Abstract
Targeted genome engineering has become an important research area for diverse disciplines, with site-specific recombinases (SSRs) being among the most popular genome engineering tools. Their ability to trigger excision, integration, inversion and translocation has made SSRs an invaluable tool to manipulate DNA in vitro and in vivo. However, sophisticated strategies that combine different SSR systems are ever increasing. Hence, the demand for additional precise and efficient recombinases is dictated by the increasing complexity of the genetic studies. Here, we describe a novel site-specific recombination system designated Vika/vox. Vika originates from a degenerate bacteriophage of Vibrio coralliilyticus and shares low sequence similarity to other tyrosine recombinases, but functionally carries out a similar type of reaction. We demonstrate that Vika is highly specific in catalyzing vox recombination without recombining target sites from other SSR systems. We also compare the recombination activity of Vika/vox with other SSR systems, providing a guideline for deciding on the most suitable enzyme for a particular application and demonstrate that Vika expression does not cause cytotoxicity in mammalian cells. Our results show that Vika/vox is a novel powerful and safe instrument in the ‘genetic toolbox’ that can be used alone or in combination with other SSRs in heterologous hosts.
INTRODUCTION
Manipulation of DNA is arguably one of the central technologies used to obtain a better molecular understanding of biology and medicine. The discoveries of restriction enzymes (1) and DNA ligases (2) were fundamental achievements that launched the era of recombinant DNA (3,4). Understanding how these enzymes work provided the opportunity to manipulate genetic material obtained from different species, and with their help, it became possible to express heterologous genes to produce valuable proteins and to perform functional studies based on cut and paste principles of genetic material. Today, we have access to several hundred restriction enzymes that are instrumental for DNA manipulations in vitro (4).
Although restriction enzymes and DNA ligases have become invaluable to manipulate DNA in vitro, they have limited utility to precisely manipulate DNA in a living system. Restriction enzymes typically recognize target sequences of around four to eight nucleotides. Hence, any recognition site is likely present many times in a whole genome, compromising the ability to precisely rearrange DNA segments in vivo. The discovery of different classes of enzymes was necessary to turn genetic manipulations into genome engineering. Rare-cutting enzymes, such as homing endonucleases, represent such a tool because they recognize longer sequences that are typically not present in genomes (5). Other systems that can be used to specifically cleave sequences in vivo include engineered enzymes combining a DNA-binding domain with a nuclease domain, such as zinc finger nucleases (6) and transcription activator-like effector nucleases (7).
Another class of enzymes that have become indispensible for genome engineering are site-specific recombinases (SSRs) (8,9). Most SSRs require additional host factors for efficient catalysis, limiting their use for in vivo applications in heterologous hosts. However, some SSRs recombine their targets efficiently without the aid of accessory proteins. Hence, they can be used to rearrange DNA in living systems (10). Because of the simplicity and efficiency, SSRs now serve as ‘molecular scissors’ for robust non-disruptive and reproducible genomic modifications. Their discovery and characterization have pushed the frontiers of molecular biology to allow conditional gene knockouts and sophisticated manipulations of whole genomes in vivo (1,9,11,12).
The tyrosine recombinases Cre and FLP are among the most popular and widely used SSRs for genome engineering. The Cre/loxP system originates from bacteriophage P1 (2,13) and FLP from 2μ plasmid of Saccharomyces cerevisiae (3,14). Both have demonstrated their utility in animal cells, carrying the 34-bp target sites loxP and FRT, respectively. Nevertheless, differences in enzymatic properties have been described for both enzymes (4,15), with Cre being more active in mammalian systems (4,16). For some experimental setups, a lower enzymatic activity of the recombinase can be beneficial, even though for most applications high activities are demanded. The efficacy of FLP has been enhanced through directed molecular evolution (5,17) and further been improved through mammalian codon optimization (6,18). However, the Cre/loxP remains the most widely used system when rapid and efficient recombination is desired. loxP and FRT sites are palindromic sequences with two 13-bp inverted repeats separated by an 8-bp spacer. Each half-site is bound by one recombinase monomer, which together catalyzes the strand exchange within the spacer region (7,19). Depending on the relative orientation of spacers, nature of the recombinase and presence and/or relative concentration of given cofactor(s), a variety of reactions can be performed, including, but not limited to, recombinase-mediated excision, integration or inversion. Further, genome engineering applications include, for instance, recombinase-mediated cassette exchange, where two heterospecific target sites placed into the genome are used for precise targeted integration (8,20). In addition to tyrosine recombinases, genetic application of serine recombinases, and their ability to promote unidirectional recombination, has found widespread use in genome engineering. In particular, their ability to mediate locus-specific and stable genome insertions using for instance phiC31 (9,10,21,22) or Bxb1 (11,23) integrases has important implications in biotechnology and biomedicine.
The growing list of applications is accompanied by sophistication in SSR-mediated genome engineering strategies (12,24). Combining several recombinases, and their ability to carry out reversible rearrangements, makes it feasible to realize sequential gene switches on different alleles or multiple inducible knockouts (9). Few additional SSRs have recently been described (25–27), and first tests have documented their utility for applied uses (28). In this study, we describe a novel SSR system Vika/vox and show that it is a valuable genome engineering instrument in bacteria and in mammalian cells.
MATERIALS AND METHODS
Identification of Vika and its target site vox
The search for potential Cre-like SSRs was performed using the position-specific iterated Basic Local Alignment Search Tool (BLAST) algorithm [PSI-BLAST/National Center for Biotechnology Information (NCBI)] on the non-redundant sequences in the public DNA database of the NCBI (http://blast.ncbi.nlm.nih.gov/, date of search: 28 October 2010). Vika (ZP_05884863) was identified from Vibrio coralliilyticus ATCC BAA-450 (GenBank: NZ_ACZN01000014.1). Initial recombinase alignments were done using ClustalW2 (29). The vox target site was identified using the SEarch for LOX-like sequences (SeLOX) algorithm (30) using the following parameters: maximum 2 asymmetry positions, maximum 13 mismatches to the input loxP sequence allowed per half-site and spacer length of 8 ± 2 bp.
Structure-based modeling
A 3D model of Vika was generated by using the crystal structure of Cre [Protein Data Bank (PDB ID: 1KBU)] (2.2 Å) as template (31) and a sequence-to-structure alignment obtained by threading the Vika sequence against a fold library containing all structures available in the PDB [ProHit Professional V 2.2.2; ProCeryon Biosciences, (32)]. A seven-residue long insertion in Vika that is not present in Cre was modeled by using the crystal structure of the lambda integrase (PDB ID: 1AE9) (1.9 Å) (33). The modeling was done with Modeler (34) embedded in Discovery Studio (Accelrys Software Inc., 2011). The model was refined with AMBER 10 (35) using the force field ff99SB (36). The model was neutralized with 13 Cl− molecules and hydrated with 14 422 TIP3P water molecules (37,38) in a truncated octahedron. The water molecules and counterions were energy minimized and equilibrated at 100 K around the fixed complex for 100 ps, at constant volume and temperature in the NVT ensemble [moles (N), volume (V) and temperature (T) are conserved]. The entire system was then heated from 100 to 300 K in 10 ps by 5-K increments with harmonic positional restraints on the solute atoms (force constant of 5.0 kcal/mol/Å2). The simulation was continued in NPT ensemble [moles (N), pressure (P) and temperature (T) are conserved]. The positional restraints were gradually removed over 250 ps and followed by 1 ns of unrestrained simulations for further equilibration. Analysis of the simulation and the image rendering was done using Tachyon software in visual molecular dynamics (39).
Construction of expression plasmids
The Vika coding sequence was synthesized (GeneArt, Life Technologies Corporation) and cloned via BsrGI and XbaI New England Biolabs (NEB) into pEVO vectors as described previously (40). Two of the recombination sites, loxP, rox, VloxP or vox, were inserted into the pEVO vector, producing pEVO-loxP, pEVO-rox, pEVO-VloxP and pEVO-vox. A mutant variant of Vika in which Tyr343 was replaced by phenylalanine was constructed by site-directed mutagenesis. Primers are listed in Supplementary Table S1. For experiments in mammalian cells, the codon-optimized recombinase coding sequences (GeneArt, Life Technologies Corporation), including a nuclear localization signal (NLS), were cloned into the expression plasmid pNPK (41). NLS-tagged recombinases were expressed from the phosphoglycerate kinase promoter. The pEVO-FLAG-Vika construct was constructed based on pEVO-Vika using primers listed in Supplementary Table S1. Western blot analysis was performed with a monoclonal mouse anti-FLAG antibody (1:2000, M2, Sigma-Aldrich) and goat anti-Mouse IgG (H + L)-HRP secondary antibody (1:5000, Bio-Rad).
Recombination reporter plasmids and recombination assays
Recombinase reporter plasmids were based on the plasmid pEVO (40) harboring the coding sequences for Cre, VCre, Dre and Vika cloned with BsrGI and XbaI (NEB) and the target sites loxP, VloxP, rox, and vox (for primer sequences see Supplementary Table S1). For comparison of the recombination activity on native target sites, three independent clones of Vika, Cre and VCre were tested on vox, loxP and VloxP, respectively. Recombinase expression was induced with L(+)-arabinose (Sigma-Aldrich) at 0, 1, 10 and 100 μg/ml.
LacZ-based reporter plasmids were constructed by inserting two of each recognition sites (loxP, VloxP and vox) into pSV-paX1 (42). Plasmid DNA from overnight cultures was isolated (Qiagen Miniprep kit) and electroporated into DH5α cells. Cells were plated on ampicillin- and X-gal- (Sigma-Aldrich) containing plates. Numbers of white or blue colonies for each reaction were counted.
For the integration assay, the host plasmids were constructed based on pEVO vectors as a result of the pEVO recombination assay in Escherichia coli, each harboring one recombinase gene and one relative recombination target site: pH-Cre-loxP, pH-VCre-VloxP and pH-Vika-vox. Donor plasmids pD-loxP, pD-VloxP and pD-vox were based on the R6K-neo vector and were constructed by inserting one recombination target site using BglII and XhoI restriction enzymes (NEB). Host plasmids were electroporated into E. coli DH5α. Cells harboring the host plasmids were treated for 2.5 h with 100 μg/ml L(+)-arabinose to induce recombinase expression, before transfection with 1 μg DNA of the donor plasmids by electroporation. After 1-h recovery, cells were plated on kanamycin plates.
The mammalian reporter plasmids were based on the plasmid pEGFP-X (enhanced green fluorescent protein) (42). The neomycin cassette was amplified with oligonucleotides harboring VloxP or vox sequences and cloned into pEGFP-X to produce pRK5-vox-EGFP and pRK5-VloxP-EGFP.
HeLa cells were plated at a density of 2 × 105 cells per well in six-well dishes and grown in 4.5 mg ml−1 glucose Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum (Invitrogen), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Gibco-Invitrogen). pRK5-vox-EGFP, pRK5-VloxP-EGFP and pRK5-loxP-EGFP reporter constructs were co-transfected (Lipofectamin 2000, Invitrogen) with recombinase expression plasmids pNPK-NLS-Vika, pNPK-NLS-VCre or pNPK-NLS-Cre. Twenty-four hours post-transfection, cells were washed with phosphate buffered saline, fixed with 4% paraformaldehyde (Sigma-Aldrich) for 10 min at room temperature, and nuclei were stained with 1 μg/ml Hoechst 33342 (Invitrogen) for 30 min. For each of three biological replicas, nine images were acquired per condition on an IX81 microscope (Olympus), and cells were examined for EGFP expression. The distributions were compared using Student’s two-tailed t-test. Primers used for the construction of plasmids are listed in Supplementary Table S1.
Viral mammalian recombinase expression constructs were based on pBabe-puro, containing an internal ribosomal entry site and GFP reporter gene (Addgene plasmid 1764). The plasmid was modified by introducing a unique XhoI restriction site upstream of the EcoRI site (pBabe-puroX). Protein coding sequences for Cre and CreR173K were amplified from LZRS-Cre and LZRS-Cre(R173K)-ERT (43), ORFs of Vika and VikaY343F from pNPK-Vika and pNPK-VikaY343F, and all four were cloned via XhoI and EcoRI into pBabe-puroX. Virus was produced in Phoenix-GP cells (provided by G. Nolan, Stanford University, Stanford, CA, USA) as described previously (http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html). Cells (1 × 107) were transfected with 18 μg of pBabe-puroX constructs containing various recombinases, 5 μg of p522 plasmid encoding ecotropic envelope and 10 μg of pR690, plasmid encoding gag-pol proteins, using Lipofectamine 2000 (Invitrogen). Viral supernatants were harvested 36 h after transfection. For infection, NIH3T3 cells (1 × 105) were plated into six-well plates 24 h before retrovirus infection and then incubated with 1 ml retroviral supernatant in the presence of 4 μg/ml polybrene and 1 mM Hepes pH 7.25 for 4 h. At various time points, cells were trypsinized, and percentages of transduced cells (GFP-positive) were determined on a FACS Calibur flow cytometer (Becton Dickinson) by using Cell Quest software. All retroviral infection experiments were repeated twice showing comparable results.
γ-H2AX assay
Induction of double-stranded DNA breaks was determined by the γ-H2AX assay. Seventy-two hours post-infection, transduced cells were fixed with 4% paraformaldehyde and stained with a primary mouse phospho-H2AX antibody (clone JBW301, 1:1000 dilution, Upstate Biotechnology) and with a donkey anti-mouse TxRed conjugated secondary antibody (1:1000 dilution, Molecular Probes). DNA was stained with 4′,6′-diamidino-2-phenylindole (1 µg/ml). Control wells were treated with the DNA-damaging agent camptothecin at 5 μM for 2 h before staining. Nine images per replica for each condition were acquired on an Olympus IX81 microscope and analyzed with the ScanR Analysis software (Olympus). Staining for each transduced cell line was repeated three times. P-values were calculated by Student’s t-test.
RESULTS
Identification and 3D model of a new tyrosine recombinase
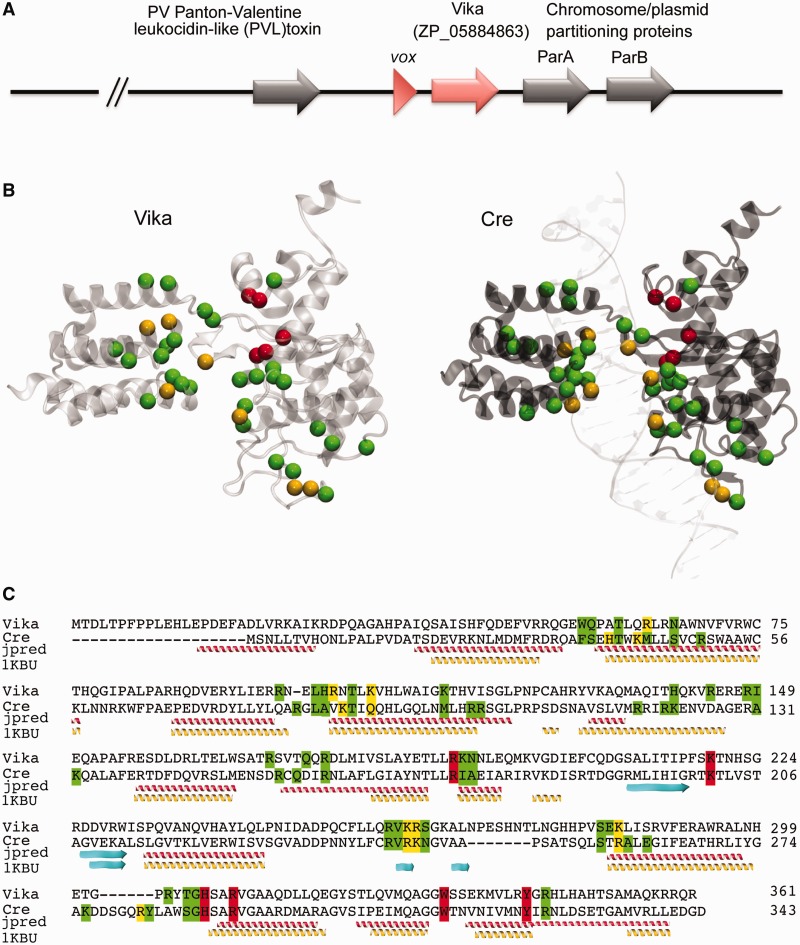
Recent sequencing efforts offer a rich resource for identifying new enzymes based on sequence homology to known proteins. To unmask putative novel Cre-like recombinases, we performed a search in the non-redundant protein database at NCBI using the PSI-BLAST algorithm (NCBI). Iterative alignment principle of the PSI-BLAST allows detecting distant sequence similarities and retrieving proteins of conserved structures. Several putative Cre-like recombinases were identified with this approach (Supplementary Table S2) with the protein ZP_05884863 (hereafter referred as Vika) emerging as a particularly promising candidate (Figure 1). Cre and many other Cre-like recombinases are bacteriophage enzymes involved in phage genome resolution (44). Vika originates from the gram-negative bacterium V. coralliilyticus ATCC BAA-450. Analysis of the genomic region surrounding the Vika sequence identified open reading frames with homologies to bacteriophage proteins, including a gp7-like capsid protein and Panton–Valentine leukocidin-like toxin along with the putative recombinase gene (Figure 1A). Hence, these sequences probably resemble a degenerate prophage present in the bacterial genome, and therefore, Vika likely originates from a bacteriophage.
Figure 1.
Sequence and structural model of Vika recombinase. (A) Localization of Vika (red) and its target site vox site (red triangle) in the genome of V. coralliilyticus ATCC BAA-450. Putative prophage genes flanking the Vika coding sequence are depicted by gray arrows. (B) Structural model of Vika and comparison with the X-ray structure of Cre (PDB ID: 1KBU). Residues known to interact with DNA in the Cre/LoxP system and their putative counterparts in the Vika/vox system are highlighted (catalytic residues in red, residues with major or minor groove contacts in yellow and residues contacting the phosphate backbone in green). (C) Sequence alignment of Vika and Cre. Residues are highlighted as mentioned earlier. A secondary structure prediction alignment using Jpred (jpred) and a structure-based alignment using threading (1KBU) are depicted. α-Helixes are in yellow and red β-sheets in cyan.
To investigate in more detail the putative Cre-like fold of Vika anticipated by its sequence similarity to Cre (26 and 49% sequence identity and similarity, respectively), and to investigate its possible substrate recognition preferences, we built a 3D atomic model of Vika. We used threading to search for the best structural template for modeling Vika; a Cre crystallographic structure (see ‘Materials and Methods’ section). The obtained Vika 3D model presented an root-mean-square deviation of 2.4 ± 0.3 Å compared with the Cre template. The sequence-to-structure alignment obtained by threading with Vika and Cre was slightly different than the sequence-based obtained by PSI-BLAST (Figure 1C). In both alignments, sequence and structure based, important residues characteristic for catalysis are preserved in Vika (Figure 1B and C). To identify residues that contribute in DNA recognition, we mapped residues involved in DNA recognition in the Cre/loxP complex (PDB ID: 1CRX) (45) onto our Vika 3D model (Figure 1B and C). Interestingly, most residues involved in recognition of the minor groove were highly conserved between Vika and Cre, whereas a number of Vika residues involved in major groove recognition could only be predicted by using the 3D model, as these residues are not conserved in sequence but in structure (Figure 1C and Supplementary Table S3). Hence, the 3D model of Vika allowed us to determine structural and functional similarities between Cre and Vika, supporting that Vika may function in a Cre-like manner. Furthermore, our 3D model helped us to identify putative residues responsible for DNA recognition and to hypothesize on their possible role in target site recognition.
Vika does not recombine target sites of known recombinase systems
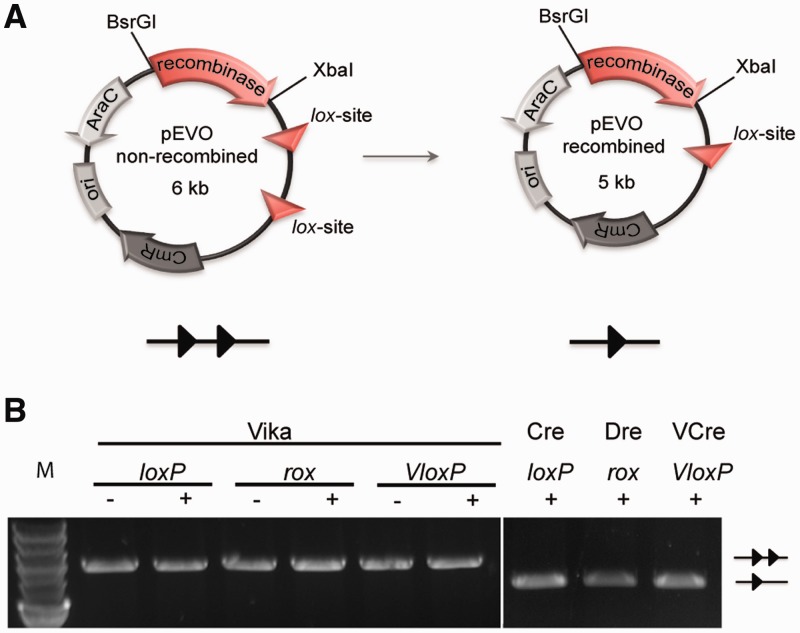
Recently, a Cre-like recombinase called VCre, and its recognition target site VloxP, has been identified on the Vibrio plasmid p0908 (26). Because both Vika and VCre originate from Vibrio strains, we compared both recombinases. An amino acid alignment between Vika and VCre showed that, like Cre, these recombinases share moderate sequence identity (37%, Supplementary Figure S1), possibly reflecting that they recognize divergent target sequences. To test whether Vika shows activity on target sites of other known recombinases, we cloned its coding sequence into E. coli recombination reporter plasmids harboring two target sites for Cre (loxP), VCre (VloxP) and Dre (rox) (25), a Cre-like recombinase from phage D6, with 23 and 37% amino acid identity to Vika and Cre, respectively (Figure 2A). In these constructs, Vika is expressed from the L-arabinose promoter, which allows inducible expression of the enzyme by addition of L(+)-arabinose to the growth medium (Supplementary Figure S2) (46). On recombination, an 1157-bp fragment is excised from the plasmid, signifying recombination activity. Isolation of the plasmid DNA after strong induction of Vika did not show a change in plasmid size of any of the used reporters (Figure 2B), demonstrating that Vika does not act on the target sites for established recombinase systems.
Figure 2.
Evaluation of Vika recombination activity on heterologous target sites. (A) Scheme of the recombination assay. Site-specific recombination excises a 1.2-kb DNA fragment, flanked by the recombinase target sites (triangles) from the plasmid. Protein coding genes are depicted by arrows and the origin of replication (ori) is shown as a box. CmR, chloramphenicol resistance gene; AraC, gene coding arabinose promoter regulator protein and recombinase, recombinase coding gene. (B) Lack of recombination activity of Vika on the loxP, rox and VloxP target sites. Site-specific recombination of native targets by Cre (loxP), Dre (rox) and VCre (VloxP) served as positive control. The − and + indicate presence or absence of L(+)-arabinose (100 μg/ml) in the growth medium. Non-recombined and recombined plasmids are denoted as two triangles and one triangle, respectively. M, marker; 2-log DNA ladder, NEB.
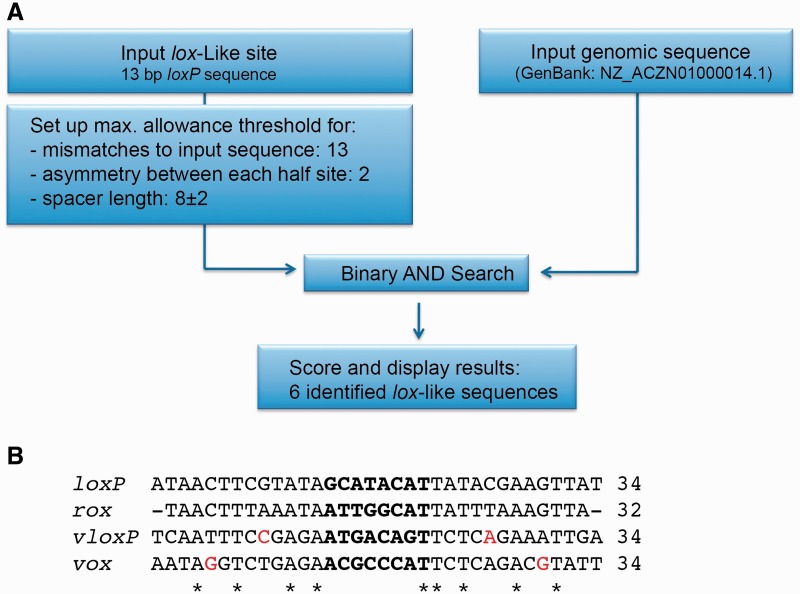
SeLOX computational target site prediction identifies the target site of Vika
Because of the structural resemblance of our Vika model to Cre-like recombinases (Figure 1), we predicted that Vika likely recognizes a similarly structured sequence. Cre and related tyrosine recombinases recognize a palindromic DNA sequence of two inverted 13-bp repeats that are typically separated by an 8-bp spacer (19). We anticipated that the genome sequence of V. coralliilyticus might contain the Vika target site. To examine the host genome to rationally predict putative targets, we used the SeLOX algorithm. SeLOX allows for a streamlined analysis of large DNA inputs for the presence of degenerate inverted repeats and is therefore ideally suited to identify putative target sites for recombinases (30). The analysis of the V. coralliilyticus ATCC BAA-450 genome predicted six palindromic sequences that matched the input criteria in a 10-kb area flanking the Vika coding sequence (see ‘Materials and Methods’ section, Supplementary Table S4). Only one of these sites was found in the vicinity of the recombinase and was called vox (Figure 3). The vox sequence is a 13-bp inverted repeat with one asymmetry separated by an 8-bp spacer and shows about 50% sequence homology to both loxP and VloxP and 33% sequence homology to rox (Figure 3B). Based on these analyses, we decided to test whether vox might be the target site of Vika.
Figure 3.
Identification and comparison of the vox target site. (A) Workflow of the SeLOX algorithm and important input parameters to predict lox-like sites in the genome of V. coralliilyticus strain NZ_ACZN01000014.1. The SeLOX results are shown in Supplementary Table S4. (B) Nucleotide alignment of vox with recombination sites for Cre (loxP), Dre (rox) and VCre (VloxP). Nucleotides that break perfect palindromic symmetry of the lox sites are colored in red. Asterisks depict conservation among all sites.
Vika specifically recombines vox sites
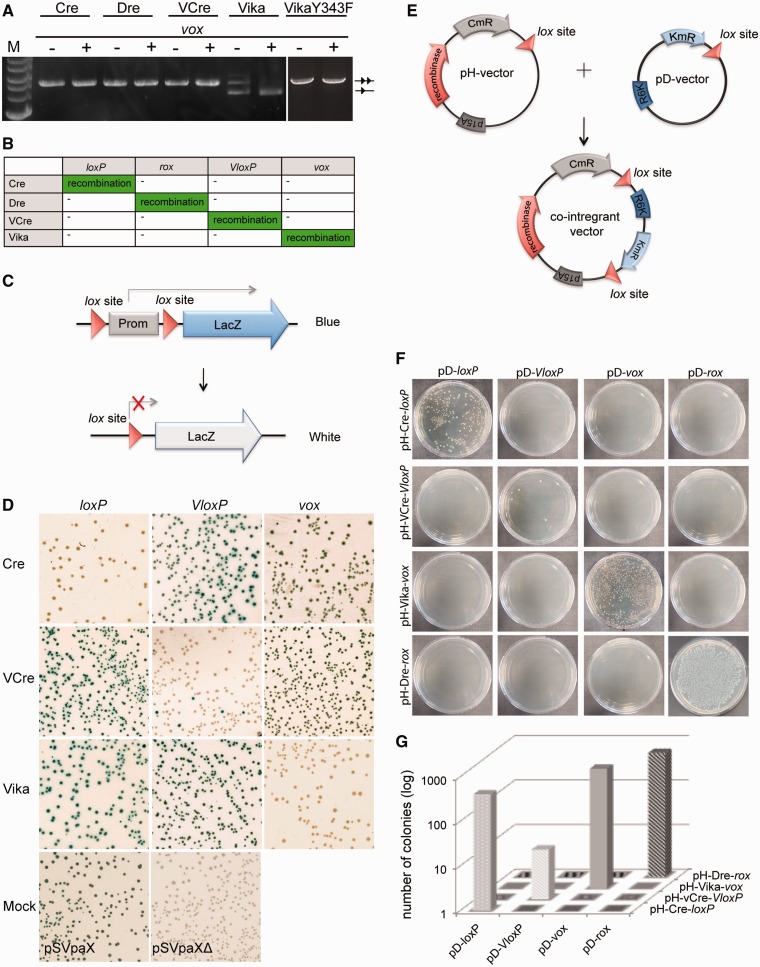
The 34-bp vox sequence was cloned into the recombination reporter vectors harboring coding sequences for Cre, Dre, VCre and Vika recombinases (Figure 2A). The reporter plasmids were transferred to E. coli, and recombinase expression was induced with L(+)-arabinose at different concentrations. None of the Cre, VCre and Dre recombinases exhibited recombination activity on vox, indicating that they do not catalyze recombination of vox sites (Figure 4A). In contrast, Vika efficiently and specifically recombined vox sites on expression of the recombinase (Figure 4A). Hence, the vox sequence is the substrate for the Vika recombinase. To confirm high specificity of the novel recombinase system, we investigated Vika, Cre and VCre in two additional assays. First, we used a LacZ reporter assay to visualize recombination activity (Figure 4C). The advantage of the LacZ-based assay lies in the possibility to detect weak/promiscuous recombination activity in order to precisely assess the cross-recombination of different recombinases and their target sites. After extended expression of the recombinases, the plasmids were harvested, and E. coli cells were retransformed and plated on X-gal plates. Each colony was evaluated by color, providing a highly sensitive assay. Non-recombined reporter plasmids express LacZ and thus form blue colonies when plated on X-gal plates, whereas cells harboring recombined plasmids appear white on the same plates. When Vika was expressed at high levels in cells with loxP or VloxP reporters, all colonies were blue, demonstrating that these sequences cannot be recombined by Vika. In contrast, almost all colonies appeared white, signifying efficient recombination, when Vika was used in conjunction with the vox reporter (Figure 4D). Corresponding results were observed for Cre and VCre on their native targets loxP and VloxP, respectively (Figure 4D). Therefore, these results confirm that Vika, Cre and VCre recognize their targets in a highly specific manner and do not cross-react with non-native sites.
Figure 4.
Evaluation of Vika recombination activity and specificity in E. coli. (A) Recombination specificity of indicated recombinases on vox. The − and + indicates absence or presence of L(+)-arabinose (100 μg /ml) in the growth medium. Non-recombined and recombined plasmids are denoted as two triangles and one triangle, respectively. M, marker; 2-log DNA ladder, NEB. (B) Matrix of the recombination specificities of indicated recombinase systems. (C) Schematic view of LacZ-based reporter assay. The non-recombined reporter plasmid constitutively expresses beta-galactosidase, inferring blue color to cells on X-gal plates. On recombination, the promoter driving LacZ expression is removed, resulting in white colonies on X-gal plates. Triangles depict lox sites. Prom = promoter. (D) Recombination activity of indicated recombinases on the loxP, vloxP and vox LacZ-based reporters. Note that white colonies, signifying recombination, only appear when a recombinase is expressed together with its corresponding reporter. Positive controls for the non-recombined (pSVpaX), and recombined form of the reporter (pSVpaXΔ), is shown as Mock. (E) Scheme of the integration assay. The host plasmids (pH) carry the coding sequence for the used recombinases, the chloramphenicol resistance gene (CmR), a p15A origin of replication (p15A) and one respective recombinase target site (triangle). The donor plasmids (pD) carry the kanamycin resistance gene (KmR), a R6K origin of replication (R6K) and also one respective recombinase target site (triangle). Only combinations of two identical target sites and their corresponding recombinases will produce co-integrant plasmids that survive kanamycin selection in pir-negative cells. (F) Colony formation assay of pir-negative E. coli cells transformed with indicated plasmid combinations. Colonies only form on site-specific integration of pD and pH plasmids. (G) Quantification of colonies carrying co-integrant plasmids after transformation with indicated plasmid combinations.
Next, we tested properties of the different enzymes for recombinase-mediated integration. For these experiments, we used a two-plasmid assay that reports integrative recombination [Figure 4E (41)]. Here, the integration property of the recombinase is tested by its ability to use two target sites located on different plasmids to merge them into one plasmid through the site-specific recombination reaction. The assay is designed with vectors based on two types of origin of replication: on p15A for the host pH plasmid (chloramphenicol resistant), and on R6K for the donor pD-plasmid (kanamycin resistant) (Figure 4E). Replication of the R6K-plasmid is pi-protein dependent, and therefore, this plasmid cannot replicate in a pir-negative bacterial strains. Hence, kanamycin resistance conveyed by the donor plasmid depends on its integration into a host plasmid through a lox site to form the co-integrant and consequent replication of the kanamycin resistance gene in pir-negative cells. When plated on medium with chloramphenicol and kanamycin, only the combinations of the recombinase and its native sites produced colonies. Vika promoted co-integration of two plasmids only if vox sites were present on both vectors, whereas abolishing integration if one of the sites was heterologous (Figure 4F). Equivalent results were obtained for Cre, Dre and VCre, demonstrating that donor and recipient plasmids have to carry the corresponding target sites to achieve co-integration. These data suggest that Vika, like Cre, Dre and VCre, is a recombinase that can use the native target site (vox) for intermolecular integration. Moreover, Vika only recognizes vox target sites, whereas Cre, Dre and VCre show no activity on vox. In conclusion, we find that Vika/vox is a novel SSR system with no cross-reactivity with other known recombinase systems.
Comparative analysis of different recombinase systems in E. coli
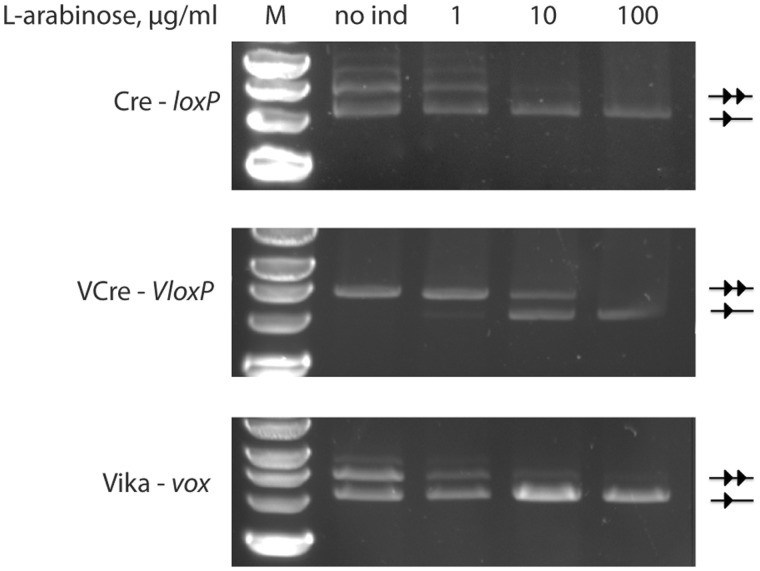
In the experiments for excision and integration, the enzymes demonstrated high target site specificity. However, we also noticed that the tested recombinases displayed different enzymatic activities. To obtain a more quantitative comparison of the activities of Cre, VCre and Vika in vivo, we used various induction levels for the recombinase expression to dissect the peculiarities of the recombination catalysis. For this purpose, we used the established recombinase reporter assay (Figure 2A) with Cre, VCre and Vika recombinases cloned into the vectors containing loxP, VloxP and vox, respectively. The plasmids were then propagated in E. coli at different L(+)-arabinose concentrations from 0 to 100 μg/ml of L(+)-arabinose. All recombinases fully recombined their corresponding target sites at 100 μg/ml of L(+)-arabinose, demonstrating that they efficiently recombine their targets when expressed at high levels (Figure 5). In contrast, differences in recombination efficiencies were evident at lower expression levels. Almost full recombination of the targets was observed for Cre and Vika already at 10 ug/ml, whereas for VCre, only ∼50% of its substrate was recombined at this concentration (Figure 5). Furthermore, no recombination was observed for VCre at 0 and 1ug/ml, whereas Cre and Vika recombined their targets to ∼50% at these concentrations (Figure 5). The apparent recombination of Cre and Vika even without the addition of L(+)-arabinose indicates the high activity of these enzymes because the ‘leakiness’ of the arabinose promoter is relatively low (46). Consistent with the excision assay, the integration assay revealed a similar activity difference. Although hundreds of colonies appeared for Vika and Cre in this assay, only few colonies were counted when VCre was co-transfected with its donor vector pD-VloxP (Figure 4F and G). Overall, these experiments identify a qualitative difference in in vivo recombination activity of the studied recombinases, with Vika and Cre being more active than VCre.
Figure 5.
Evaluation of the recombination activity of indicated recombination systems in E. coli. One and two triangles mark the size of the recombined and non-recombined plasmids, respectively. M = marker, 2-log DNA ladder, NEB, no ind = no addition of L(+)-arabinose to the growth medium, 1, 10 and 100 = growth of the cells in the presence of 1 μg/ml, 10 μg/ml and 100 μg/ml L(+)-arabinose, respectively.
Vika efficiently and specifically recombines vox in mammalian cells
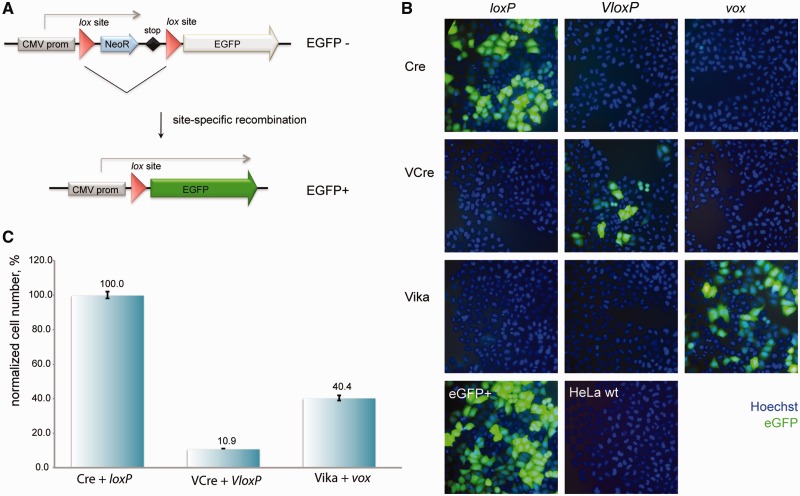
For the applied use in genome engineering, the recombinase needs to work in heterologous organisms. To investigate whether Vika can be used in organisms other then the natural host and E. coli, we tested the recombinase in human cells. HeLa cells were co-transfected with GFP reporter plasmids and recombinase expression plasmids (Figure 6A). Twenty-four hours post-transfection, the cells were monitored for GFP expression. Many GFP-positive cells were observed, when the Vika expression plasmid was co-transfected with the vox reporter (Figure 6B), signifying that Vika efficiently functions in mammalian cells. In contrast, no green cells were observed with the reporters for loxP and VloxP (Figure 6B), demonstrating Vika’s specificity also in mammalian cells. Corresponding results were seen with Cre and VCre, where these enzymes showed activity only on their target sites without recombining the target sites of the other recombinases (Figure 6B). Hence, these enzymes can be used in combination without cross-activities. To compare the activities of the different recombinases also in mammalian cells, we quantified the number of GFP-positive cells from the co-transfection experiments. Similar to the activity differences measured in E. coli, we also observed a difference in activity in HeLa cells. In this host, Cre showed the highest activity followed by Vika that exhibited about half of the activity of Cre. As in E. coli, the number of GFP-positive cells for VCre was markedly lower (∼10%), indicating that this recombinase also possesses less activity in mammalian cells (Figure 6C).
Figure 6.
Comparative recombination activity of Vika in mammalian cells. (A) Scheme of the GFP-based recombination reporter assay. The non-recombined reporter plasmid constitutively expresses the neomycin resistance gene (NeoR). On recombination, NeoR is removed, resulting in expression of the GFP driven from the cytomegalovirus promoter (CMV prom). Triangles depict lox sites. (B) Recombination assay of Cre, VCre and Vika in HeLa cells. Images show cells stained with Hoechst 33342 (blue) and imaged for EGFP expression (green), 24 h after co-transfection with indicated recombinase expression and reporter plasmids. (C) Quantification of EGFP-positive cells for indicated recombinase expression and reporter plasmids from B, n = 3. Error bars indicate standard deviation of the mean value.
Vika expression does not cause cytopathic effects in human and mouse cells
The successful use of recombinases in heterologous hosts rests on their restricted activity on the introduced target sites. Important concerns are cryptic (pseudo) recombination sites in the genome, which could potentially have cytopathic consequences. Active pseudo-loxP sites have indeed been described in human and mouse (47). As a consequence, expression of Cre in human and mouse cells can cause cytotoxicity (43,48–51).
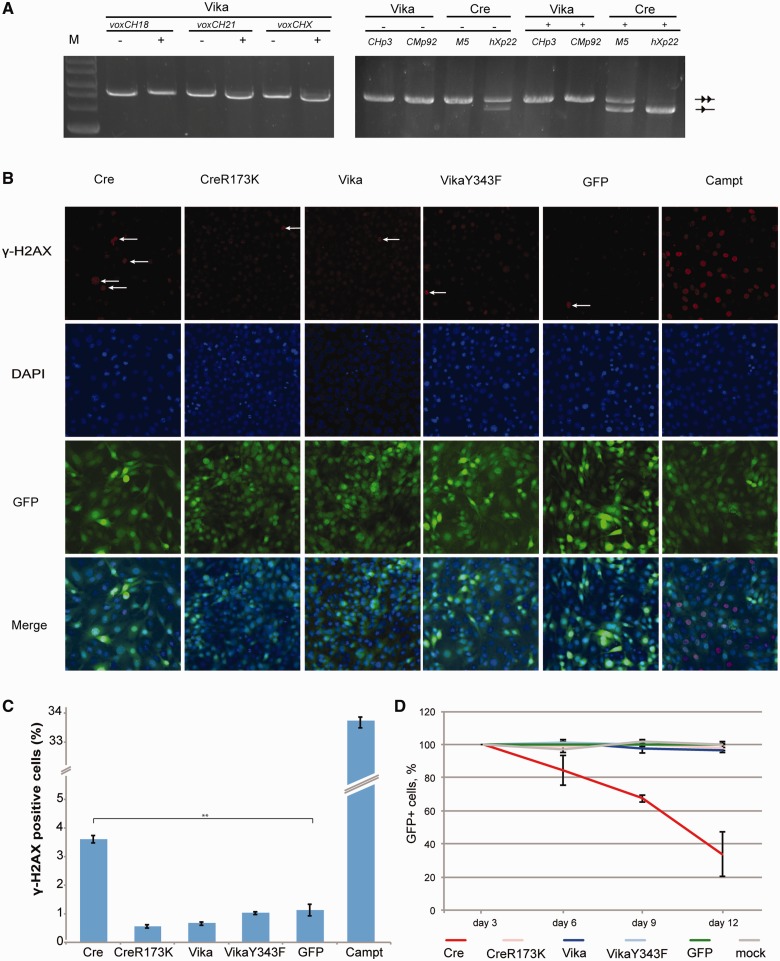
To investigate potential site effects of Vika, we searched for pseudo-vox sites in the mouse and human genome using the SeLOX algorithm (30). In both species, lower overall numbers of pseudo-vox sites were uncovered than pseudo-loxP sites (Supplementary Figure S3). We then tested pseudo-vox sites that most closely resembled vox sites (Supplementary Figure S4) experimentally. In contrast to Cre, which showed prominent activity on pseudo-loxP sites, Vika did not display measurable activity on pseudo-vox sites in these assays (Figure 7A). To examine the influence of prolonged expression of Vika in primary cells, we generated mouse embryonic stem (ES) cells stably expressing Vika. No effect on cell growth and morphology was observed, indicating that Vika is active when selected for stable expression in mouse ES cells and is well tolerated (Supplementary Figure S5). Next, we investigated the potential impact of recombinase expression on DNA damage. NIH3T3 cells were infected with GFP-bicistronic retroviral particles encoding Cre, Vika or controls. Three days post-infection, γ-H2AX signals were investigated in fixed cells. Cre expression caused a marked increase in γ-H2AX signals signifying induction of DNA damage. In contrast, Vika had no influence on the amount of γ-H2AX counted (Figure 7B and C), indicating that Vika expression does not lead to increased DNA damage in these cells. Finally, we monitored the percentage of GFP-positive cells over time in the populations as an indicator for cytotoxicity. As published previously (43), Cre expression led to a rapid decline of GFP-positive cells in the population (Figure 7D and Supplementary Figure S6), whereas no decline in GFP-positive cells was observed in Vika-expressing cells. These experiments indicate that high levels of Cre are cytotoxic, whereas Vika expression is well tolerated in the same setting.
Figure 7.
Evaluation of genotoxicity and cytotoxicity of Vika. (A) Recombination specificity of Vika and Cre on the respective cryptic human and mouse chromosomal target sites. The − and + indicates absence or presence of L(+)-arabinose (100 μg/ml) in the growth medium. Non-recombined and recombined plasmids are denoted as two triangles and one triangle, respectively. M, marker; 2-log DNA ladder, NEB. (B) Evaluation of DNA damage induction by γ-H2AX assay 72 h after infection with indicated recombinase-expressing virus, n = 3. (C) Quantification of the γ-H2AX-positive cells either infected with virus for respective recombinase expression or treated with campthothecin for 2 h. Statistically significant increase of γ-H2AX signals is indicated by asterisks. Error bars indicate standard deviation of the mean value (** indicate P = 0.01). (D) Proliferation effects on expression of indicated recombinases in mouse NIH3T3 cells. Cells were infected with bicistronic retroviruses expressing respective recombinase linked to GFP. Every 72 h, cells were analyzed by flow cytometry. Error bars indicate standard deviation of the mean value, n = 2.
We conclude that Vika is an active and specific SSR that should be useful for genome engineering purposes in mammalian cells.
DISCUSSION
In this study, we describe the discovery of Vika/vox, a new tyrosine family site-specific recombination system. Vika was identified based on sequence alignments and protein fold predictions from sequences and structures of known SSRs. Based on known structures of SSRs, we generated a structural model, which predicted that Vika has Cre-like structural properties. We then implemented a rational approach for the search of Vika’s target site and identified its native recombination site, vox. We demonstrate that Vika/vox does not cross-recombine with either the Cre, VCre or Dre recombinase systems in bacteria or in human cultured cells. Therefore, these SSR systems can be combined for advanced genome engineering approaches.
To optimally use the different SSR systems, it is essential to characterize their applied properties. The determination of the in vivo recombinase reactivity and maximum reachable recombination rate may be crucial for planning an experiment. Sophisticated genetic schemes that include simultaneous modifications (e.g. double knockouts) will require recombinases of comparable activity, whereas for sequential DNA rearrangements (e.g. recombinase-mediated cassette exchange), it may be more important to design the experiment in a manner that intermediate recombination events could be selected for. We compared recombination efficiencies of the new recombinase Vika with Cre and another recently reported Cre-like recombinase VCre. Although VCre was described to display efficient recombination rates (26), our results indicate that VCre recombines its target VloxP at considerably lower rate when directly compared with Cre/loxP and Vika/vox. In our work, we used assays that detect various recombination stages based on induction levels. We observed a consistent trend for differences between the tested recombinases. Cre and Vika were consistently more active then VCre. The comparison of Vika with Cre and VCre should be useful for considerations when planning experiments with several independent recombinases. For instance, in development of a double conditional strategy, described recombinases can be combined in efficiency correlated ways, e.g. Cre + FLPe and Vika + VCre, where Cre and Vika are dedicated to conditional knockouts, whereas FLPe and VCre are reserved for excision of selection marker cassettes. Alternatively, molecular evolution strategies could be used to enhance the activity of recombinases, as has been shown for the FLP recombinase (17), if a multitude of highly efficient recombinases is required.
To integrate Vika recombinase for the applied use in higher organisms, we have addressed the question of cytotoxicity. Vika, as other SSRs, acts on DNA, and therefore, undesired binding to non-target sequences may cause undesired events. With the full genome sequences available for many organisms, it is possible to analyze beforehand whether there are potential binding sites for DNA-binding proteins. We have analyzed and tested pseudo-vox sites for human and mouse in this study and did not observe recombination on these sites. Similar analyses can be carried out for other sequenced species to evaluate the suitability of Vika in a particular organism. We have also compared high-level expression of Vika and Cre in mouse cells via retroviral gene delivery and observed that cells expressing Vika recombinase, unlike Cre, did not exhibit growth inhibition. In addition, in contrast to Cre, Vika did not cause DNA damage when expressed from the retroviral constructs. Therefore, Vika might become an additional tool or even a valuable substitute for experimental settings where Cre is not applicable.
To expand the utility of SSRs, enzymes with novel properties have been developed using directed molecula evolution. Several studies have reported alteration of substrate specificity for both Cre and FLP (52–56) Molecular evolution approaches provide new perspectives for potential therapeutic applications, as these approaches offer design of customized recombinases. Indeed, eradication of a Human immunodeficiency virus provirus from infected human cells has already been demonstrated with the evolved Cre-like recombinase Tre, which recombines a defined sequence within the Human immunodeficiency virus-1 long terminal repeat (54). However, the development of such recombinases remains tedious and time consuming. The discovery of novel SSR systems should be helpful in two ways to accelerate the development of therapeutically relevant enzymes. First, the detailed investigation of DNA binding should lead to a better understanding of how these enzymes specifically recognize their targets. With this knowledge, rational design of enzymes with novel specificities might become possible. Second, different naturally occurring recombinases could be useful to accelerate the directed evolution process. It has been demonstrated that shuffling of related genes can speed up the evolution process (57). Hence, new recombinases with desired properties might become available in shorter time through family shuffling.
Up to date, several enzymatic systems have been successfully developed for genome engineering purposes (58,59). However, the rapid growth and scale of genetic studies impose the need for a broad spectrum of genetic instruments. Nature is a rich source for discovering DNA-modifying enzymes with desired properties. Large-scale sequencing projects, in particular of environmental samples, provide a rich source for such discoveries (60). With the identification of the Vika/vox system and the proposed recombinase systems in Supplementary Table S2, we demonstrate the utility of these resources if appropriate bioinformatics tools are available. We predict that many more useful genome engineering tools, including additional SSRs with unique specificities, will be discovered using similar approaches in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–6.
FUNDING
J Klaus Tschira Stiftung gGmbH (to A.-G.); European Union’s Seventh Framework Programme for Research (EU FP7) SyBoSS [242129]; Deutsche Forschungsgemeinschaft (German Research Foundation) [SPP1356 BU1400/3-1, SFB655]. Funding for open access charge: Medical Systems Biology, University Hospital and Medical Faculty, Carl Gustav Carus, University of Technology, Dresden, Germany.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Francis Stewart and Jun Fu, BioTec, Dresden, Germany, for providing the R6K-neo vector and Anton Berns and Jos Jonkers, Netherlands Cancer Institute, Amsterdam, The Netherlands, for sharing retrovirus plasmids (LZRS-Cre, LZRS-Cre173 and LZRS-GFP). They also thank Ralf Gey for technical support and the ZIH at the Technische Universität Dresden for computational resources and assistance and the members of the Buchholz and Pisabarro groups for helpful discussions.
REFERENCES
- 1.Roberts RJ. Restriction endonucleases. CRC Crit. Rev. Biochem. 1976;4:123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- 2.Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 3.Dawid IB, Wahli W. Application of recombinant DNA technology to questions of developmental biology: a review. Dev. Biol. 1979;69:305–328. doi: 10.1016/0012-1606(79)90294-x. [DOI] [PubMed] [Google Scholar]
- 4.Williams RJ. Restriction endonucleases: classification, properties, and applications. Mol. Biotechnol. 2003;23:225–243. doi: 10.1385/mb:23:3:225. [DOI] [PubMed] [Google Scholar]
- 5.Marcaida MJ, Muñoz IG, Blanco FJ, Prieto J, Montoya G. Homing endonucleases: from basics to therapeutic applications. Cell. Mol. Life Sci. 2009;67:727–748. doi: 10.1007/s00018-009-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 7.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 8.Brown WRA, Lee NCO, Xu Z, Smith MCM. Serine recombinases as tools for genome engineering. Methods. 2010;53:372–379. doi: 10.1016/j.ymeth.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat. Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 10.Kolb AF. Genome engineering using site-specific recombinases. Cloning Stem Cells. 2002;4:65–80. doi: 10.1089/153623002753632066. [DOI] [PubMed] [Google Scholar]
- 11.Rossant J, Nagy A. Genome engineering: the new mouse genetics. Nat. Med. 1995;1:592–594. doi: 10.1038/nm0695-592. [DOI] [PubMed] [Google Scholar]
- 12.Branda CS, Dymecki SM. Talking about a revolution: review the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 13.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination: part I. Recombination between loxP sites. J. Mol. Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 14.Andrews BJ, Proteau GA, Beatty LG, Sadowski PD. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985;40:795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 15.Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 17.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 18.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Duyne GD. A structural view of cre-loxp site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 20.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J. Mol. Biol. 2011;2:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, Hoelters J, Calos MP. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 22.Karow M, Chavez CL, Farruggio AP, Geisinger JM, Keravala A, Jung WE, Lan F, Wu JC, Chen-Tsai Y, Calos MP. Site-specific recombinase strategy to create induced pluripotent stem cells efficiently with plasmid DNA. Stem Cells. 2011;29:1696–1704. doi: 10.1002/stem.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Ghosh P, Hatfull GF, Hong Y. Successive and targeted DNA integrations in the Drosophila genome by Bxb1 and phiC31 integrases. Genetics. 2011;189:391–395. doi: 10.1534/genetics.111.129247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol. 2010;477:109–123. doi: 10.1016/S0076-6879(10)77007-5. [DOI] [PubMed] [Google Scholar]
- 25.Sauer B. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 2004;32:6086–6095. doi: 10.1093/nar/gkh941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki E, Nakayama M. VCre/VloxP and SCre/SloxP: new site-specific recombination systems for genome engineering. Nucleic Acids Res. 2011;39:e49–e49. doi: 10.1093/nar/gkq1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nern A, Pfeiffer BD, Svoboda K, Rubin GM. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl Acad. Sci. USA. 2011;108:14198–14203. doi: 10.1073/pnas.1111704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model. Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 29.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Surendranath V, Chusainow J, Hauber J, Buchholz F, Habermann BH. SeLOX–a locus of recombination site search tool for the detection and directed evolution of site-specific recombination systems. Nucleic Acids Res. 2010;38:W293–W298. doi: 10.1093/nar/gkq523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin SS, Pulido E, Chu VC, Lechner TS, Baldwin EP. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J. Mol. Biol. 2002;319:107–127. doi: 10.1016/S0022-2836(02)00246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sippl MJ. Boltzmann's principle, knowledge-based mean fields and protein folding. An approach to the computational determination of protein structures. J. Comput. Aided Mol. Des. 1993;7:473–501. doi: 10.1007/BF02337562. [DOI] [PubMed] [Google Scholar]
- 33.Kwon HJ, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 35.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 38.Mahoney MW, Jorgensen WL. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112:8910–8922. [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Buchholz F, Stewart AF. Alteration of Cre 30 recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 41.Kellendonk C, Tronche F, Monaghan AP, Angrand PO, Stewart F, Schütz G. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. 1996;24:1404–1411. doi: 10.1093/nar/24.8.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchholz F, Bishop J. loxP-directed cloning: Use Cre recombinase as a universal restriction enzyme. Biotechniques. 2001;31:906–918. doi: 10.2144/01314rr02. [DOI] [PubMed] [Google Scholar]
- 43.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25:729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- 45.Guo F, Gopaul DN, Van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 46.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thyagarajan B, Guimarães MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 48.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 49.Forni PE. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J. Neurosci. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higashi AY, Ikawa T, Muramatsu M, Economides AN, Niwa A, Okuda T, Murphy AJ, Rojas J, Heike T, Nakahata T, et al. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J. Immunol. 2009;182:5633–5640. doi: 10.4049/jimmunol.0802413. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl Acad. Sci. USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voziyanov Y, Konieczka JH, Stewart AF, Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J. Mol. Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 53.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 55.Buchholz F, Hauber J. In vitro evolution and analysis of HIV-1 LTR-specific recombinases. Methods. 2011;53:102–109. doi: 10.1016/j.ymeth.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 58.Buchholz F. Engineering DNA processing enzymes for the postgenomic era. Curr. Opin. Biotechnol. 2009;20:383–389. doi: 10.1016/j.copbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307–307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16–e466. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.