PTB) and AD-PTB served as the positive control for the protein–protein interaction (+). (C) In vitro GST pull-down assays. Escherichia coli lysates expressing GST, GST-SLBP or GST-AMSH (negative control) were mixed with purified recombinant His-CTIF(365-598). After GST pull-down, the purified proteins were analyzed by western blotting (WB) using α-GST antibody (top) or α-6xHis antibody (bottom). The locations of molecular weight (MW) markers are indicated on the left. (D) IPs of SLBP and SLBP(W75A). The extracts of HeLa cells transiently expressing either FLAG-SLBP or FLAG-SLBP(W75A) were either untreated or treated with RNase A and then subjected to IPs using α-FLAG-conjugated agarose beads. WB was carried out to detect the indicated proteins (top). To confirm that cellular RNAs were completely removed by RNase A treatment, cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was analyzed using RT-PCR (bottom). The relative amounts of cell extracts used before IP compared with after IP are provided in Supplementary Table S1. (E–G) In vivo bimolecular fluorescence complementation (BiFC) analysis for CTIF–SLBP interaction. HeLa cells were transiently transfected with plasmid expressing either N-terminal fragment (1–173 amino acids) of yellow Venus-enhanced fluorescent proteins (VN) or CTIF-VN and plasmid expressing C-terminal fragment (155–238 amino acids) of yellow Venus-enhanced fluorescent proteins (VC), SLBP-VC, or SLBP(W75A)-VC. After 8 h, the cells were fixed and observed by confocal microscopy. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). All results are representative of at least two independently performed experiments.
The specific interaction between CTIF and SLBP was further confirmed by in vitro GST pull-down assay, IP and bimolecular fluorescence complementation (BiFC) technology (23,24). First, the results of the pull-down assay showed that recombinant His-CTIF(365–598), which is the 6xHis-tagged N-terminally deleted form of CTIF, co-precipitated with GST-SLBP, but not GST and GST-AMSH (an associated molecule with the SH3 domain of signal transducing adaptor molecule (STAM); the molecular weight of AMSH is similar to that of SLBP), both of which served as negative control proteins (Figure 1C). Further in vitro GST pull-down assay (Supplementary Figure S1A) and IPs (Supplementary Figure S1B) revealed that the C-terminal half of CTIF, CTIF(365–598), and the N-terminal half of SLBP, SLBP(1–127), which contains a translation-activation domain (TAD) (22,25,26), are sufficient for interaction.
Second, we generated the mutated version of human SLBP, SLBP(W75A), which contains a tryptophan to alanine amino acid substitution at residue 75 in the TAD (Figure 1A). The corresponding mutant of SLBP1, a Xenopus homolog of human SLBP, lacks the ability to enhance the translation of histone mRNAs without affecting histone pre-mRNA processing and cell-cycle regulation (22,25,26). The IP results using wild-type (WT) and mutant SLBP showed that although SLBP(WT) was associated with CTIF, SLBP(W75A) failed to associate with CTIF (Figure 1D). Considering that a single amino acid substitution at this residue causes SLBP to lose its ability to enhance the translation of histone mRNAs (22,25,26), it is plausible that SLBP(W75A) fails to enhance translation of histone mRNA owing to a lack of ability to associate with CTIF.
Third, to clearly determine the in vivo interaction between CTIF and SLBP, we used BiFC technology, which allows for the determination of in vivo interaction and cellular localization where an interaction between two tested proteins occurs (Figure 1E–G). To this end, HeLa cells were transiently transfected with plasmid expressing either N-terminal fragment (1–173 amino acids) of yellow Venus-enhanced fluorescent protein (VN) or CTIF-fused VN, and plasmid expressing C-terminal fragment (155–238 amino acids) of yellow Venus-enhanced fluorescent protein (VC), SLBP-fused VC or SLBP(W75A)-fused VC. Interaction between fused proteins brings the fluorescent fragments VN and VC in proximity, resulting in the reformation of the Venus-enhanced fluorescent protein and the emission of a fluorescence signal (23,24). The BiFC results showed that, although fluorescent signals were mainly detected at the perinuclear region of cells expressing CTIF-VN and SLBP-VC (Figure 1F), no fluorescent signals were observed from cells expressing CTIF-VN and SLBP(W75A)-VC (Figure 1G) or cells expressing VN and VC (Figure 1E). The comparable fluorescent signals were detected when reciprocal combinations, either SLBP(WT)-VN or SLBP(W75A)-VN and CTIF-VC, were transiently expressed (Supplementary Figure S1C–E). These results suggest that (i) CTIF-VN interacts with SLBP(WT)-VC and (ii) an interaction between CTIF-VN and SLBP(WT)-VC mainly occurs at the perinuclear region. Considering that CTIF is mainly localized to the perinuclear region (5), our observations suggest that CTIF interacts with SLBP in vivo, during, or right after, the export of histone mRNAs via nuclear pore complex.
The collective results indicate that SLBP directly interacts with CTIF in vitro and in vivo and that amino acid residue 75 in human SLBP is critical for CTIF binding.
SLBP preferentially associates with the CT complex
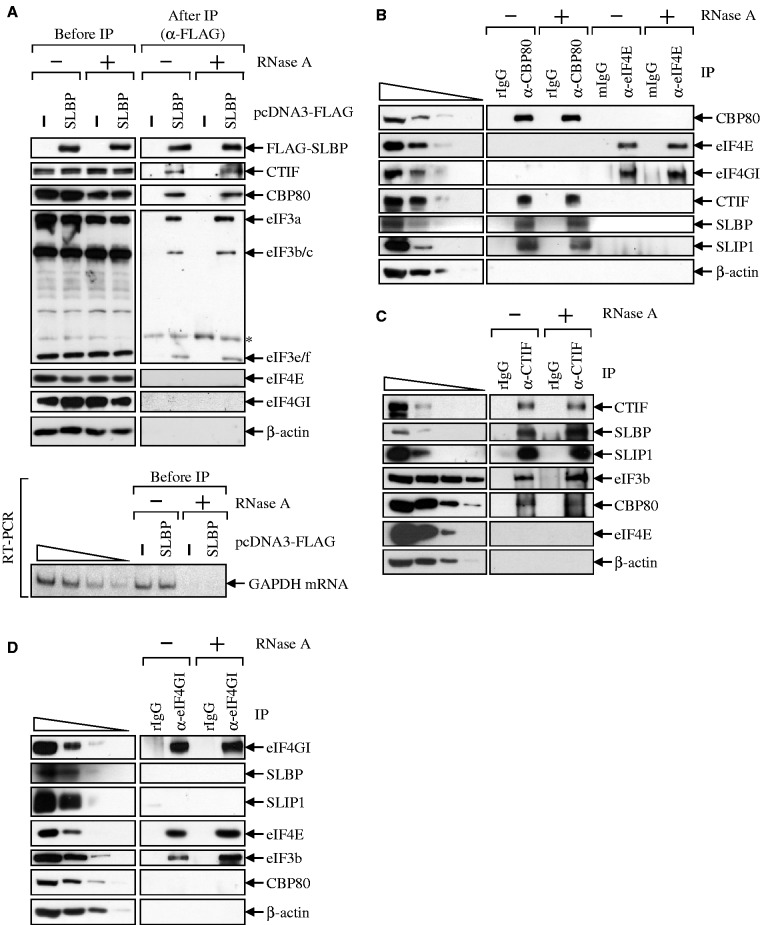
Given that CTIF is preferentially involved in the CT (5), our findings demonstrating a CTIF–SLBP interaction (Figure 1) led us to test whether SLBP selectively associates with the CT complex, but not with ET complex. Previous studies showed that, whereas the CT complex contains CBP80, CTIF and eIF3, the ET complex contains eIF4E, eIF4GI/II and eIF3 (4–8). Several lines of evidence support the preferential association of SLBP with the CT complex. First, CBP80, CTIF and eIF3 subunits, but not eIF4E and eIF4GI, co-immunopurified with FLAG-SLBP in a ribonuclease (RNase) A-resistant manner (Figure 2A). Second, the IP results using either α-CBP80 antibody or α-eIF4E antibody revealed that CTIF, SLBP and SLIP1, which is required for histone mRNA translation triggered by SLBP (22), but not eIF4E and eIF4GI, co-immunopurified with CBP80 in an RNase A-resistant manner (Figure 2B). In contrast, eIF4GI, but not CBP80, CTIF, SLBP and SLIP1, co-immunopurified with eIF4E in an RNase A-resistant manner (Figure 2B). Third, the results of IPs using α-CTIF antibody showed that endogenous SLBP, SLIP1, eIF3b (a subunit of eIF3) and CBP80, but not eIF4E and β-actin, co-immunopurified with endogenous CTIF in an RNase A-resistant manner (Figure 2C). Fourth, IPs using α-eIF4GI antibody showed that eIF4E and eIF3b, but not SLBP, SLIP1 and CBP80, co-immunopurified with endogenous eIF4GI in an RNase A-resistant manner (Figure 2D). All of these results suggest that SLBP preferentially associates with the CT complex by a direct interaction with CTIF.
Figure 2.
SLBP associates with CT initiation complex. (A) IP of FLAG-SLBP. HEK293T cells were transiently transfected with plasmid expressing either FLAG or FLAG-SLBP. Total-cell extracts were obtained 2 days after transfection and either untreated or treated with RNase A before IP. Each extract was subjected to IP using α-FLAG-conjugated agarose beads. WB was carried out to detect the indicated proteins (top). To confirm that cellular RNAs were completely removed by RNase A treatment, cellular GAPDH mRNA was analyzed using RT-PCR (bottom). To show that RT-PCR used in this study was sufficiently semi-quantitative, 2-fold serial dilutions of total-cell RNAs were presented in the four left-most lanes. (B) IPs of endogenous CBP80 and eIF4E. The extracts of HeLa cells were either untreated or treated with RNase A and then subjected to IPs using α-CBP80 antibody, rabbit IgG (rIgG) as a control for α-CBP80 antibody, α-eIF4E antibody or mouse IgG (mIgG) as a control for α-eIF4E antibody. To show that WB used in this study was sufficiently semi-quantitative, 3-fold serial dilutions of total-cell extracts are represented in the four left-most lanes. (C) IP of endogenous CTIF. Total-cell extracts were subjected to IP using α-CTIF antibody or, as a control, α-rabbit IgG (rIgG) before or after treatment with RNase A. (D) IP of endogenous eIF4GI. All results are representative of at least two independently performed experiments. The relative amounts of cell extracts used before IP compared with after IP are provided in Supplementary Table S1.
Downregulation of endogenous CTIF or overexpression of SLBP(W75A) inhibits the rapid degradation of replication-dependent histone mRNA
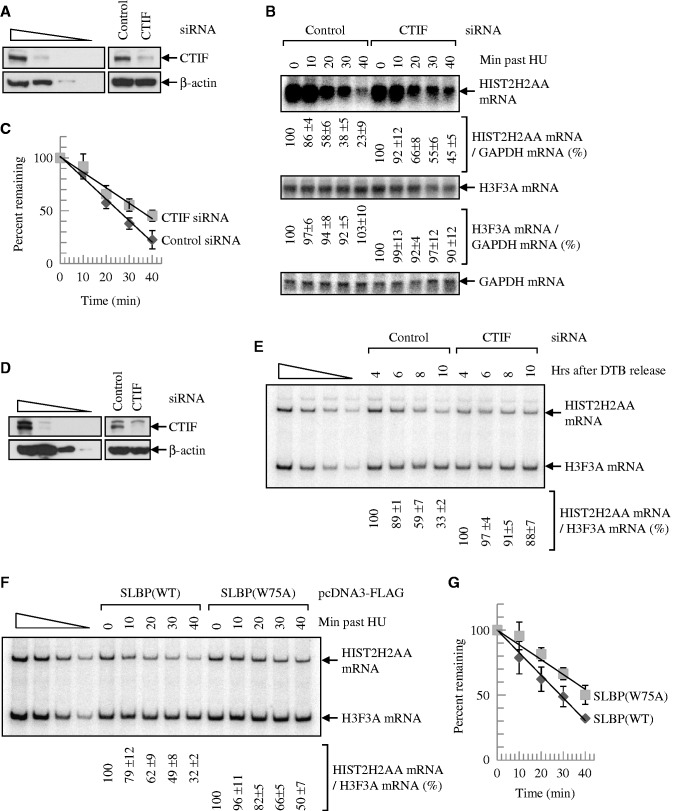
Given that (i) rapid degradation of histone mRNAs on the inhibition of DNA replication or at the end of S phase requires translation (26–28); (ii) SLBP enhances both translation and degradation of histone mRNAs (16,17,29,30); and (iii) SLBP associates with CTIF (Figures 1 and 2), it is likely that CTIF is involved in the translation-dependent degradation of histone mRNAs. To address this possibility, endogenous CTIF was downregulated using small interfering RNA (siRNA), and then the levels of cellular histone mRNAs were monitored on the inhibition of DNA replication (Figure 3A–C) or at the end of S phase (Figure 3D and E).
Figure 3.
Downregulation of CTIF or overexpression of SLBP(W75A) abolishes the rapid degradation of replication-dependent histone mRNA on the inhibition of DNA replication or at the end of S phase. (A–C) HeLa cells were transiently transfected with CTIF siRNA or non-specific Control siRNA. Three days later, cells were treated with HU. Total-cell proteins and RNAs were purified at the indicated time points. (A) WB demonstrating efficient downregulation of CTIF. (B) Northern blotting of HIST2H2AA mRNAs and H3F3A mRNAs. The levels of HIST2H2AA mRNAs were normalized to the level of GAPDH mRNAs. Normalized levels of HIST2H2AA mRNAs obtained at 0 min after HU treatment in the presence of each siRNA were set to 100%. (C) Half-life of HIST2H2AA mRNAs. The normalized levels of HIST2H2AA mRNAs obtained from Figure 3B were plotted as a function of time after HU treatment. (D and E) HEK293T cells were transiently transfected with CTIF siRNA or non-specific control siRNA. One day after transfection, cells were synchronized by DTB. Total-cell RNAs and proteins were prepared at the indicated time points after the release from DTB until these cells exited S phase completely. (D) WB demonstrating efficient downregulation of CTIF. (E) RT-PCR of HIST2H2AA mRNAs. The levels of HIST2H2AA mRNAs were normalized to the level of H3F3A mRNAs. Normalized levels of HIST2H2AA mRNAs obtained at 4 h after DTB release in the presence of each siRNA were set to 100%. The means and standard deviations of results obtained from three independent transfections and RT-PCRs are presented below the panel. (F and G) HeLa cells were transiently transfected with plasmid expressing either SLBP(WT) or SLBP(W75A). Two days later, cells were treated with HU and total-cell proteins and RNAs were purified at the indicated time points. (F) RT-PCRs of HIST2H2AA mRNAs and H3F3A mRNAs. The levels of HIST2H2AA mRNAs were normalized to the level of H3F3A mRNAs. Normalized levels of HIST2H2AA mRNAs obtained at 0 min after HU treatment were set to 100%. (G) Half-life of HIST2H2AA mRNAs. The normalized levels of HIST2H2AA mRNAs obtained from Figure 3F were plotted as a function of time after HU treatment.
Western blotting results revealed that the level of endogenous CTIF was downregulated to 33% of normal (Figure 3A). Northern blotting results showed that downregulation of CTIF significantly increased the abundance and the half-life of cellular HIST2H2AA mRNA (Figure 3B and C), which is a replication-dependent histone mRNA (31,32). On the other hand, the level of H3F3A mRNA, which is a replication-independent and poly(A)-containing histone mRNA (31,32) and served to control for variations in this study, was not affected by CTIF downregulation. The results of reverse transcription-polymerase chain reaction (RT-PCR) using α-[32P]-dATP and quantitative real-time RT-PCR (qRT-PCR) using the same samples were comparable with the results obtained from northern blotting (Supplementary Figure S2A and B), demonstrating that the methodologies used in this study are sufficiently quantitative. Of note, the level of uridylated HIST2H2AA mRNA, which is a pre-requisite intermediate form for rapid degradation of histone mRNA (30), was increased by 2.7-fold 20 min after hydroxyurea (HU) treatment. On the other hand, downregulation of CTIF blocked the formation of uridylated HIST2H2AA mRNA under the same conditions (Supplementary Figure S3). All these results indicate that downregulation of CTIF abrogates a rapid degradation of replication-dependent histone mRNA induced by HU treatment.
We next compared the levels of replication-dependent histone mRNA during the cell cycle on CTIF downregulation (Figure 3D and E). The synchronization and release of cell cycle by double thymidine block (DTB) were demonstrated by flow cytometry (Supplementary Figure S2C), and the selective downregulation of CTIF was confirmed by western blotting (Figure 3D). RT-PCR results using α-[32P]-dATP revealed that the downregulation of CTIF increased the abundance of HIST2H2AA mRNA (Figure 3E). Collectively, the data suggest that CTIF is involved in degradation of replication-dependent histone mRNAs on the inhibition of DNA replication or at the end of S phase.
To further clarify the role of CTIF–SLBP interaction in histone mRNA stability, we used SLBP(W75A), which lacks the ability to bind to CTIF (Figure 1D–G). Overexpression of SLBP(W75A), but not SLBP(WT), significantly increased the abundance and the half-life of cellular HIST2H2AA mRNA (Figure 3F and G). In addition, simple overexpression of SLBP(WT) had no significant effect on histone mRNA stability (Supplementary Figure S4). All of the results in Figure 3 suggest that the CTIF–SLBP interaction is important for degradation of histone mRNAs.
Rapid degradation of replication-dependent histone mRNAs largely occurs on CBP80/20-bound mRNAs
Considering that (i) rapid degradation of histone mRNAs requires translation (26–28); (ii) CTIF is a specific factor involved in efficient CT (5),; and (iii) CTIF downregulation or overexpression of SLBP(W75A) stabilizes histone mRNAs (Figure 3), it is most likely that degradation of histone mRNAs largely occurs during CT (i.e. on CBP80/20-bound mRNAs).
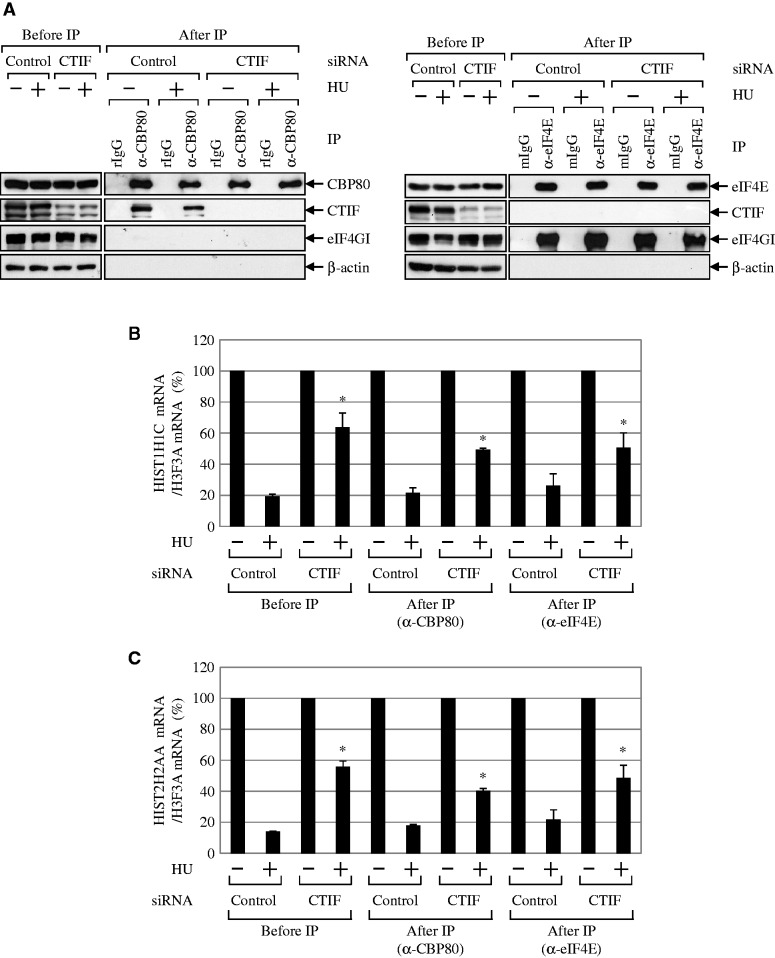
To address this possibility, we used a similar approach with some modifications that was previously used to demonstrate that NMD occurs on CBP80/20-bound mRNAs (4). CTIF-undepleted or -depleted HEK293T cells were either untreated or treated with HU. Total-cell extracts were then subjected to IP for the immunopurification of endogenous CBP80 or eIF4E. The levels of co-immunopurified proteins and histone mRNAs were then analyzed by western blotting and qRT-PCR (Figure 4). The specificity of each IP was demonstrated by western blotting using the indicated antibodies (Figure 4A). The levels of two replication-dependent histone mRNAs (HIST1H1C mRNA and HIST2H2AA mRNA) and a replication-independent histone mRNA (H3F3A mRNA) were analyzed by qRT-PCR (Figure 4B and C).
Figure 4.
Histone mRNA degradation occurs on CBP80/20-bound mRNAs. HEK293T cells were transiently transfected with CTIF siRNA or non-specific control siRNA. Three days later, cells were treated with HU for 1 h before cell harvest. Cell extracts were subjected to IP using α-CBP80 antibody or α-eIF4E antibody. Co-immunopurified proteins and RNAs were analyzed by WB and qRT-PCRs. (A) WB results to demonstrate the specific IPs of CBP80 (left) and eIF4E (right). The relative amounts of cell extracts used before IP compared with after IP are provided in Supplementary Table S1. (B) qRT-PCR of HIST1H1C mRNAs. The levels of HIST1H1C mRNAs were normalized to the level of H3F3A mRNAs. The normalized levels of HIST1H1C mRNAs that were obtained from cells untreated with HU before or after IP were then set to 100%. *P < 0.05 as determined by Student’s t-tests. (C) As in Figure 4B, except that the levels of endogenous HIST2H2AA mRNAs were analyzed by qRT-PCRs. The columns and bars in each panel represent the mean and standard deviation of at least two independently performed transfections, IPs and qRT-PCRs. *P < 0.05.
Based on previous reports (4,8,33), we can offer several speculations. First, if histone mRNA degradation were preferentially coupled to ET, reductions in the levels of co-immunopurified histone mRNAs would only be observed in the IP of eIF4E in a CTIF-independent manner. Second, if histone mRNA degradation were preferentially coupled to CT, comparable reductions in levels of co-immunopurified histone mRNAs would be observed in the IPs of both CBP80 and eIF4E. In addition, downregulation of CTIF would result in comparable increases in the levels of co-immunopurified histone mRNAs in the IP of both CBP80 and eIF4E. Third, if histone mRNA degradation were coupled to both CT and ET, gradual reductions of co-immunopurified histone mRNAs would be observed in the IPs of CBP80 and eIF4E, respectively. In this case, CTIF downregulation would cause an increase in the levels of co-immunopurified histone mRNAs only in the IP of CBP80.
The results of qRT-PCR using the samples before IPs showed that the levels of HIST1H1C mRNAs and HIST2H2AA mRNAs on HU treatment were reduced to 17–21% and 14% of untreated levels, respectively (Figure 4B and C, Before IP). Furthermore, CTIF downregulation increased the amount of histone mRNAs by 3-fold (Figure 4B and C, Before IP).
Next, the levels of co-immunopurified HIST1H1C mRNAs and HIST2H2AA mRNAs after CBP80 IP or eIF4E IP were measured using qRT-PCR. The results revealed comparable reductions in the levels of co-immunopurified HIST1H1C mRNAs and HIST2H2AA mRNAs in the IPs of CBP80 and eIF4E (Figure 4B and C, After IP). Intriguingly, ∼2–3-fold greater amounts of co-immunopurified histone mRNAs were enriched in both CBP80 IP and eIF4E IP when CTIF was downregulated (Figure 4B and C, After IP). Given that CBP80/20-bound messenger ribonucleoprotein (mRNP) is a precursor form of eIF4E-bound mRNP (4,8) and histone mRNA degradation requires translation (26–28), these observations suggest that the degradation of replication-dependent histone mRNAs largely takes place during CT (i.e. on CBP80/20-bound mRNAs). In support of this suggestion, the degradation of replication-dependent histone mRNAs was not significantly affected by the selective inhibition of ET by the overexpression of eIF4E-BP1 (Supplementary Figure S5), which competes with eIF4G for binding to eIF4E (8,14), and by the overexpression of eIF4E-T (Supplementary Figure S6), which causes nuclear accumulation of eIF4E (34,35).
A greater fraction of replication-dependent HIST1H1C mRNAs and HIST2H2AA mRNAs binds to CBP80/20 than eIF4E, compared with poly(A)-containing β-actin mRNAs and eEF2 mRNAs
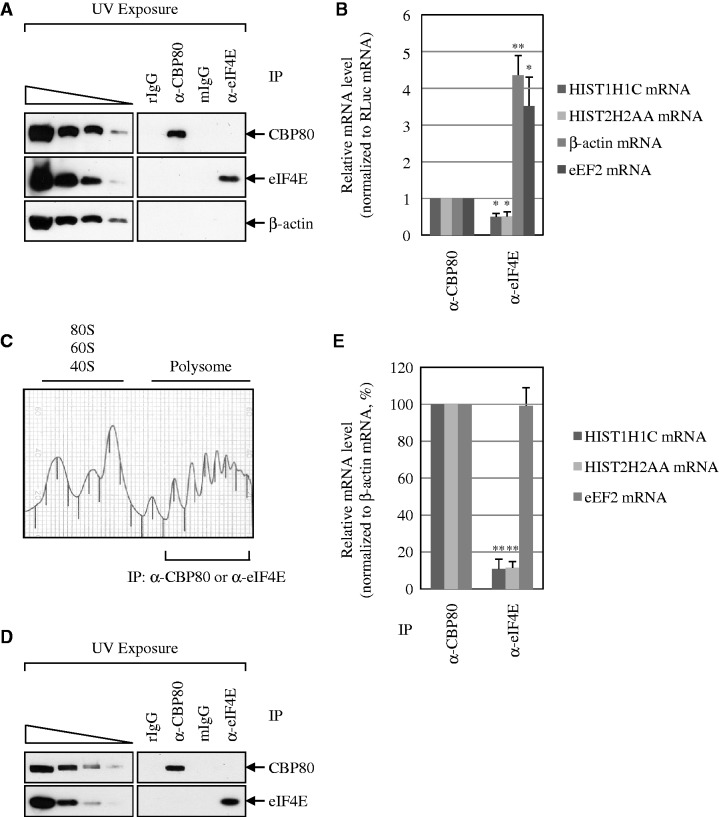
Considering that histone mRNAs are rapidly degraded on the inhibition of DNA replication or at the end of S phase in a translation-dependent manner (26–28), and that the rapid degradation of histone mRNAs occurs on CBP80/20-bound mRNAs (Figures 3 and 4), it is likely that a larger amount of histone mRNA binds to CBP80/20 than eIF4E at the steady state. To address this idea, we performed IPs using α-CBP80 antibody or α-eIF4E antibody. To rule out a possible mRNA rearrangement during IP, cells were ultraviolet (UV) irradiated before harvest. In addition, to control for variations of RNA extraction and qRT-PCR, in vitro-transcribed Renilla luciferase (RLuc) mRNAs were added to samples after IPs.
The specific IPs were confirmed by western blotting (Figure 5A). IP efficiencies of CBP80 and eIF4E were 12 ± 0% and 15 ± 6%, respectively. qRT-PCR results showed that the amounts of β-actin and eEF2 mRNAs associated with eIF4E were 4- and 3-fold more, respectively, than the amounts associated with CBP80 (Figure 5B). On the contrary, the amounts of HIST1H1C mRNAs and HIST2H2AA mRNAs associated with eIF4E were 2-fold less than the amounts associated with CBP80 (Figure 5B). These results suggest that replication-dependent histone mRNAs bind to CBP80/20 by 6- to 8-fold more than to eIF4E at the steady state, compared with the poly(A)-containing cellular mRNAs.
Figure 5.
A larger amount of replication-dependent histone mRNAs bind to CBP80/20 than eIF4E at the steady state, compared with poly(A)-containing mRNAs. (A and B) IPs were performed using either α-CBP80 antibody or α-eIF4E antibody and the extracts of UV-irradiated HEK293T cells. To control for variations of RNA extraction and qRT-PCR, in vitro-transcribed Renilla luciferase (RLuc) mRNAs were added to samples after IPs. (A) WB results to demonstrate the specific IPs of CBP80 and eIF4E. (B) qRT-PCR of co-immunoprecipitated mRNAs. The level of each tested mRNA was normalized to the level of RLuc mRNA. The normalized level of each mRNA was further normalized to the IP efficiency. The normalized level of each mRNA obtained in CBP80 IP was then set to 1. The columns and bars in each panel represent the mean and standard deviation of at least two independently performed transfections, IPs and qRT-PCRs. *P < 0.05, **P < 0.01. (C–E) IPs of polysome fractions. The cytosolic extracts were obtained and then fractionated on 10–50% sucrose gradients to resolve the polysomes. Polysome-fractionated protein samples were pooled, subjected to the exposure to UV, and IPs using either α-CBP80 antibody or α-eIF4E antibody. Rabbit (r) and mouse (m) IgGs were used for negative controls for α-CBP80 antibody and α-eIF4E antibody, respectively. (C) The polysome profile of cytosolic extracts. (D) WB results to demonstrate the specific IPs of CBP80 and eIF4E from polysome fractionated samples. (E) qRT-PCRs of co-immunoprecipitated mRNAs. The levels of co-immunoprecipitated histone mRNAs and eEF2 mRNAs were normalized to the level of β-actin mRNA. The normalized levels in CBP80 IP were set to 100%. The relative amounts of cell extracts used before IP compared with after IP are provided in Supplementary Table S1. **P < 0.01.
To further test whether a larger amount of histone mRNA being translated binds to CBP80/20 than eIF4E, we used a polysome-fractionation approach followed by IPs (Figure 5C–E). To rule out the possibility that the previously described results reflect a difference in nuclear and cytoplasmic distributions of the tested mRNAs, the cytosolic extracts were obtained and then fractionated by sucrose gradients (Figure 5C). The polysome fractions were pooled and subjected to the exposure to UV and IP using either α-CBP80 or α-eIF4E antibody (Figure 5D). The co-immunopurified mRNAs were analyzed using qRT-PCRs (Figure 5E).
In support of the previously described findings (Figure 5A and B), the relative ratio of co-immunopurified HIST1H1C and HIST2H2AA mRNAs to β-actin mRNAs in CBP80 IP was 9-fold higher than the relative ratio in eIF4E IP (Figure 5E). On the other hand, comparable levels of the relative ratio of co-immunopurified eEF2 mRNAs to β-actin mRNAs were observed in IPs of CBP80 and eIF4E.
All these results suggest that a majority of histone mRNAs being translated in the cytoplasm bind to CBP80/20 than eIF4E, compared with poly(A)-containing mRNAs.
DISCUSSION
Here, we identify an SLBP as a CTIF-interacting protein and show that histone mRNA degradation largely occurs on CBP80/20-bound mRNA. Based on these results, we propose a model in which the histone mRNPs are remodeled from CT complex to either ET complex or degradation complex (Supplementary Figure S7). Histone mRNA is exported from the nucleus to the cytoplasm via NPC with CBP80/20 bound to the 5′-end of the cap structure and with SLBP bound to the 3′-end. During export, CTIF, which is largely localized to the cytoplasmic side of the nuclear envelope (5), would be loaded onto the 5′-end of the histone mRNA by virtue of its interaction with CBP80 (5). Alternatively, CTIF could be loaded onto the 5′-end of the histone mRNA in the nucleus because a minor fraction of CTIF is localized to the nucleus (5). The loaded CTIF recruits eIF3 via CTIF–eIF3g interaction (7) and, in turn, recruits the 40S ribosome subunit to initiate CT (5). When the 3′-end of histone mRNA is completely exported, SLBP at the 3′-end might directly interact with CBP80 at the 5′-end (36), probably generating circularized histone mRNP.
After or during CT, CBP80/20 is replaced by eIF4E in a translation-independent manner by importin-α and importin-β (12). Notably, we found that a larger amount of histone mRNAs binds to CBP80/20 than eIF4E at steady state (Figure 5). Thus, it is plausible that an interaction between 5′-untranslated region (UTR) and 3′-UTR via CTIF–SLBP interaction stabilizes CBP80/20-bound histone mRNAs, blocking the replacement of CBP80/20 with eIF4E. The stabilized histone mRNAs would be subject to multiple rounds of CT. A minor fraction of histone mRNAs would undergo mRNP remodeling from CT complex to ET complex. The eIF4E-bound histone mRNA would be stable because histone mRNA degradation largely occurs on CBP80/20-bound mRNA (Figure 4).
Although our group showed that CBP80 preferentially associates with CTIF (5) and that SLBP and SLIP1 preferentially associate with CT complex (Figure 2), several alternative CT complexes of replication-dependent histone mRNAs could be formed based on previous reports: (i) an interaction between CBP80 and eIF4GI (13,37) and (ii) an interaction between eIF4GI and SLIP1 (22). The formation of different CT complexes of histone mRNA may vary by cellular conditions, tissues or cell types (1). However, alternative complexes including interactions of CBP80-eIF4GI/II, SLIP1-eIF4GI/II or SLBP-eIF4GI/II may be weak or transient (5,22). In support of this idea, SLIP1-eIF4GI interaction is abolished by RNase treatment (22). In addition, although SLBP-eIF4GI interaction was not detected in IPs using non-cross-linked cell extracts (Figure 2), a significant SLBP–eIF4GI interaction was observed in the IP using the extracts of cells treated with formaldehyde (Supplementary Figure S8), which helps cross-link protein to protein.
A previous report showed that introduction of the W73A mutation in Xenopus SLBP1 abolishes the interaction between Xenopus SLBP1 and human SLIP1 (22). However, the interaction of FLAG-SLBP and Myc-SLIP1 was not affected by the W75A mutation in human SLBP, although the human CTIF–SLBP interaction was drastically inhibited by the W75A mutation under our conditions (Supplementary Figure S9). This may be due to the difference in amino acid sequences among human SLBP and Xenopus SLBP1 (71% similarity and 56% identity in amino acid sequences).
When cells encounter DNA replication stress or reach the end of S phase, histone mRNPs may undergo remodeling, in which a terminating ribosome on a stop codon may recruit Upf1 more actively (29). In turn, Upf1 may recruit terminal uridylyl transferases (TUTases) and general mRNA degrading enzymes to remove the entire histone mRNA (30). Future studies need to address what signaling pathway causes the remodeling of CBP80/20-bound histone mRNP on DNA replication stress or at the end of S phase. Also, the molecular details of CBP80/20-bound histone mRNP remodeling from a CT complex to a degradation-competent complex should be addressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–9, Supplementary Manuscript, Supplementary Materials and Methods and Supplementary References [38–40].
FUNDING
Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [2012002469] and by Korea University Grant (to Y.K.K.); National Research Foundation of Korea funded by the Korean Government (Ministry of Education, Science and Technology) [NRF-35B-2011-1-C00026 to K.M.K.]. Funding for open access charge: NRF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank C.D. Hu for providing BiFC plasmids, B. Müller for α-SLBP antibody, S.K. Jang for α-eIF4GI antibody and W.F. Marzluff for α-SLIP1 antibody and also D. Morris, W.F. Marzluff and L.E. Maquat for scientific comments.
REFERENCES
- 1.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durand S, Lykke-Andersen J. SnapShot: nonsense-mediated mRNA decay. Cell. 2011;145:324–324. doi: 10.1016/j.cell.2011.03.038. e2. [DOI] [PubMed] [Google Scholar]
- 4.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 5.Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV, Lorsch JR, Hellen CU. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 7.Choe J, Oh N, Park S, Lee YK, Song OK, Locker N, Chi SG, Kim YK. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J. Biol. Chem. 2012;287:18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apcher S, Daskalogianni C, Lejeune F, Manoury B, Imhoos G, Heslop L, Fahraeus R. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc. Natl Acad. Sci. USA. 2011;108:11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009;23:2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 14.Choe J, Cho H, Lee HC, Kim YK. microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 2010;11:380–386. doi: 10.1038/embor.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe J, Cho H, Chi SG, Kim YK. Ago2/miRISC-mediated inhibition of CBP80/20-dependent translation and thereby abrogation of nonsense-mediated mRNA decay require the cap-associating activity of Ago2. FEBS Lett. 2011;585:2682–2687. doi: 10.1016/j.febslet.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 16.Jaeger S, Barends S, Giege R, Eriani G, Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. doi: 10.1016/j.biochi.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS, Kim YK. Staufen1-mediated mRNA decay functions in adipogenesis. Mol. Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 21.Zhao X, McKillop-Smith S, Muller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 2004;117:6043–6051. doi: 10.1242/jcs.01523. [DOI] [PubMed] [Google Scholar]
- 22.Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol. Cell Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez R, Marzluff WF. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graves RA, Pandey NB, Chodchoy N, Marzluff WF. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 28.Baumbach LL, Marashi F, Plumb M, Stein G, Stein J. Inhibition of DNA replication coordinately reduces cellular levels of core and H1 histone mRNAs: requirement for protein synthesis. Biochemistry. 1984;23:1618–1625. doi: 10.1021/bi00303a006. [DOI] [PubMed] [Google Scholar]
- 29.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 30.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells D, Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc. Natl Acad. Sci. USA. 1985;82:2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brush D, Dodgson JB, Choi OR, Stevens PW, Engel JD. Replacement variant histone genes contain intervening sequences. Mol. Cell. Biol. 1985;5:1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HC, Cho H, Kim YK. Ectopic expression of eIF4E-transporter triggers the movement of eIF4E into P-bodies, inhibiting steady-state translation but not the pioneer round of translation. Biochem. Biophys. Res. Commun. 2008;369:1160–1165. doi: 10.1016/j.bbrc.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 37.McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh YL, Hahm B, Kim YK, Lee HK, Lee JW, Song O, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang SK. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 1998;331(Pt 1):169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 40.Streit S, Michalski CW, Erkan M, Kleeff J, Friess H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat. Protoc. 2009;4:37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.