Abstract
Ribosomal subunit biogenesis in eukaryotes is a complex multistep process. Mrd1 is an essential and conserved small (40S) ribosomal subunit synthesis factor that is required for early cleavages in the 35S pre-ribosomal RNA (rRNA). Yeast Mrd1 contains five RNA-binding domains (RBDs), all of which are necessary for optimal function of the protein. Proteomic data showed that Mrd1 is part of the early pre-ribosomal complexes, and deletion of individual RBDs perturbs the pre-ribosomal structure. In vivo ultraviolet cross-linking showed that Mrd1 binds to the pre-rRNA at two sites within the 18S region, in helix 27 (h27) and helix 28. The major binding site lies in h27, and mutational analyses shows that this interaction requires the RBD1-3 region of Mrd1. RBD2 plays the dominant role in h27 binding, but other RBDs also contribute directly. h27 and helix 28 are located close to the sequences that form the central pseudoknot, a key structural feature of the mature 40S subunit. We speculate that the modular structure of Mrd1 coordinates pseudoknot formation with pre-rRNA processing and subunit assembly.
INTRODUCTION
Biogenesis of the eukaryotic small (40S) and large (60S) ribosomal subunits is a complex multistep process that requires a large number of non-ribosomal proteins and many small nucleolar RNAs (snoRNAs) (1–3). The 5S ribosomal RNA (rRNA) component of the 60S subunit is transcribed by RNA polymerase III. The large precursor-rRNA (pre-rRNA), transcribed by RNA polymerase I, contains the three remaining rRNAs, the 18S rRNA of the 40S subunit and the 5.8S and 25/28S rRNAs of the 60S subunit. The processing of the 5′ part of the pre-rRNA into a pre-40S subunit and the processing of the 3′ part into a pre-60S subunit are largely, but not completely, independent (4).
In growing yeast cells, roughly 70% of the pre-40S subunits are released co-transcriptionally from the earliest detectable precursor (90S) (5,6). Separation of the pre-40S and pre-60S subunits occurs when the RNA polymerase I has transcribed well into the 25S region of the gene, ∼100 s after initiation of transcription (5,6). During this time window, the pre-rRNA is continuously synthesized and folded, as well as assembled with many proteins and some ribosomal proteins. In addition, the many small nucleolar ribonucleoproteins (snoRNPs) find their specific locations within the pre-rRNA and perform their modifications of the pre-rRNA (5). It is still unknown how all these co-transcriptional events take place within a dynamic pre-rRNP. However, it is evident that the pre-rRNA has to fold in sequential intermediate folds and assemble into intermediate pre-rRNP complexes. Processing subcomplexes have been identified and their order of association with the pre-rRNA studied (7,8), and different sequential structures have been observed in the electron microscope (6). Presumably, the pre-rRNA has evolved to optimize the specific folds and assemblies, and it is furthermore reasonable to assume that processing factors facilitate these structures to allow coordination, minimize non-productive structures and allow for quality control.
The box C/D U3 snoRNP has an essential role in pre-40S processing, being required for three early cleavages of the pre-rRNA, at positions A0–A2. These cleavages define and liberate the 20S rRNA, contained in the pre-40S subunit (9). The U3 snoRNP appears to be involved in coordinating essential pre-rRNP structures via its base pairing at several positions within the 5′ external transcribed spacer (ETS) and at the 5′ end of the 18S rRNA (10–14).
Mrd1 is an essential RNA-binding protein involved in the maturation of the small ribosomal subunits in Saccharomyces cerevisiae (15). It contains five RNA-binding domains (RBDs), whereas higher eukaryotic homologues have six such domains (16). Mrd1 binds co-transcriptionally to the pre-rRNA, and together with many other factors, it is involved in processing of the 90S pre-ribosome, which will result in mature 40S subunits (15,17). All of the individual RBDs are required for optimal protein function, but they are of different importance (18). RBD3 and RBD5 are essential for protein function, whereas RBD1, RBD2 and RBD4 are non-essential, but growth defects of the individual deletions range from mild (RBD4) to severe (RBD1) (18). The presence of multiple RBDs and previous experimental findings suggested that Mrd1 associates directly with the pre-rRNA and participates in formation of processing competent pre-rRNP structures. To assess the function of Mrd1 further, we used in vivo ultraviolet (UV) cross-linking and complementary DNA (cDNA) cloning (19) to map potential RNA interactions of Mrd1. Mrd1 was found to be associated with the pre-rRNA at two sites within the 18S sequence, located close to the 3′ side of the universally conserved pseudoknot within the mature 18S rRNA structure. The location of the binding sites in the pre-ribosome indicates that Mrd1 is involved in the folding and/or restructuring of the pre-rRNA, leading to formation of the central pseudoknot. This structure is conserved throughout evolution and is a key feature of the small ribosomal subunit. Analyses of RBD deletions revealed that the N-terminal half of Mrd1 associates with the major interaction site within the 18S sequence, and that RBD2 is important for the efficiency of this interaction.
MATERIALS AND METHODS
Strains and plasmids
The MRD1 gene and the MRD1 RBD deletion genes were genomically tagged at their 3′ ends with either the Calmodulin-TEV-Protein A (TAP) or the His6-TEV-Protein A (HTP) epitopes by a one-step polymerase chain reaction (PCR)-based gene integration (20). The PCR fragments were made by amplification of the TAP tag from pBS1539 and the HTP tag from the pBS1539/HTP or pBS1479/HTP vectors (19) flanked by MRD1- or RRP5-specific sequences. Strains were verified by PCR and western blot. Conditionally mutant strains (HisMX6-PGAL1-3HA-MRD1 and HisMX6-PGAL1-3HA-RRP5) and 3HA epitope-tagged strains (UTP10-3HA-His3MX6, UTP22-3HA-His3MX6, UTP22-3HA-KanMX6 and RRP5-3HA-KanMX6) were created by transformation with a PCR construct containing either the His3MX6-PGAL1-3HA, 3HA-His3MX6 or 3HA-KanMX6 gene cassettes with flanking regions of the target gene, and tested for glucose sensitivity or histidine prototrophy and/or G418 resistance. The genotypes of the strains used in this study are listed in Supplementary Table S1.
To integrate an additional genomic MRD1 wt allele under control of the GAL1 promoter into the ΔRBD1, ΔRBD3 and ΔRBD5 strains, the plasmid pRS406-PGAL-HA-MRD1 was cut with StuI in the URA3 open reading frame. Transformants were selected for uracil prototrophy and tested for galactose-dependent HA-MRD1 protein expression.
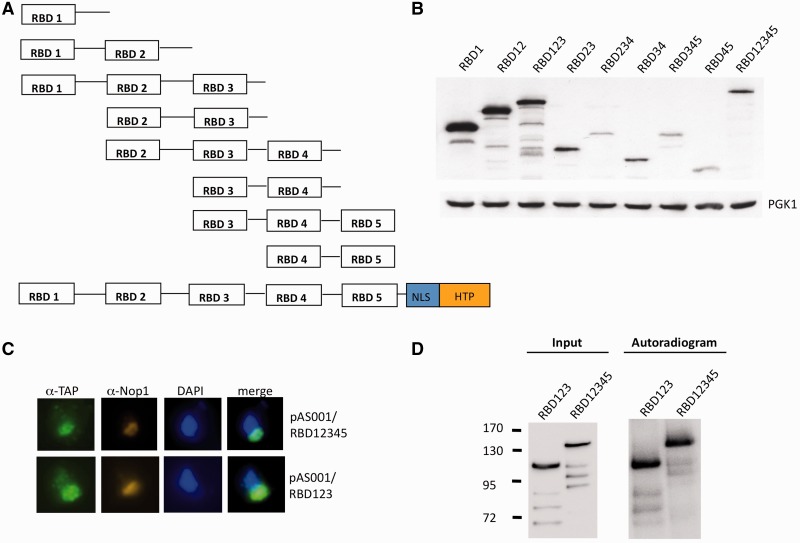
A PCR construct with 500 nt upstream the transcription start site for MRD1, a BssHII and a BclI restriction site, the HTP tag-klURA3 and a nuclear localization signal was transformed together with SalI and KpnI cleaved pRS316 to construct plasmid pAS001 via homologous recombination. The pAS001 plasmid was digested with BssHII and BclI, and transformed into yeast together with various MRD1-RBD PCR constructs flanked by pAS001-specific sequence creating pAS001/RBD1, RBD12, RBD123, RBD23, RBD234, RBD34, RBD345, RBD45 and RBD12345 (Figure 4A). The plasmids were sequenced, and expression of HTP-tagged RBD domains was verified by western blots.
Figure 4.
The N-terminal part of Mrd1 is responsible for interaction of Mrd1 at h27. (A) Schematic figure of the RBD constructs in pRS316. All constructs contained the nuclear localization signal and HTP regions at their C-terminus as shown for RBD12345. (B) Western blot analysis of the protein expression of the RBD constructs visualized with Peroxidase-Anti-Peroxidase Soluble Complex (upper). Anti-phosphoglycerokinase 1 (lower) served as a loading control. (C) Immunocytochemistry of RBD12345-HTP (top) and RBD123-HTP (bottom) constructs with anti-TAP (green) or anti-Nop1 (red) antibodies and DAPI. (D) CRAC analysis of RBD12345-HTP and RBD123-HTP constructs.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde at room temperature for 30 min. The cells were treated with zymolase, adhered to poly-L-lysine-coated coverslips and dehydrated over night in 70% ethanol at −20°C. HTP-tagged Mrd1 protein segments were visualized using rabbit anti-TAP (CAB1001, 1:200, Open Biosystems) and anti-rabbit Alexafluor-488-conjugated secondary antibodies (Molecular Probes). The nucleolus was identified using a mouse anti-Nop1 antibody (MCA-38F3, 1:800, EnCor Biotech), and a secondary goat anti-mouse antibody conjugated to Alexafluor-647 (Molecular Probes). The DNA was visualized with DAPI, which was present in the mounting agent.
UV cross-linking and cDNA cloning (CRAC)
Cross-linking and cloning of cDNAs was performed as described previously (19). UV-cross-linking was either performed in vivo with cells in Petri dishes in a Stratalinker 1800 (Stratagene) or in vivo in culture medium in the Megatron (UVO3) as described previously (21). UV cross-linking and analysis of cDNAs (CRAC) cDNA libraries were generated using adapter sequences as described previously (21).
Bioinformatics analysis of CRAC data
CRAC cDNA sequence libraries were generated using Illumina HiSeq and MiSeq systems according to the manufacturer’s procedures.
After quality filtering, PCR duplicates were removed using fastx_collapser (http://hannonlab.cshl.edu/fastx_toolkit/), and random nucleotide information was stored in the 5′ adapter sequences of the reads. Reads were aligned to the yeast genomic reference sequence (ENSEMBL) using Novoalign 2.0.7 (www.novocraft.com). Downstream analyses, including the pile-ups presented here, were performed using the pyCRAC tool suite (available on request: sgrannem@staffmail.ed.ac.uk; manuscript in preparation).
Sucrose gradient centrifugations
Sucrose gradient centrifugations were made essentially as described previously (17). ASY082, 094, 111 and PLY178 with the plasmid pAS001/RBD123 were depleted for either Mrd1 or Rrp5 in glucose for 7 h before extraction.
Tandem affinity purification of Mrd1-TAP/HTP, ΔRBD1-TAP/HTP, ΔRBD3-TAP/HTP and ΔRBD5-TAP/HTP
The TAP purifications were performed essentially as described previously (22). The ΔRBD1-TAP/HTP, ΔRBD3-TAP/HTP and ΔRBD5-TAP/HTP strains were depleted for endogenous Mrd1 for 6 h in glucose before harvest. Cells were grown to OD600 ∼1, washed in 1X phosphate buffered saline and frozen in liquid nitrogen. Cell pellets corresponding to 6 × 1010 cells each were resuspended in 1 volume lysis buffer, TMN150 [50 mM Tris–HCl (pH 7.8), 150 mM NaCl, 1.5 mM MgCl2, 0.1% Nonidet-P40 and 5 mM beta-mercaptoethanol], containing 1X Complete Protease Inhibitor Cocktail, EDTA free, (Roche) and lysed with 3 volumes Zirconia beads (0.5 mm, Thistle Scientific). The cell extracts were resuspended in 3 volumes lysis buffer and clarified by centrifugation (20 min at 4600 r.p.m. and 20 min at 14 000r.p.m.). The pH was adjusted to 7.5 by 1 M Tris–HCl (pH 8.8). Extracts were incubated with 500 µl pre-equilibrated IgG Sepharose 6 Fast Flow beads (GE Healthcare) for 1 h at 4°C. Beads were washed three times in 10 ml of TMN150, resuspended in 500 µl of TMN150 buffer with 30 U of AcTEV (Invitrogen) and incubated for 2 h at 18°C. The solution was transferred to micro bio-spin chromatography columns (BioRad) and the eluate collected. The eluated samples were mixed with 500 µl of TMN150 containing 4 mM CaCl2 and incubated with 150 µl pre-equilibrated Calmodulin Sepharose 4B beads (GE Healthcare) for 2 h at 4°C. The beads were washed three times with 1 ml of TMN150 with 2 mM CaCl2. The washed beads were transferred to micro bio-spin chromatography columns and eluted with EGTA buffer [5 mM EGTA, 10 mM Tris–HCl (pH 7.8) and 50 mM NaCl]. The proteins were precipitated with TCA and 20 µg glycogen, and half of the precipitated material was run on a 4–12% NuPAGE gel and silver stained according to the protocol for SilverQuest staining kit (Invitrogen). The other half was subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis, in-gel digested using trypsin (23) and analysed using LC-MS on a LTQ-Orbitrap (Thermo Fisher Scientific) coupled to high pressure liquid chromatography via a nanoelectrospray ion source. The MS data were analysed using MaxQuant (24) and proteins identified by searching MS and MS/MS data using the MASCOT search engine (Matrix Science, UK).
Immunoprecipitations (IPs)
Extracts from 25 OD600 units of growing cells were used for tandem affinity purifications. Strains ASY080-084 and ASY105-110 were grown to OD600 ∼1, and for each strain, 25 OD600 units of cells were used for tandem affinity purification as described in the previous section. For strain ASY082-084, ASY107 and ASY110, expression of endogenous Mrd1 was shut off in glucose for 6 h before harvest. Purified proteins were resuspended in loading buffer, separated in 4–12% NuPAGE gels and transferred to Immobilon PVDF membranes (Millipore).
Immunoprecipitation (IP) of ribonuclease (RNase)-treated protein complexes was done as described previously on extracts that were incubated with 50 U of RNaceIt (Stratagene) for 15 min at 37°C before centrifugation to clear the extracts.
For analysis of affinity-purified U3 snoRNA and 35S pre-rRNA, 25 OD600 units of cells from strains ASY053-056 and PLY094 (wt Mrd1) were harvested, extracted and purified on IgG Sepharose 6 Fast Flow beads as described previously. RNA was extracted with a mixture of Guanidine thiocyanate and phenol, followed by ethanol precipitation. The purified RNA was analysed by northern blot hybridization.
Western blot analysis
Primary antibodies used were polyclonal rabbit anti-TAP antibody (CAB1001, 1:5000, Open Biosystems), anti-HA peroxidase monoclonal antibody (3F10, 1:500–1:1000, Roche), polyclonal anti-Mpp10 antibody (1:10 000) (25), Peroxidase-Anti-Peroxidase Soluble Complex (P1291, 1:5000, Sigma) and monoclonal anti-PGK1 antibody (anti-phosphoglycerokinase 1: 22C5, 1:10 000, Molecular Probes). Secondary antibodies used were polyclonal goat anti-mouse- and swine anti-rabbit HRP-coupled antibodies (1:1000–1:10 000, DakoCytomation), and immunodetection was performed with ECL solution (Amersham Biosciences).
Northern blot analysis
RNA was separated on 1.5% agarose-formaldehyde gels, blotted onto Zeta-probe membranes (BioRad), UV cross-linked and hybridized with 32P-labelled oligonucleotide probes as described previously (17).
RNA–protein binding in vitro
The complete Mrd1 was expressed in Escherichia coli as a GST fusion protein and purified on Glutathione-Sepharose 4B beads (GE Healthcare). Specified parts of the MRD1 gene were obtained by PCR using specific primers and genomic DNA as template. The gene fragments were cloned into the pET-46 Ek/LIC vector (Novagen) and expressed in E. coli. His-tagged proteins were purified by affinity chromatography (Ni-NTA agarose, Qiagen), followed by gel chromatography on Hi Load 16/60, size S-75 or S-200, Superdex (GE Healthcare).
Defined parts of the 35S pre-rRNA were obtained by PCR, introducing a T7 promoter at the 5′ end of each PCR fragment. Central regions of 18S rRNA were synthesized and cloned (Eurofins MWG Operon), before PCR. 32P-labelled RNA was synthesized by T7 polymerase in vitro. The RNA was purified on 4% denaturing polyacrylamide gels. For in vitro binding, performed essentially as described previously (26), 2–3 fmol of RNA was heated at 60°C for 15 min in RNA buffer [20 mM Tris–HCl (pH 7.5) 200 mM KCl and 5 mM MgCl2] and allowed to cool to 20°C during 5 min. The RNA was then incubated with different concentrations of purified protein in 20 mM Tris–HCl (pH 7.5) 200 mM KCl, 5 mM MgCl2, 20% glycerol, 50 µg/ml transfer RNA and 10 µg/ml bovine serum albumin, for 30 min at 20°C. The RNA–protein mixtures were filtered through nitrocellulose filters (0.45 µm HA, Millipore), followed by three washes of 300 µl of RNA buffer. The fraction of bound RNA was measured by Cerenkov counting.
RESULTS
Mrd1 is part of pre-ribosomal complexes, and RBD deletions perturb these complexes
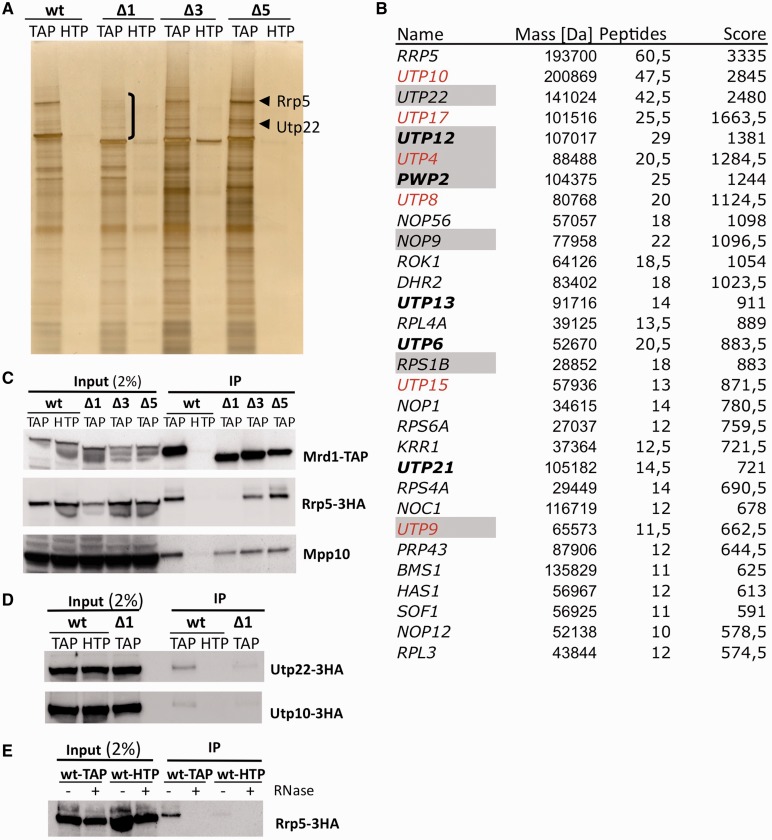
To assess whether Mrd1 is part of pre-ribosomal complexes, we performed TAP purifications of C-terminally TAP-tagged wt Mrd1 (Figure 1A). Mrd1-HTP, which lacks the calmodulin-binding peptide used in the second step of TAP purification, served as a control. Mass spectrometry analysis of complexes purified from two individual experiments with wt Mrd1-TAP showed enrichment for many known components of the early 90S pre-ribosome or small subunit processome. Figure 1B shows a list of the top 30 purified proteins listed after average peptide scoring from the two experiments. The top hit was Rrp5, which is involved in both 40S and 60S subunit maturation (27). Six of the seven characterized UTP-A/t-Utp proteins (Figure 1B, red), five of the six characterized UTP-B proteins (Figure 1B, bold) and the UTP-C protein Utp22 were included among the 30 top proteins. In addition, we found four RNA helicases. Seven of the proteins (Figure 1B, grey shadow) have previously been identified as Mrd1-interacting proteins [Utp22, Rps1B, Utp9 (28), Utp12 (29), Utp4, Pwp2 (17) and Nop9 (30)]. We conclude that Mrd1 is part of the 90S pre-ribosomal complex.
Figure 1.
Mrd1 is part of pre-ribosomes, and deletion of RBDs perturbs the pre-ribosome. (A) Tandem affinity purification of Mrd1-TAP/-HTP (wt), ΔRBD1-TAP/-HTP, ΔRBD3-TAP/-HTP and ΔRBD5-TAP/-HTP cells resolved on a 4–12% Bis-Tris NuPAGE gel and were silver stained. Bracketed region shows where ΔRBD1-TAP exhibits reduced protein interaction. Arrows indicate position of Rrp5 and Utp22. (B) Table of the 30 top hit proteins purified with Mrd1-TAP based on average peptide scoring number after mass spectrometry from two sets of Mrd1-TAP purifications. Grey boxes indicate previously identified Mrd1-interacting components [Utp22, Rps1B, Utp9 (28), Utp12 (29), Utp4, Pwp2 (17) and Nop9 (30)]. Red indicates t-UTPs/UTP-A complex proteins and bold UTP-B complex proteins. Proteins purified also with Mrd1-HTP were removed from the list. (C) Anti-TAP IPs of Mrd1-TAP/Rrp5-3HA (wt), Mrd1-HTP/Rrp5-3HA (wt), ΔRBD1-TAP/Rrp5-3HA, ΔRBD3-TAP/Rrp5-3HA and ΔRBD5-TAP/Rrp5-3HA cells. Western blot analysis with anti-TAP (top), anti-HA peroxidase (middle) and anti-Mpp10 (bottom) antibodies. (D) Anti-TAP IPs of Mrd1-TAP/Utp22-3HA (wt), Mrd1-HTP/Utp22-3HA (wt), ΔRBD1-TAP/Utp22-3HA cells (top), Mrd1-TAP/Utp10-3HA (wt), Mrd1-HTP/Utp10-3HA (wt) and ΔRBD1-TAP/Utp10-3HA cells (bottom), and western blot analysis with anti-HA peroxidase antibodies. (E) Anti-TAP IPs of Mrd1-TAP/Rrp5-3HA (wt-TAP) and Mrd1-HTP/Rrp5-3HA (wt-HTP) cells in the presence (+) or absence (−) of RNase treatment of cell extracts before IP. Western blot analysis with anti-HA peroxidase antibodies.
Deletion of RBD3 or RBD5 (named ΔRBD3 or ΔRBD5) has previously shown that RBD3 and RBD5 are essential for Mrd1 function, while deletion of RBD1 (named ΔRBD1) results in severe growth defect and lethality at 37°C (18). TAP purifications of the ΔRBD3, ΔRBD5 and ΔRBD1 mutants were compared with wt Mrd1 (Figure 1A). The ΔRBD1 protein showed distinctly reduced association with high-molecular weight-interacting proteins, including the prominent Rrp5 and Utp22 (Figure 1A, compare wt lane with Δ1). Minor changes were observed in the protein composition of pre-ribosomes purified with the ΔRBD3 or ΔRBD5 Mrd1 proteins (Figure 1A). These results show that the ΔRBD1, ΔRBD3 and ΔRBD5 Mrd1 proteins are associated with pre-ribosomes. Consistent with this, all three deletion versions of Mrd1 were associated with U3 snoRNA and 35S pre-rRNA as shown by co-purification (Supplementary Figure S1). The composition of the pre-ribosomes containing ΔRBD1, ΔRBD3 and ΔRBD5 is, however, perturbed.
The TAP-purified pre-ribosomes were further analysed by western blotting in strains where wt Mrd1, ΔRBD1, ΔRBD3 or ΔRBD5 were TAP tagged, and Rrp5 was C-terminally tagged with a 3HA epitope. The co-purification of Rrp5-3HA was dramatically reduced in ΔRBD1, as compared with wt, ΔRBD3 and ΔRBD5 (Figure 1C). The loss of Rrp5-3HA was not because of a general collapse of pre-ribosomes in the presence of ΔRBD1, as there was no significant change in co-precipitation of Mpp10 with the ΔRBD1 mutant Mrd1 (Figure 1C). In agreement with the pattern of migration for co-purified proteins (Figure 1A), the large Utp22-3HA and Utp10-3HA also showed reduced association with ΔRBD1, as compared with wt Mrd1 (Figure 1D).
We could not detect reciprocal loss of Mrd1 ΔRBD1-13-Myc in western blot analyses of Rrp5 TAP-purified proteins (data not shown). This may result from differences in pre-ribosome stability in combination with which TAP-tagged protein was used for precipitation of pre-ribosomes. However, ΔRBD1 clearly influences the pre-ribosomes such that Rrp5-3HA is not associated with the ΔRBD1-TAP-purified complexes.
We investigated whether Mrd1 or ΔRBD1 influences recruitment of Rrp5 to pre-ribosomes. Sucrose gradient centrifugation experiments showed that depletion of Mrd1 did not influence the association of Rrp5-TAP with pre-ribosomal complexes, and the reciprocal depletion of Rrp5 did not lead to defects in association of Mrd1-TAP with the pre-ribosome complexes (Supplementary Figure S2). We conclude that Mrd1 and Rrp5 are present in the same pre-ribosomal complexes but are recruited independently. In addition, we could not detect a defect in recruitment of Rrp5-3HA to 90S pre-ribosomes in the presence of only ΔRBD1 (Supplementary Figure S2, lower panel). This indicates that Rrp5 and ΔRBD1 are in the same pre-ribosomal complex, but deletion of RBD1 reduced the stability of pre-ribosomal complexes resulting in loss of Rrp5 during purification (Figure 1).
To test whether Mrd1 and Rrp5 interact directly or through indirect protein–protein interactions within pre-ribosomes, extracts were treated with RNase before precipitation. The interaction between Mrd1 and Rrp5 was lost, showing it to be dependent on RNA (Figure 1E). In addition, Mrd1 and Rrp5 did not interact in a yeast two-hybrid test (data not shown), further arguing that their interaction is mediated by pre-ribosomes.
Mrd1 interacts mainly with two regions of the pre-rRNA within the 18S rRNA sequence
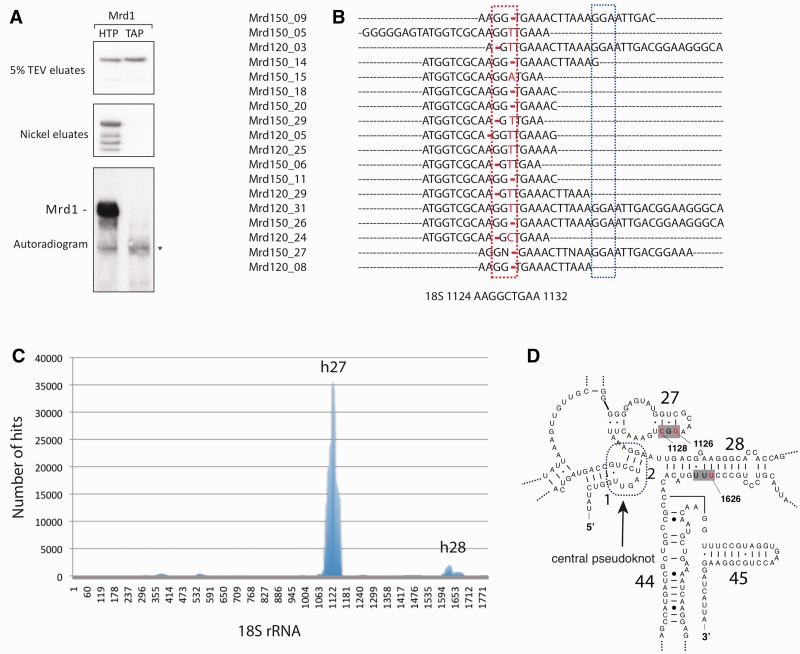
We wished to identify the pre-RNA sequences that interact with Mrd1 in vivo and which Mrd1 domains are responsible for the interactions. We therefore applied CRAC (19). This technique accurately maps the RNA binding site(s) for target proteins via tandem affinity purification of the tagged protein and the identification of short cross-linked RNA fragments by cDNA sequencing. Intact cells expressing Mrd1 with a C-terminal HTP tag (His6-TEV-ProtA) or a conventional TAP tag (CBP-TEV-ProtA) were rapidly chilled and harvested before UV irradiation at 254 nm in a Stratalinker. During the CRAC procedure, RNA fragments are 5′ labelled with [32P] allowing the cross-linking efficiency to be assessed. Mrd1-HTP showed strong cross-linking to RNA and good recovery during purification (Figure 2A). In comparison, the TAP-tagged Mrd1 that bound to IgG sepharose beads was, as expected, not retained on the nickel column, showing that the labelled band indeed corresponds to Mrd1-HTP. RNA bound to purified Mrd1-HTP was reverse transcribed and cloned, and random clones were sequenced.
Figure 2.
UV cross-linking of Mrd1 to pre-rRNA revealed cross-linking of Mrd1 to two regions within the 18S sequence. (A) Five percent of the TEV eluates and nickel eluates from Mrd1-HTP and Mrd1-TAP-tagged cells, detected by western blot analysis (upper panels). Autoradiogram (lower panel) shows Mrd1-HTP cross-linked to radiolabelled RNA. The asterisk indicates frequently detected contaminants. (B) Alignment of the 18 cDNA clones to the 18S sequence nucleotides 1124–1132. Red box indicates the Mrd1 binding sites and blue box the 18S sequence involved in base pairing at the 3′ side of the central pseudoknot. Substitutions and deletions are marked with red. (C) High-throughput sequencing revealed cross-linking of Mrd1 to h27 and h28. Histogram showing the location of the Mrd1-HTP associated RNA fragment hits (y-axis) mapped to the 18S rRNA (x-axis). (D) The Mrd1 binding sites (red within grey boxes) in h27 and h28 are shown for the mature secondary structure of the central region of 18S rRNA. The location of the central pseudoknot (blue) is indicated. PDB numbers are 3U5B (RNA) and 3U5C (rproteins).
Twenty-seven of 28 cloned cDNAs were 18S rRNA sequences. A total of 18 of the 27 sequences aligned at one single region corresponding to helix 27 (h27) in the mature RNA structure (Figure 2B and D). Each of the 18 sequences contained base alterations, either substitutions or deletions, at positions 1126 and/or 1128 in the 18S rRNA (Figure 2B), which identified the location of the direct protein–RNA contacts (19). To identify potential additional interactions, Solexa deep sequencing was performed on CRAC-purified cDNA. This too revealed that the dominant in vivo cross-linking site of Mrd1 is the h27 region (data not shown).
To search for further, less abundant or transient RNA interaction sites for Mrd1, UV cross-linking was performed on cells actively growing in culture medium (21). After removal of PCR duplicates using random barcode sequence information, a total of 141 422 unique cDNAs were identified. Ninety-two percent of the reads could be aligned to genomic features, and of these 105 956 reads mapped to the rRNA (corresponding to 81%), and of those, 85 790 were found to align to the 18S (81% of rRNA reads) (Figure 2C). The vast majority of 18S reads (∼90%) again aligned to the established h27 and contained deletions/substitutions at positions 1126 and 1128 (44% at G1126 and 67% at C1128).
Additional, minor interaction sites were also identified (Figure 2C). The most prominent of these covered a region centred around nucleotide 1635 within the 18S rRNA. Hits to this region amounted to ∼6% of the frequency at h27. The probable direct interaction site in this region is within helix 28 (h28), at U1626 (44% of the reads had substitutions/deletions at U1626). Although the binding of Mrd1 at positions 1126–1128 and 1626 is separated by ∼500 nt in the primary sequence, h27 and h28 are located in close proximity to the mature 18S secondary structure (Figure 2D).
RBD1, 3 and 5 of Mrd1 are individually dispensable for 18S rRNA h27 interaction
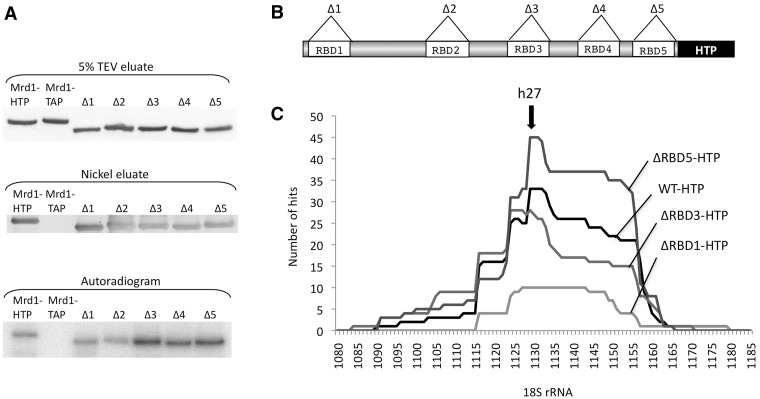
We next wished to establish which, if any, of the RBDs in Mrd1 specifically mediate the pre-rRNA interactions. All five RBD deletion constructs (ΔRBD1–5) were genomically C-terminally HTP tagged and tested for RNA cross-linking (Figure 3). For the lethal and severely growth-defective RBD deletions (ΔRBD1, ΔRBD3 and ΔRBD5), a PGAL1-HA-MRD1 gene was inserted into the URA3 locus with a yeast integrating plasmid containing the MRD1 allele (pRS406/PGAL1-HA-MRD1). Before UV cross-linking of the cells, the strains were transferred to glucose for 6 h to deplete wt Mrd1. The HTP-tagged Mrd1 RBD deletions were expressed at similar levels (Figure 3A, 5% TEV input). The ΔRBD1, 3, 4 and 5 mutant proteins interacted with RNA with efficiencies similar to, or greater than, intact Mrd1-HTP (Figure 3A, autoradiogram). The efficient cross-linking observed especially for the ΔRBD3-HTP, ΔRBD4-HTP and ΔRBD5-HTP mutants is probably a consequence of the stalled pre-ribosomal complexes that these deletions induce (18).
Figure 3.
UV cross-linking properties of Mrd1 deletion mutants, lacking RBD1, 3 or 5. (A) Five percent of the TEV eluates and nickel eluates from Mrd1-HTP, Mrd1-TAP, ΔRBD1-HTP, ΔRBD2-HTP, ΔRBD3-HTP, ΔRBD4-HTP and ΔRBD5-HTP cells detected by western blot analyses. The autoradiogram (lower panel) shows the proteins cross-linked to radiolabelled RNA. (B) Schematic representation of Mrd1-HTP and the five individual RBD deletion mutants indicated (C) Histogram of the RNA hits associated with Mrd1-HTP, ΔRBD1-HTP, ΔRBD3-HTP and ΔRBD5-HTP (y-axis) mapped to the 18S sequence, nucleotides 1080–1185 (x-axis). The Mrd1 binding site at h27 is indicated with an arrow.
We analysed the lethal or severely growth affected ΔRBD1-HTP, ΔRBD3-HTP and ΔRBD5-HTP mutant strains with in vivo CRAC analysis. cDNA sequencing from ΔRBD1-HTP (20 clones), ΔRBD3-HTP (70 clones) or ΔRBD5-HTP (70 clones) revealed that in each case, 70–80% of all hits aligned to the rRNA. The majority of these hits (62, 56 and 83% in ΔRBD1, 3 and 5, respectively) were located in 18S rRNA h27 (Figure 3C), and also contained the substitutions or deletions at positions 1126 and 1128. We conclude that none of the functionally important RBD1, RBD3 or RBD5 in Mrd1 is individually responsible for interacting with the h27 sequence in 18S rRNA. This is probably also the case for RBD4, as the RNA cross-linking efficiency remained high for ΔRBD4. Cross-linking for ΔRBD2 was weak and studied in detail later. We were not able to establish whether any of the RBDs are responsible for h28 interaction, as we did not perform high-throughput sequencing on the cDNA libraries from the deletion mutants.
The N-terminal half of Mrd1 is responsible for interaction with the 18S rRNA h27
We wished to further analyse the RNA binding ability of Mrd1 and what part of the protein that is responsible for the 18S rRNA sequence interaction. For this purpose, the protein was divided into fragments that included various RBDs and a C-terminal HTP epitope. We constructed plasmids containing the MRD1 promoter (a 500-bp fragment upstream of the transcription start site), various regions of the MRD1 ORF, a nuclear localization signal and the HTP tag. Figure 4A shows schematically the parts of MRD1 included in each construct. The HTP-tagged parts of Mrd1 were all expressed, but at variable levels (Figure 4B). All Mrd1 constructs containing RBD1 were more highly expressed than constructs lacking RBD1 (Figure 4B compare lanes 1–3 with lanes 4–8). Immunofluorescence localization of the expressed Mrd1 parts revealed that only RBD123 and wt full-length Mrd1 (RBD12345) were localized predominantly to the nucleolus as judged by co-localization with Nop1 (Figure 4C). All the other constructs (RBD1, RBD12, RBD23, RBD234, RBD34, RBD345 and RBD45) showed widespread immunolocalization signals in the nucleus and cytoplasm (data not shown). RBD123 was incorporated into pre-ribosomal complexes (Supplementary Figure S3A) but did not support cleavage of the pre-rRNA at sites A0–A2 (Supplementary Figure S3B) or cell growth (data not shown).
We tested the in vivo RNA binding capacity of the more highly expressed constructs containing RBD1 (RBD12-HTP and RBD123-HTP) together with the full-length RBD12345-HTP. RBD123-HTP was UV cross-linked to RNA with the same efficiency as the full-length construct (Figure 4D). A total of 10 cDNA clones were sequenced from the RBD123 construct, of which eight contained the 18S rRNA h27 sequence with substitutions/deletions at positions 1126 and 1128. In contrast, RBD12-HTP cross-linked to RNA with lower efficiency, and none of 10 clones sequenced could be mapped to the pre-rRNA (data not shown). The lack of binding of RBD12-HTP to rRNA could be because of disturbed localization of the construct in the cell.
RBD2 is required for efficient cross-linking of Mrd1 to h27
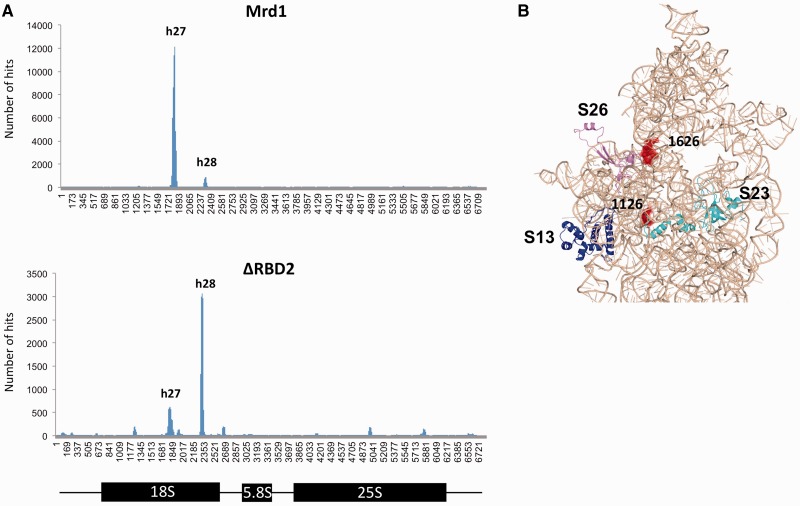
In Figure 3A, it was suggested that UV cross-linking to RNA was reduced as a result of deletion of RBD2. To analyse the importance of RBD2 in more detail, cells expressing wt Mrd1-HTP or ΔRBD2-HTP were UV irradiated while actively growing in SD growth medium, and cross-linked RNAs were analysed by high-throughput sequencing. For wt Mrd1, as in Figure 2C, the vast majority of the interactions were at h27 with minor interaction at h28 (Figure 5A). Approximately 20-fold fewer sequences were recovered for h27 in ΔRBD2-HTP than for wt Mrd1-HTP. This apparently reflects a specific influence on the interactions at h27, which were greatly reduced, whereas recovery of sequences from h28 was slightly increased (3-fold), as compared with wt Mrd1 (Figure 5A). In consequence, the relative frequency of interaction at h27 and h28 was reversed between wt and ΔRBD2-HTP. Among the residual h27 sequences recovered with ΔRBD2-HTP, mutations at positions 1126 and 1128 were less frequent (28 and 32% for ΔRBD2 compared with 42 and 76% in wt Mrd1), strongly indicating that the residual interaction between ΔRBD2 and h27 differs in the specific RNA–protein contacts compared with wt Mrd1. In contrast, the mutation frequency at position 1626 in h28 was 52% for ΔRBD2 and 57% for wt Mrd1, consistent with unaltered contacts in this region.
Figure 5.
RBD2 is important for Mrd1 association with h27. (A) High-throughput sequencing of CRAC cDNA library for wt Mrd1 and ΔRBD2. Histogram shows hits (y-axis) versus the position in 35S pre-rRNA (x-axis). h27 and h28 are denoted. (B) The modelled Mrd1 binding sites (red) of nucleotides 1126 and 1626 are located and highlighted in the mature secondary structure of S. cerevisiae 18S rRNA. The location of small ribosomal subunit proteins S13 (blue), S23 (light blue) and S26 (purple) are indicated as references.
We conclude that RBD2 is required for efficient cross-linking of Mrd1 to h27, but is not responsible for interaction at h28. In the absence of RBD2, cross-linking at h27 is however not completely absent. This indicates that while RBD2 plays an important role in the interactions at h27, other regions of Mrd1 also interact at this site.
The results for ΔRBD2 underscore the positioning of the Mrd1 interactions within the 18S region of the pre-rRNA. In Figures 2D and 5B, the spatial relationship between the interactions at h27 and h28 are highlighted in relation to the mature fold of 18S rRNA. The interaction sites in h27 and h28 are close to each other, and to the central pseudoknot and the decoding centre.
In vitro RNA binding of Mrd1, RBD123 and RBD45
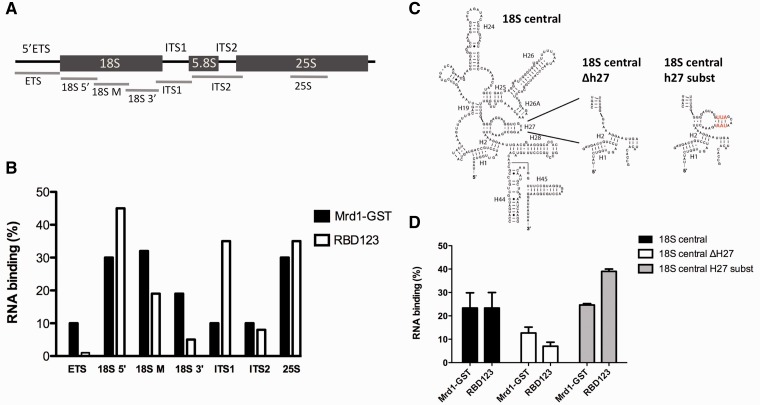
To complement the in vivo RNA binding results, we performed RNA–protein binding in vitro, using RNA fragments corresponding to regions of the pre-rRNA (Figure 6A), and full-length Mrd1 or parts of Mrd1 expressed in E. coli (see Supplementary Figure S4). Full-length Mrd1 bound equally well to the 5′ and middle regions of 18S rRNA (nucleotides 1–600 of 18S and nucleotides 577–1256 of 18S, respectively) with 30–35% total binding and a Kd value of 20–30 nM. Significantly lower binding for full-length Mrd1 was seen for the 3′ end of 18S (nucleotides 1233–1796 of 18S), 5′ETS, ITS1 and ITS2 (4–19%), while a region in 25S bound with similar affinity as the first two-thirds of 18S (Figure 6B). As full-length Mrd1 was GST tagged, we tested GST alone for binding. GST did not bind any of the RNA fragments (data not shown). To analyse whether the central region of 18S, including h27 and the central pseudoknot sequences, confer binding to Mrd1 in vitro, we tested an RNA construct containing only the central region of 18S rRNA (18S central in Figure 6C). In this RNA, h27 and h28 are present, but most of the surrounding 5′ domain, 3′ major domain and part of the central domain of 18S rRNA were not included. Mrd1 bound to this construct to a similar level as to the middle region of 18S (also containing h27) and with similar Kd value (Figure 6B and D). We tested two mutant versions of the central region of 18S (Figure 6C). Deletion of h27 (18S central Δh27) resulted in reduced binding to Mrd1, whereas nucleotide substitutions in the stem of h27 (18S central h27 substitution) did not alter binding (Figure 6D). The substitutions changed the GGC nucleotides at position 1126–1128 to UAA, while compensatory mutations allowed the h27 stem to form. We tested a short RNA fragment corresponding to only h27, and this did not bind to Mrd1 (data not shown). In addition to the pre-rRNA fragments, we tested whether full-length Mrd1 could bind in vitro to any of the three snoRNAs, U3, U14 and snR30, which are essential for processing of the 40S ribosomal subunit. Neither the CRAC data nor our in vitro binding studies convincingly showed binding to these snoRNAs (data not shown). We conclude that in vitro, Mrd1 shows higher affinity for the first two-thirds of 18S, including h27, relative to other RNAs tested.
Figure 6.
The N-terminal part of Mrd1 binds to pre-rRNA in vitro. (A) Schematic figure of the 35S pre-rRNA. The lines indicate the RNA regions used in the protein–RNA filter binding assays. (B) Percent RNA binding of Mrd1-GST and RBD123 to the indicated RNA regions. (C) Schematic drawing of the synthetic RNA constructs of the 18S central domain, the h27 deletion (Δh27) and helix 27 substitution (h27 substitution). (D) Percent RNA binding of Mrd1-GST and RBD123 to the 18S central constructs.
We analysed in vitro RNA binding of two separate domains of Mrd1, an N-terminal domain containing RBD1, 2 and 3 (RBD123) and a C-terminal domain containing RBD 4 and 5 (RBD45). We also analysed binding of the five individual RBDs. Binding of RBD123, as compared to binding of RBD45, showed that the in vitro binding specificity of Mrd1 is located within the N-terminus of Mrd1 (Figure 6B). RBD123 exhibited approximately the same binding properties as full-length Mrd1 (Figure 6B), including level of binding and Kd values, but it also bound to the ITS1. Binding of RBD123 to the 18S central constructs mimicked that of full-length Mrd1. RBD123 did not bind to the h27 fragment (data not shown). We could not detect any significant binding of RBD45 or of any of the individual RBDs to the tested RNA fragments.
We conclude that none of the isolated individual RBDs binds RNA in vitro. However, in vitro RNA binding by Mrd1 is observed for the first three RBDs, including linkers 1 and 2. This is in agreement with in vivo UV cross-linking data for the RBD123 construct (Figure 4D) and consistent with cooperative binding to the central region of 18S rRNA.
DISCUSSION
Mrd1 is essential for synthesis of the pre-40S subunit and is required for the A0–A2 cleavages of 35S pre-rRNA. The unusual domain structure of Mrd1, with five RBDs, suggested that it might play an important role in coordinating events at different sites in the pre-rRNA during the early steps in ribosome synthesis. Previous data revealed that Mrd1 localizes to the nucleolus, co-precipitates 35S pre-rRNA and co-sediments with 80–90S pre-ribosomes (17,18). Our tandem affinity purifications and proteomic analyses demonstrated that Mrd1 is indeed incorporated into early 90S pre-ribosomes. Analyses of deletion mutants showed that Mrd1 lacking RBD1, 3 or 5 retains the ability to bind pre-ribosomes; however, changes in co-precipitated proteins indicate that these particles are structurally abnormal and/or less stable than complexes formed with wilt-type Mrd1. We conclude that Mrd1 indeed plays a role in the correct assembly of the early pre-ribosomes.
Cross-linking (CRAC) data revealed two Mrd1 binding sites, both located within the 18S rRNA region. No interactions in the transcribed spacer regions were identified. There are no indications that Mrd1 is associated with mature 18S rRNA or 40S ribosomal subunits (data not shown), and we therefore conclude that the CRAC results revealed interactions between Mrd1 and pre-rRNA. The precise sites of interaction in the 18S region were mapped based on substitutions/deletions, to nucleotides 1126 and 1128 in h27 and nucleotide 1626 in h28. Both of these 18S rRNA regions are functionally important in the mature 18S rRNA, and both are in close proximity to the central pseudoknot (Figure 2D). The pseudoknot is a universal long-range interaction that is required for small ribosomal subunit function (31,32). We therefore speculate that Mrd1 plays a role in promoting or regulating formation of the pseudoknot within early pre-ribosomes, acting in cooperation with other factors, including the U3 snoRNP. The early cleavages of the pre-rRNA, releasing the pre-40S ribosome, are predominantly co-transcriptional and are preceded by compaction of the nascent transcript (5,6). The location of the Mrd1 binding sites is in agreement with its requirement for the cleavages and for a role in compaction of the pre-ribosome before cleavage (18).
Binding sites for pre-40S subunit synthesis factors have been mapped using CRAC (33). This revealed binding of Tsr1 at regions surrounding h27, but, in contrast to Mrd1, Tsr1 cross-linking did not generate mutations within h27. Instead, nucleotides 1143–1146 show mutations in cDNAs recovered with Tsr1, which is present in the nucleolus but acts in pre-40S maturation after formation of 20S pre-rRNA (34). Tsr1 is, therefore, likely to associate with the pre-40S particles following dissociation of Mrd1. Another factor, Dim1, associates with both the 90S and 40S pre-ribosomes. CRAC analysis located Dim1 binding to the h28 region, but on the opposite side of the duplex from the Mrd1 binding site. Dim1 also binds across the central pseudoknot (h2 and h28, nucleotides 1137–1156), interaction sites that suggest a role in pseudoknot formation (33).
The U3 snoRNP is required for the A0–A2 cleavages of 35S pre-rRNA (9) and base pairs with two sites in the pre-rRNA 5′ ETS (14,35), the 5′ side of the pseudoknot (10,36) and two sequences close to the 3′ side of the pseudoknot (37). The pseudoknot forms between helix 1 in the 5′ end of 18S rRNA, forming the 5′ side of the pseudoknot, and the 3′ side is located about 1140 nt downstream of helix 1. The pseudoknot involves only 3 bp; therefore, for its formation, the two sides of the pseudoknot probably need to be initially aligned by other factors within the pre-ribosome. Little is known about when and how this occurs, but the pseudoknot fold is a key, and most likely irreversible, step during subunit maturation, and the timing of its formation must therefore be strictly controlled. U3 snoRNA may participate in interactions that bring the 5′ and 3′ sides of the pseudoknot into contact, but would need to be released before pseudoknot formation.
The CRAC results demonstrated that Mrd1 is positioned close to the 3′ side of the central pseudoknot, potentially facilitating its formation. In the absence of Mrd1, U3 snoRNA is recruited to pre-ribosomes (17), but its release from base pairing with 35S pre-rRNA is inhibited (18). CRAC experiments did not reveal direct association between Mrd1 and U3, but deletion of individual RBDs altered the association of U3-associated proteins Utp10 and Utp22 with the pre-ribosomes. This suggests that the role of Mrd1 in U3 release is either indirect, perhaps owing to alterations in pre-ribosome structure, or involves interactions only with the protein components of the U3 snoRNP.
It is likely that the RBDs within Mrd1 are functionally coordinated as suggested by the fact that they all contribute to the essential function of Mrd1 (18). The individual RBDs in Mrd1 exhibited low RNA binding affinity to pre-rRNA sequences suggesting that, as for other RNA-binding proteins built from multiple domains, the modular structure is important for regulating binding specificity and activity (38). The deletion versions of Mrd1 were notably stable, in particular when RBD1 is retained (Figure 4B). Each RBD in Mrd1 is likely to have its specific version of the conserved typical RBD structure. During evolution, the number of RBDs in Mrd1 homologues has changed between four and six (39), but the identity of individual RBDs has been conserved (39). Functional analyses show the importance of each RBD (18), supporting the model that each RBD plays a specific role, although with some coupling. Together with the observed stability of the deletion versions of Mrd1, this indicates a modular design in which functionally distinct RBDs are folded independent of each other in the typical RBD fold, while the linker domains are unstructured.
RBD123 is sufficient for positioning Mrd1 close to the 3′ side of the pseudoknot. Although RBD1 and RBD3 are dispensable for interaction at h27, RBD2 is required for efficient interaction at h27 but not at h28. In the absence of RBD2, Mrd1 interacts with h27 to a reduced extent, and shows changes in the mutation frequency that indicates an altered mode of binding. This implies that other parts of Mrd1 can interact with h27 in the absence of RBD2, although less efficiently. We postulate that RBD2 is normally responsible for direct recognition of h27. In the absence of RBD2, other RBDs continue to interact with h28 and still position the protein in association with h27. Under these conditions, the interaction with h27 may be more transient, or less tight leading to the observed reduction in cross-linking, but is apparently sufficient to support ribosome synthesis, as the ΔRBD2 mutant is viable. RBD2 is not required for the association of Mrd1 with h28, but our analyses did not identify the specific domain(s) of Mrd1 responsible for this interaction. As deletion of RBD2 leads to reduced cleavages at A0–A2 and reduced synthesis of mature 18S rRNA, the interaction at h27 appears functionally important.
Mrd1 must, however, have other functionally important features, as ΔRBD3 and ΔRBD5 Mrd1 proteins still interact at h27 but do not support productive ribosome synthesis (18). Mrd1 that lacks RBD3 or especially RBD5 locks base paired U3 snoRNA into stalled pre-rRNP complexes (18). This suggests that RBD3 and RBD5 participate in structural rearrangements in the pre-ribosome and/or interactions with the U3 snoRNP that lead to U3 eviction and pseudoknot formation. These additional interactions are likely to coordinate structures and/or factors at sites in close proximity in space in the pre-ribosome that are crucial for pseudoknot formation and pre-rRNA cleavage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Swedish Research Fund, Carl Tryggers Stiftelse, an EMBO short-term fellowship (to Å.S.); Wellcome Trust (to S.G. and D.T.). Funding for open access charge: Swedish Research Fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the GenePool (Edinburgh) and the Cambridge University sequencing facilities for performing the high-throughput sequencing on Illumina HiSeq and MiSeq platforms.
REFERENCES
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 3.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 5.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Fernandez J, Martin-Marcos P, Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res. 2011;39:8105–8121. doi: 10.1093/nar/gkr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutca LM, Gallagher JE, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011;39:5164–5180. doi: 10.1093/nar/gkr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin SB, Zhao J, Björk P, Schmekel K, Ljungdahl PO, Wieslander L. Mrd1p is required for processing of pre-rRNA and for maintenance of steady-state levels of 40 S ribosomal subunits in yeast. J. Biol. Chem. 2002;277:18431–18439. doi: 10.1074/jbc.M112395200. [DOI] [PubMed] [Google Scholar]
- 16.Björk P, Baurén G, Jin S, Tong YG, Bürglin TR, Hellman U, Wieslander L. A novel conserved RNA-binding domain protein, RBD-1, is essential for ribosome biogenesis. Mol. Biol. Cell. 2002;13:3683–3695. doi: 10.1091/mbc.E02-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segerstolpe A, Lundkvist P, Osheim YN, Beyer AL, Wieslander L. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res. 2008;36:4364–4380. doi: 10.1093/nar/gkn384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundkvist P, Jupiter S, Segerstolpe A, Osheim YN, Beyer AL, Wieslander L. Mrd1p is required for release of base-paired U3 snoRNA within the preribosomal complex. Mol. Cell Biol. 2009;29:5763–5774. doi: 10.1128/MCB.00428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad Sci. USA. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Granneman S, Petfalski E, Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Dunbar DA, Wormsley S, Agentis TM, Baserga SJ. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- 27.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 28.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 29.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neefs JM, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marmier-Gourrier N, Clery A, Schlotter F, Senty-Segault V, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 2011;39:9731–9745. doi: 10.1093/nar/gkr675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 37.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl Acad. Sci. USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallberg Y, Segerstolpe Å, Lackmann F, Persson B, Wieslander L. Extensive evolutionary conservation of the ribosomal biogenesis factor Rbm19/Mrd1: implications for structure and function. Plos One. 2012;7:e43786. doi: 10.1371/journal.pone.0043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.