Abstract
Gene expression profiles can be used to infer previously unknown transcriptional regulatory interaction among thousands of genes, via systems biology ‘reverse engineering’ approaches. We ‘reverse engineered’ an embryonic stem (ES)-specific transcriptional network from 171 gene expression profiles, measured in ES cells, to identify master regulators of gene expression (‘hubs’). We discovered that E130012A19Rik (E13), highly expressed in mouse ES cells as compared with differentiated cells, was a central ‘hub’ of the network. We demonstrated that E13 is a protein-coding gene implicated in regulating the commitment towards the different neuronal subtypes and glia cells. The overexpression and knock-down of E13 in ES cell lines, undergoing differentiation into neurons and glia cells, caused a strong up-regulation of the glutamatergic neurons marker Vglut2 and a strong down-regulation of the GABAergic neurons marker GAD65 and of the radial glia marker Blbp. We confirmed E13 expression in the cerebral cortex of adult mice and during development. By immuno-based affinity purification, we characterized protein partners of E13, involved in the Polycomb complex. Our results suggest a role of E13 in regulating the division between glutamatergic projection neurons and GABAergic interneurons and glia cells possibly by epigenetic-mediated transcriptional regulation.
INTRODUCTION
Embryonic stem (ES) cells derive from the inner cell mass of blastocyst-stage embryos (1,2). The ES properties to self-renew (3) and differentiate in all three germ layers both in vitro and in vivo (4,5) have made these cells a unique in vitro system for studying the molecular mechanisms that regulate lineage specification. High-throughput experimental techniques, combined to the use of systems biology approaches to infer gene regulatory networks (reverse engineering), have shown promise in the elucidation of stem cell renewal and differentiation (6).
In this work, starting from a collection of ∼200 gene expression profiles (GEPs) generated in mouse ES cells following overexpression of single genes (7), we ‘reverse engineered’ a transcriptional network encompassing ES-specific genes to identify master regulators of gene expression in ES cells (‘hubs’). We discovered that a previously uncharacterized gene, E130012A19Rik (E13), highly expressed in mouse ES cells as compared with differentiated cells, is a central ‘hub’ of the network. We generated E13-overexpressing and E13 knock-down ES clones. We performed transcriptome analysis of these clones and demonstrated an enrichment of differentially expressed genes involved in axon guidance and neuronal differentiation. By immune-based affinity purification, we identified protein–protein interactions of E13 with components of the Polycomb chromatin remodelling complex and proteins involved in transcriptional regulation. We found that ES cells overexpressing E13 and differentiated into neurons and glial cells show up-regulation of the glutamatergic neurons marker Vglut2 (8) and down-regulation of both the γ-aminobutyric acid (GABA)ergic neuron marker GAD65 (9,10) and of the radial glia marker Blbp (11,12), as compared with wild-type ES clones. We further demonstrated that E13 is specifically expressed in the developing and adult cerebral cortex. Taken together our results show that E13 has a role in regulating the commitment towards the different neuronal subtypes and glia cells.
MATERIALS AND METHODS
Data analysis of differentially expressed genes in ES cells versus differentiated cells
We compared our collection of 171 ES-specific GEPs (GSE19836 and GSE32015) to a collection of 180 GEPs derived from normal mouse tissues and differentiated cell lines (GSE10246) (13). The two data sets were first normalized together using the RMA algorithm (14). The median was chosen as measure of the expression values for each probe set within each data set. The variability of the data was taken into account by dividing this measure by a pooled variance given by the sum median absolute deviation of the genes expression values in the two data collections. Each probe set was thus associated with two coordinates representing median expression in the ES-specific data set and in the differentiated data set, and thus represented as a dot in Figure 1. The distance from the diagonal was computed, and an empirical P-value and a corresponding false discovery rate (FDR) were estimated to identify ES-specific transcripts.
Figure 1.
Scatter plot of the median gene expression level in mouse ES cells versus differentiated cells. The median level of expression of each gene in mouse ES cells (x-axis) and in differentiated cells (y-axis) is represented as a dot. The diagonal line corresponds to the set of genes with the same expression in ES cells versus differentiated cells. Genes corresponding to dots significantly off the diagonal (in magenta, genes with FDR <0.025 and in green FDR <0.05) represent either genes whose expression is lower in ES cells than in differentiated cells (above the diagonal), or genes whose expression higher in ES cells than in differentiated cells (below the diagonal). Some ES-specific markers (Oct4, Nanog and Sox2) and the novel ES differentially expressed gene E130012A19Rik are highlighted.
Regulatory network inference
We used ARACNe (15) on 171 microarray experiments (GSE19836 and GSE32015) to reconstruct the transcriptional regulatory network (Supplementary File S1) in mouse ES cells, following the steps shown in Supplementary Figure S1. The gene network among the 45 101 transcripts (probe sets) was inferred using as a significance threshold for the mutual information (MI) a P < 0.001 and setting the data processing inequality threshold to 0.01. The expression value of each probe set was averaged across biological replicates before ARACNe analysis, and a low-entropy filter was applied to remove probe sets whose changes were not significant across the data set, thus removing 4511 probe sets. The low-entropy filter removes non-informative probe sets by computing the entropy of each probe set across the data set as described in (16). Probe sets with entropy values less than the 10th percentile were removed from further analysis.
We validated the inferred network by computing the positive predictive value [PPV = TP/(TP + FP)] against two different 'Golden Standards’ (GS): (i) the Reactome database: containing experimentally validated interactions from the literature (Supplementary File S2); and (ii) the ESCAPE (Embryonic Stem Cell Atlas from Pluripotency Evidence) database: containing putative transcription factor (TF)–messenger RNA (mRNA) regulatory interactions predicted from gene expression profiling in mouse ES cells (Supplementary File S3). The PPV represents the percentage of correctly inferred interactions, i.e. those interactions confirmed by one of the two GS. To compute the PPV, we first converted transcripts to genes and then selected only those genes present also in the ‘Golden Standard’ (and their inferred interactions).
The number of predicted interactions in the inferred transcript-wise network is 299 610 among 40 590 transcripts, whereas the gene-wise network has 131 587 interactions among 17 645 genes.
The ESCAPE GS and the inferred gene-wise network have in common 14 151 of 17 645 genes. Among these 14 151 genes, there are 107 663 interactions in the ESCAPE GS, and 91 925 interactions in the inferred network. Therefore, the random PPV for the ESCAPE GS is equal to 107 663/[(14 151^2−14 151)/2] = 0.0011.
The Reactome GS and the inferred gene-wise network have in common 3087 genes of 17 645 genes. Among those 3087 genes, there are 53 933 in the Reactome GS, and 4973 interactions in the inferred network. Therefore, the random PPV for the Reactome GS is equal to 53 933/[(3087^2−3087)/2] = 0.0113.
We also built a smaller ES-specific subnetwork by selecting only the 543 ES-specific genes (FDR < 0.025) and the genes they were connected to in the network (gene neighbours). To identify ES-specific ‘hub genes’, we first ranked the 5863 probe sets in the ES-specific sub-network according to their degree (i.e. the number of interactions a probe set has in the network) and retained only the top 100 probe sets with the highest degree. We then ranked the 5863 probe sets according to their ES-specific expression, i.e. with the smallest FDR (as detailed in the ‘Identification of genes prevalently expressed in mouse ES cells’ section) and retained only the top 100 probe sets with the most specific expression. Finally to identify ES-specific ‘hub genes’, we intersected the two lists of probe sets (highest degree versus the most specific expression) to obtain the 14 probe sets in Figure 2.
Figure 2.
ES-specific subnetwork inferred from the analysis of 171 GEPs. A reverse engineering algorithm was applied to the set of 171 GEPs from ES cells comprising >45 000 transcripts. The resulting network was used to obtain an ES-specific subnetwork by selecting only the 543 genes with ES-specific expression and the genes they were connected to. We then identified ‘hub’ genes in the subnetwork (numbered from 1 to 14) by ranking the 543 genes according to the number of connections.
Generation of ES clones
E14Tg2a.4 [E14 (17)] and EBRTcH3 [EB3 (18)] parental cell lines were used. The EB3 cell line (18) was obtained from the laboratory of Dr Hitoshi Niwa as previously described in (7). Mouse ES cells were grown as previously described (7). The two E13-inducible cell lines (not-tagged and 3xFLAG-tagged) were derived from the EB3 cell line. For the generation of two exchange vectors (pPTHC-E13 and pPTHC-E13-3xFLAG), we used the vector pPthC-Oct-3/4 obtained from the laboratory of Dr Hitoshi Niwa (18) and modified it as in (7). The primer pair for the generation of the two E13-inducible cell lines and for the selection of positive clones, performed as previously described, (7) are reported in Supplementary Table S1.
The knock-down control (shCTL) clones and the E13 knock-down (shE13) clones were derived from the E14 cell line. For the generation of the pSuper.neo-shE13 and pSuper.neo-shGFP plasmids, the pSuper.neo vector (Oligoengine, Seattle, WA, USA) was used. The knock-down of E13 mRNA was verified by quantitative real-time polymerase chain reaction (q-PCR) on total RNA extracted from three shE13 clones (shE13 A7, shE13 C1 and shE13 C4), as compared with three shCTL clones (shC B6, shC C1 and shC C3). The primer pair used for q-PCR was the E13-affy primer pair reported in Supplementary Table S1. Full details of the protocol can be found in (7).
Induction of transgene expression in E13-inducible cell lines
Three inducible non-tagged clones (C1, C3 and C6) and two inducible 3xFLAG-tagged clones (B5 and B8) were thawed, amplified and tested for transgene induction as previously described (7). q-PCR experiments were performed using LightCycler 480 II (Roche) for signal detection. Primers are reported in Supplementary Table S1.
Microarray hybridization and differential gene expression analysis
For the analysis of E13-inducible cell clones, microarray hybridization experiments were performed on three biological replicates of non-tagged clones (C1, C3 and C6) induced in mouse ES media deprived of tetracycline (Tc) for 48 h. As control, the same clones were grown in mouse ES media containing 1 μg/ml of Tc. Similarly, for the E13 knock-down cell line, experiments were performed on three biological replicates for the shE13 clones (shE13 A7, shE13 C1 and shE13 C4) and on two biological replicates for the shCTL clones (shC C1 and shC C3). Three micrograms of total RNA from each clone were used and hybridized to the Affymetrix GeneChip Mouse Genome 430_2 array (Mouse 430_2) using standard protocols. Differentially expressed genes were detected by a Bayesian t-test method [Cyber-t (19)] followed by FDR correction. The thresholds used were FDR < 0.05 for the induction set and FDR < 0.10 for the knock-down set (different FDR thresholds were selected to have a comparable number of false positives in the two data sets).
For the analysis of the Sugino microarray data set [(20) GSE2882], data were normalized using the RMA method (14). The normalized microarray data relative to E13 were extracted and a one-way analysis of variance (21) was carried out. The Tukey multiple comparison procedure was then performed to identify cell types displaying a statistical difference in the expression of E13.
ES cell differentiation protocol and data analysis
Mouse ES cells were differentiated towards neurons and glial cells using the one-step differentiation method (22). The differentiation procedure was applied (i) to two E13-inducible non-tagged (C1 and C3) and to two 3xFLAG-tagged clones (B5 and B8); and (ii) to two shE13 knock-down clones (shE13 A7 and shE13 C4) and to two shCTL knock-down clones (shC C1 and shC C3). The morphology of differentiated cells was followed using a stereomicroscope (MZ16FA, Leica Microsystems, Wetzlar, Germany); images were acquired on a DFC 320 camera (Leica). For each time course expression profile of the selected markers, we used a statistical modelling approach based on Gaussian processes (GPs) to identify those markers that were significantly affected by E13 overexpression or knock-down as described in (23). GPs enable to quantify the true signal and noise embedded in a GEP over time and moreover provide a ranking of the genes according to their differential expression. The method estimates the continuous trajectory of the gene expression by means of GP regression. In particular, given an observed GEP, two different hypothesis H1 and H2 are compared: either the gene is truly differential expressed (H1) or the observed profile is just the effect of random noise (H2). The log-ratio of the marginal likelihoods (llr) measures which of the two hypotheses is more likely, with positive values indicating that hypothesis H1 is more likely, and vice versa for negative values.
Western blotting and immunofluorescence analysis
Fractioned cell lysates from EB3 (CTL) cells and from two 3xFLAG-tagged clones (B5 and B8) induced for 24 and 48 h were prepared. Cytoplasmic (Cyt) and nuclear (Nuc) fractions were obtained using standard protocol (24). Forty micrograms of total protein extracts were fractionated on 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, whereas 15 µg of fractioned protein extracts were separated on a 10% SDS-PAGE gel. Western blotting was then performed as previously described (7), and the following primary antibodies were used: a mouse monoclonal anti-Flag M2-peroxidase antibody (Sigma) and a mouse monoclonal anti-β-Tubulin (Sigma).
For immunofluorescence analysis, ES cell clones were plated at a density of 1000 cells/cm2 on gelatine-coated 24-well plates and induced for 48 h in mouse ES media and in differentiation medium, respectively, deprived of Tc. The same clones grown in medium containing Tc were used as control. The following primary antibodies were used: an anti-Vglut2 (Abcam, ab79157), an anti-GAD65 (G1166, Sigma) and an anti-Blbp antibody (Abcam, ab32423). As secondary antibodies, we used AlexaFluor594 goat anti-mouse (1:400, Molecular Probes, Invitrogen) or AlexaFluor594 goat anti-rabbit (1:400, Molecular Probes, Invitrogen). In all immunofluorescence analysis performed, the 4′,6-diamidino-2-phenylindole (10 µg/ml, Sigma) in phosphate buffered saline was used to stain the nucleus. Labelling was detected by fluorescent illumination using an inverted microscope (DMIRB, Leica Microsystems, Wetzelar, DE); images were acquired on a DC 350 FX camera (Leica).
Co-immunoprecipitation (co-IP), mass spectrometry and liquid chromatography
Co-immunoprecipitation (co-IP) was performed using the Sigma FLAG Immunoprecipitation Kit (Sigma Aldrich) according to the manufacturer’s instructions. A total of 107 cells from two inducible 3xFLAG-tagged clones (B5 and B8) and two control clones from EB3 parental cell line (EB3/1 and EB3/2) were lysed for co-IP experiment. A cell-free negative control and a positive control with 50 ng of FLAG-tagged bacterial alkaline phosphatase (BAP) protein were applied. After the elution of immuno-precipitated proteins, the samples were fractionated on SDS-PAGE (Bio-Rad mini format) followed by coomassie staining for the visualization of the quality of the samples. Each gel strip was cut into 20 equal-sized gel slices and subjected to MS database-based protein identification after tryptic digestion. Liquid chromatography was performed on an Easy-nLC device (Proxeon, Denmark), which is directly coupled to ESI-MS analysis. Liquid chromatography (LC)/electrospray ionization mass spectrometry (ESI-MS)/MS was performed on a LCQ Deca XP ion trap instrument (Thermo Finnigan, Waltham, MA, USA). ESI-MS data acquisition was performed throughout the LC run. Three scan events, (i) full scan; (ii) zoom scan of most intense ion in (i); and (iii) MS/MS scan of most intense ion in (i) were applied sequentially. No MS/MS scan was performed on single charged ions. The raw data were extracted by TurboSEQUEST algorithm; trypsin autolytic fragments and known keratin peptides were filtered out. All DTA files of the same original sample (20 gel slices) generated by Sequest were merged and converted to mascot generic format files. The mascot generic format files were searched using our Mascot Version 2.1 in-house license. The MS/MS ion searches against Expasy UniProt were performed with the following parameters: taxonomy Mus musculus, one trypsin miss-cleavage accepted, monoisotopic mass values, peptide and fragment mass tolerance ± 0.8 Da. Oxidation of methionine and acrylamide adducts on cystein was expected as variable modification. Protein hits corresponding to P < 0.05 (Mowse score >28) were considered as significant protein identifications. FDRs were estimated based on matches to reversed sequences in the concatenated target–decoy database. A maximum FDR of 0.01 at both peptide and protein levels was tolerated. Protein–protein interaction data were submitted directly to Database of Interacting Proteins (DIP) database.
To confirm some of the protein interactions identified by MS/MS, we proceeded as follows. Polyclonal rabbit IgGs against the following proteins were used: embryonic ectoderm development (EED) (Anti-EED, Millipore, 09-774), suppressor of zeste 12 (Anti-SUZ12 C-terminal region, Aviva Systems Biology, ARP39190_P050), general TF IIE beta (Anti-GTF2E2, Proteintech, 11 596-1-AP). The FLAG peptide tag (rabbit IgG against FLAG peptide epitope, Sigma) was used to detect E13 expression in the E13-inducible ES clones with the 3xFLAG before and after IP experiment. Forty micrograms of protein extracts of the following eight samples were first separated by SDS-PAGE using the Laemmli system (Bio-Rad mini electrophoresis running chamber, 12% Polyacrylamide, gel format: 7.5 × 8 cm): E13-inducible clone B5 (B5) after FLAG-Tag IP, E13-inducible clone B8 (B8) after IP, control cell clone Eb3/A2 (K1) after IP, control cell clone Eb3/B2 (K2) after IP, E13-inducible clone B5 before IP, E13-inducible clone B8 before IP, control cell clone Eb3/A2 before IP and control cell clone Eb3/B2 before IP. For the western blotting against FLAG-tag, a 49-kDa FLAG-BAP fusion protein from Escherichia coli (Sigma-Aldrich, F7425) was used as the positive control (PK) for the immunoblotting methodology. Subsequently, proteins were transferred onto the supported nitrocellulose membrane using the standard procedure (semi-dry transfer chamber, 40 mA, 1.5 h). The One-Hour Western standard kit, including secondary rabbit antibody and TMB substrate, (GenScript L00204T, according to the manufacturer’s instruction) was used for the immunodetection and visualization.
Immunohistochemistry (IHC) and in situ hybridization (ISH)
Mouse tissue sections were prepared using standard protocols (25) Antisense RNA probes were labelled using a DIG-RNA labelling kit (Roche). The following probes were used: Gad67 and E13 (1100-nt length, containing the E13 complete coding DNA sequence). RNA in situ hybridization (ISH) hybridization procedures, combined or not with standard immunohistochemistry (IHC), were performed as previously described (26). For IHC, the following primary antibodies were used:anti-Blbp, rabbit polyclonal antibody (1:100, Abcam); anti-GFAP rabbit polyclonal antibody (1:400, Dako); anti-Tbr2, rabbit polyclonal antibody (1:1000, Chemicon); anti-NeuN, mouse monoclonal antibody, (1:200, Chemicon); anti-Vglut1, mouse monoclonal antibody, (1:100, Millipore); anti-Calbindin D-28k, rabbit polyclonal antibody (1:2500, Swant) and anti-TH, rabbit polyclonal antibody (1:200, Cell Signaling). All experiments in mice were conducted following guidelines of the Institutional Animal Care and Use Committee, Cardarelli Hospital (Naples, Italy).
RESULTS
Identification of genes prevalently expressed in mouse ES cells
In a previous study, we generated a collection of 120 GEPs measured using microarrays in mouse ES cells by overexpressing 20 mouse orthologous of human chromosome 21 genes [Gene Expression Omnibus (GEO) accession number: GSE19836] (7). We have now extended this collection by overexpressing eight more genes, comprising mostly TFs (Supplementary Table S2), thus generating 51 additional GEPs (GEO accesion number: GSE32015).
We analysed the entire data set of 171 GEPs to identify genes whose expression is enriched in mouse ES cells, as compared with differentiated cells. To this end, we compared our collection of ES-specific GEPs (GSE19836 and GSE32015) with a collection of 180 GEPs derived from normal mouse tissues and differentiated cell lines (GSE10246) (13). Both sets were obtained with the same microarray platform (Affymetrix Mouse 430_2), enabling a homogeneous comparison.
To identify genes that are significantly more expressed in ES cells versus differentiated cells, we computed the ‘median’ expression level of each probe set in the two gene expression data set (ES cells and differentiated cells), which is shown as a dot in Figure 1. We then computed the distance from the diagonal (corresponding to the set of probe sets with the same expression levels in both ES and differentiated cells). We retained for further analysis only those probe sets whose distance from the diagonal was determined to be statistically significant. This approach enabled the identification of 543 genes that were significantly more expressed in ES cells versus non-ES cells (FDR < 0.025), independently of their absolute level of expression (‘ES-specific genes’), among which many known ‘stemness’ genes, such as Oct4 [Pou5f1 (27)] and Nanog (28) (Supplementary Table S3). The gene ontology enrichment analysis (GOEA) (29,30) performed on the list of 543 genes confirms that this set is strongly enriched for stemness-related processes (Supplementary Table S4), such as stem cell differentiation (GO:0048863 – p = 9.6e-10), stem cell maintenance (GO:0019827 – p = 3.8e-7) and stem cell development (GO:0048864 – p = 5.4e-7), thus confirming the validity of our approach and its potential for the discovery of novel genes involved in ES cell fate regulation.
Identification of a novel mouse gene as central ‘hub’ of an ES-specific gene regulatory network
‘Reverse engineering’ approaches allow to infer gene–gene regulatory interactions by computational analysis of GEPs (31,32). We applied an information-theoretic approach, named ARACNe (15), which uses a generalization of pairwise correlation coefficient, called MI, to identify co-regulated genes from GEPs. ARACNe was run on the collection 171 ES-specific GEPs. The resulting gene network consists of 299 610 predicted interactions among 40 590 transcripts. To measure the reliability of the inferred network, we used two GS data sets: (i) a collection of 153 920 putative TF–mRNA regulatory interactions obtained from studies of TF loss- or gain-of-function followed by mRNA microarray profiling in mammalian ES cells (ESCAPE, http://www.maayanlab.net/ESCAPE/index.php); and (ii) a smaller set of high-quality experimentally verified functional interactions found in the Reactome database, an open-source, open-access, manually curated and peer-reviewed interaction database (33). We then computed the percentage of correctly inferred interactions (PPV) by ranking interactions by the value of their MI (‘Materials and Methods’ section). As shown in Supplementary Figure S2 for the ESCAPE database and Supplementary Figure S3 for the Reactome database, the inferred interactions are significantly enriched for functional interactions.
We then applied a hierarchical clustering algorithm based on the Jaccard distance to discover communities within the network (34–36). A community is defined as a set of genes strongly co-regulated with each other, but with few interactions with other genes in the network. We thus identified 53 communities (Supplementary Table S5). For each community, we performed GOEA to identify biological functions significantly enriched among genes within the community (Supplementary Table S6).
To better analyse this large network, we built a smaller subnetwork, as shown in Figure 2, by selecting the 543 ES-enriched genes (FDR < 0.025) described earlier and the genes they were connected to in the network (gene neighbours). This subnetwork comprises 5863 transcripts and 12 944 interactions among them. A graph containing the complete workflow, data sources and cut-offs threshold used to obtain the final ES-specific subnetwork is reported in Supplementary Figure S1.
We identified ‘hub genes’ in this subnetwork, by selecting genes with the highest number of inferred regulatory interactions, which were specifically expressed in ES cells (‘Materials and Methods’ section). As shown in Figure 2, hub genes comprise the sal-like 4 (Sall4) TF, a known regulator of stem cells pluripotency (37); the genes of the Dppa family, known be involved in pluripotency and stemness (38); the zinc finger protein of the cerebellum 3 (Zic3), which is required for maintainance of pluripotency in ES cells (39) and the undifferentiated embryonic cell TF 1 [UTF1,(40–41)]. Interestingly, we found that one of the ES-specific ‘hubs’ was a gene with unknown function, E130012A19Rik (hereafter abbreviated as E13).
To gain initial insight into its function, we performed GOEA on the 26 genes connected to E13 in the network (Supplementary Table S7C). Such a ‘guilty-by-association’ approach has already been successfully applied to predict gene function (42). The results of this analysis suggested a role of E13 in transcriptional regulation and nucleic acid metabolism (Supplementary Table S7C). Moreover, E13 belongs to network community 2 (Supplementary Table S5), which is significantly enriched for biological process such as: pattern specification process (GO:0007389 – p = 7e-4), axon guidance (GO:0007411 – p = 0.02), embryonic morphogenesis (GO:0048598 – p = 0.02) and embryonic development (GO:0009790 – p = 0.03) (Supplementary Table S6).
We also performed a bioinformatic analysis of the E13 sequence. This gene is predicted to encode a hypothetical protein product (LOC103551, http://www.ncbi.nlm.nih.gov/gene/) that is highly conserved across vertebrates (Supplementary Figure S4A), and contains a proline-rich domain [PROSITE database (43)] (Supplementary Figure S4B). Analysis of the predicted 3D structure suggests that part of E13 protein has similarity to the DNA-directed RNA polymerase II. However, it differs from other RNA polymerases because of the presence of additional peripheral 3D structures not present in normal RNA polymerases. Three putative phosphorylation sites (S15, S19 and S22) were annotated in PhosphoSite database (44) (Supplementary Figure S4C). In addition, E13 may be regulated by KLF4, MYC, NANOG, OCT4, REST SOX2, TCF3 and TRIM28 according to chromatin immunoprecipation experiments reported in (45).
Generation of E13 transgenic mouse ES cell lines
We generated three stable mouse ES cell lines: two inducible cell lines overexpressing E13, which only differ for the presence in one of them of a 3xFLAG epitope at the C-terminus of the transgene-coding sequence, and one cell line in which E13 was stably knocked-down (Supplementary Figure S5).
q-PCR analysis (‘Materials and Methods’ section) confirmed that the expression of the E13 mRNA was induced on the removal of Tc from the medium in both the overexpressing clones (Supplementary Figure S5A and B). Moreover, we verified the correct induction of the E13 protein product and determined its intracellular localization by western blot (Supplementary Figure S6) with a FLAG-specific monoclonal antibody in the inducible 3xFLAG-tagged cell line (‘Materials and Methods’ section). As shown in Supplementary Figure S6A, we detected an ∼43-kDa band corresponding to the E13-3xFLAG expected protein product (the E13 molecular weight is about 41 kDa plus 2.4 kDa of the 3xFLAG peptide), confirming that E13 is a protein-coding gene. The E13 protein appears to be present in both cytoplasmic and nuclear fractions (Supplementary Figure S6B).
The knock-down cell line (shE13 clones) was generated by stably expressing a specific short hairpin RNA against the E13 sequence, thus knocking-down E13 expression in the parental mouse ES cell line [E14 (17)]. As control, we selected a short hairpin RNA against the green fluorescent protein reporter, thus generating control knock-down clones (shCTL clones) (‘Materials and Methods’ section). The extent of inhibition of E13 expression was efficient (∼90%) and comparable in the three different shE13 clones generated (Supplementary Figure S5C).
Transcriptome analysis of E13 transgenic mouse ES cell lines
We performed gene expression profiling experiments using Affymetrix microarrays in both inducible not-tagged clones (n = 3) and in shE13 clones (n = 3) (GSE31701). We found that both the overexpression and knock-down of E13 perturbed the transcriptome in a statistically significant manner (‘Materials and Methods’ section). In Supplementary Table S8 and in Supplementary Table S9, we report the complete list of differentially expressed genes in the two sets of experiments.
To obtain a high-confidence set of genes responsive to E13, we selected only those genes (n = 221) whose expression levels changed consistently (i.e. increased in E13 overexpressing clones and decreased in knock-down shE13 clones or vice versa—‘Materials and Methods’ section and Supplementary Table S10). A GOEA of the 221 high-confidence genes revealed a significant enrichment for biological processes related to axon guidance, axonogenesis and other processes involved in neurogenesis (Table 1).
Table 1.
Significantly enriched GOEA terms for the 221 high-confidence genes
| Gene onthology terms | FDR | Fold enrichment | P-value |
|---|---|---|---|
| Axon guidance | 0.052 | 7.5 | 3.2E-4 |
| Axonogenesis | 0.069 | 4.5 | 4.5E-3 |
| Anatomical structure development | 0.086 | 1.6 | 5.6E-3 |
| Neuron projection morphogenesis | 0.098 | 4.2 | 6.4E-3 |
| Cell morphogenesis involved in neuron differentiation | 0.11 | 4.1 | 7.5E-3 |
Significant GOEA terms, the FDR, the fold enrichment and the P-values for each term. GOEA was performed with the DAVID online tool restricting the output to all biological process terms with a significance threshold of FDR ≤0.1 and fold enrichment ≥1.5.
To better characterize the role of E13, we subdivided the list of 221 genes in two sublists: the first list (UP) consists of 145 genes up-regulated by the induction of E13 and down-regulated by its knock-down (Supplementary Table S11); the second list (DOWN) includes 76 genes down-regulated by the induction of E13 and up-regulated by its knock-down (Supplementary Table S12). GOEA revealed that the UP list contains genes involved in anatomical structure development, axon guidance and other related processes, which suggest a possible involvement of E13 in neurogenesis (Supplementary Table S13). On the other hand, the DOWN list consists of genes involved in the regulation of biosynthetic process and regulation of gene expression, suggesting a role of E13 on global transcriptional regulation (Supplementary Table S14), and confirming the results obtained by the ‘guilty-by-association’ approach on the ES network.
We also explored the network of the genes surrounding E13, i.e. all the nodes at a Distance 2 from E13 (Figure 3), and obtained a subnetwork composed by 106 genes and 128 connections. We found that 63 of 106 genes were indeed perturbed in their expression level by E13 (Supplementary Table S15).
Figure 3.
Subnetwork of genes surrounding E13. The network comprises all of the nodes at a Distance 2 from E13. It consists of 106 genes and 128 connections. Nodes are coloured according to their differential expression (squares if significant, circles if not) following E13 overexpression (inner square/circle) or knock-down (outer square/circle).
Identification of protein interaction partners of E13
We performed immuno-based affinity purification experiments followed by mass spectrometric protein identification (46) using the E13-inducible ES clones with the 3xFLAG (‘Materials and Methods’ section). We identified 23 potential protein partners of E13 with high-confidence (Table 2), among which there are two TFs [Gtf2e2, Btf3 (47)], several mRNA processing proteins and two components of the Polycomb complex [Eed, Suz12 (48) and the retinoblastoma binding protein Rbbp4, which may be involved in pluripotent stem cell maintenance and neuronal differentiation (49)]. Twelve of 23 transcripts corresponding to this subset of proteins were also differentially expressed following E13 overexpression (FDR < 0.1) (Supplementary Table S16).
Table 2.
Proteins identified from the co-IP experiment as potential partners of E13
| UniProt ID | Gene symbol | Protein name | Mowse score | Molecular weight (Da) | Sample name* |
|---|---|---|---|---|---|
| BTF3_MOUSE | Btf3 | Basic TF 3 | 47 | 22 017 | IC |
| C1QBP_MOUSE | C1qbp | Complement component 1 Q subcomponent-binding protein | 53 | 30 994 | IC |
| CPSF6_MOUSE | Cpsf6 | Cleavage and polyadenylation specificity factor subunit | 31 | 59 116 | IC |
| CQ096_MOUSE | E130012A19Rik | Uncharacterized protein C17orf96 homologue | 103 | 38 071 | IC |
| DDX5_MOUSE | Ddx5 | Probable ATP-dependent RNA helicase DDX | 57 | 69 277 | IC |
| EED_MOUSE | Eed | Polycomb protein EED | 53 | 50 166 | IC |
| H12_MOUSE | Hist1h1c | Histone H1.2 | 76 | 21 254 | IC |
| HS90A_MOUSE | Hsp90aa1 | Heat shock protein HSP 90-alpha | 35 | 84 735 | IC |
| IGH1M_MOUSE | Ighg1 | Ig gamma-1 chain C region, membrane-bound form | 601 | 43 359 | IC |
| IGKC_MOUSE | Igkc | Ig kappa chain C region | 843 | 11 771 | IC |
| INT3_MOUSE | Ints3 | Integrator complex subunit 3 | 28 | 117 862 | IC |
| KPRA_MOUSE | Prpsap1 | Phosphoribosyl pyrophosphate synthetase-associated protein 1 | 142 | 39 407 | IC |
| KV2A7_MOUSE | LOC636468 | Ig kappa chain V-II region 26-10 | 796 | 12 265 | IC |
| ML12B_MOUSE | Myl12b | Myosin regulatory light chain 12B | 776 | 19 767 | IC |
| MTF2_MOUSE | Mtf2 | Metal-response element-binding TF 2 | 42 | 66 882 | IC |
| MYH9_MOUSE | Myh9 | Myosin-9 | 2487 | 226 232 | IC |
| NEK9_MOUSE | Nek9 | Serine/threonine-protein kinase Nek9 | 32 | 107 015 | IC |
| NKTR_MOUSE | Nktr | NK-tumour recognition protein | 32 | 163 341 | IC |
| PCNA_MOUSE | Pcna | Proliferating cell nuclear antigen | 72 | 28 766 | IC |
| RBBP4_MOUSE | Rbbp4 | Histone-binding protein RBBP4 | 38 | 47 625 | IC |
| RL23A_MOUSE | Rpl23a | 60S ribosomal protein L23a | 240 | 17 684 | IC |
| RL28_MOUSE | Rpl28 | 60S ribosomal protein L28 | 98 | 15 724 | IC |
| RS11_MOUSE | Rps11 | 40S ribosomal protein S11 | 82 | 18 419 | IC |
| RS13_MOUSE | Rps13 | 40S ribosomal protein S13 | 105 | 17 212 | IC |
| SETX_MOUSE | Setx | Probable helicase senataxin | 53 | 297 401 | IC |
| SPTA2_MOUSE | Sptan1 | Spectrin alpha chain, brain | 658 | 284 422 | IC |
| SPTB2_MOUSE | Sptbn1 | Spectrin beta chain, brain 1 | 607 | 274 052 | IC |
| SUZ12_MOUSE | Suz12 | Polycomb protein Suz12 | 78 | 82 974 | IC |
| T2EB_MOUSE | Gtf2e2 | General TF IIE subunit 2 | 42 | 33 026 | IC |
| THOC4_MOUSE | Thoc4 | THO complex subunit 4 | 84 | 26 924 | IC |
| UBIQ_MOUSE | Rps27a | Ubiquitin | 105 | 8560 | IC |
| ACTB_MOUSE | Actb | Actin, cytoplasmic 1 | 247 | 41 710 | CTL |
| HNRPF_MOUSE | Hnrnpf | Heterogeneous nuclear ribonucleoprotein F | 170 | 45 701 | CTL |
| IGH1M_MOUSE | Ighg1 | Ig gamma-1 chain C region, membrane-bound form | 699 | 43 359 | CTL |
| IGKC_MOUSE | Igkc | Ig kappa chain C region | 306 | 11 771 | CTL |
| KV2A7_MOUSE | LOC636468 | Ig kappa chain V-II region 26-10 | 796 | 12 265 | CTL |
| MYH10_MOUSE | Myh10 | Myosin-10 | 1883 | 228 855 | CTL |
| PRPS1_MOUSE | Prps1 | Ribose-phosphate pyrophosphokinase 1 | 123 | 34 826 | CTL |
| RS5_MOUSE | Rps5 | 40S ribosomal protein S5 | 84 | 22 875 | CTL |
| IGH1M_MOUSE | Ighg1 | Ig gamma-1 chain C region, membrane-bound form | 699 | 43 359 | P-CTL |
| IGKC_MOUSE | Igkc | Ig kappa chain C region | 310 | 11 771 | P-CTL |
| KV2A7_MOUSE | LOC636468 | Ig kappa chain V-II region 26-10 | 368 | 12 265 | P-CTL |
| PPB_ECOLI | phoA | Alkaline phosphatase OS = E. coli (strain K12) | 6665 | 49 408 | P-CTL |
| IGH1M_MOUSE | Ighg1 | Ig gamma-1 chain C region, membrane-bound form | 625 | 43 359 | N-CTL |
| IGKC_MOUSE | Igkc | Ig kappa chain C region | 735 | 11 771 | N-CTL |
| KV2A7_MOUSE | LOC636468 | Ig kappa chain V-II region 26-10 | 535 | 12 265 | N-CTL |
IC = inducible cell line; CTL = control cell line; P-CTL = positive control with flag-tagged BAP protein; N-CTL = negative control without protein.
Two inducible cell samples, two control cell samples, one negative control and one positive control were processed simultaneously.
Protein hits corresponding to P < 0.05 (Mowse score >28) were considered as significant protein identifications.
To further confirm these protein interactions, we selected 3 of the 23 proteins, taking into account the availability of antibodies and their biological functions: Suz12 and EED (part of the Polycomb complex) and the general TF GTF2E2. We then immune-precipitated E13 using the anti-3xFLAG antibody in E13 overexpressing clones followed by western blot analysis with the three antibodies against the selected proteins (the experiment was performed in duplicate) (‘Materials and Methods’ section). For all the three proteins, we were able confirm the interaction of E13 with 3 of 23 potential protein partners, as shown in Supplementary Figure S7.
The interaction of E13 with these proteins further suggests a role of E13 in mRNA processing and epigenetic regulation (Supplementary Table S17).
E13 is a modulator of neuronal differentiation
Results of the bioinformatic analysis and of the transcriptomics and proteomics experiments suggested a role of E13 in regulating gene expression and neurogenesis. To confirm a role of E13 in this process, we specifically differentiated both E13-inducible and knock-down ES clones into neurons and glia cells using the one-step differentiation method (22).
We first verified, by western blot analysis, the expression of the E13 protein, at Days 10 and 15 of the differentiation protocol (Supplementary Figure S8). We then verified by q-PCR the potential of the inducible and of the knock-down E13 clones to differentiate along the neuronal and glial cell lineages. To this purpose, we collected RNA samples at different time points and analysed the expression profiles of E13 and of the markers of undifferentiated ES cells [Oct4 (27)], and of neuronal precursors [Nestin (50) and Neurog1 (51)].
Figure 4 shows the expression profiles of E13 (Figure 4A), Oct4 (Figure 4B), Nestin (Figure 4C) and Neurog1 (Figure 4D) in the inducible clones (left panels) and in the knock-down clones (right panels). All of the clones displayed the expected down-regulation of the pluripotency gene Oct4. We observed that during differentiation the expression profiles of the neuronal precursor markers, Nestin and Neurog1 were significantly, albeit transiently, affected by E13 knock-down.
Figure 4.
Expression profile of E13, Oct4, Nestin and Neurog1 during neuronal differentiation. The expression profiles of E13 (A), Oct4 (B), Nestin (C) and Neurog1 (D) transcripts were evaluated by q-PCR in two inducible not-tagged clones derived from parental ES cell line EB3 (left panels) and in two knock-down clones derived from parental ES cell line E14 (right panels). For E13 transcript, E13-3xFLAG primer pair and E13-affy primer pair were used in inducible and in knock-down clones, respectively. The red bar represents the control, the green bar represents the expression profile of each transgene in the inducible and in the knock-down clones, respectively. For each expression graph, we reported on the x-axis the days of the differentiation protocol and on the y-axis the relative expression of the transcript expressed as 2-dCT. For each expression profile, the llr was calculated, and the value was reported on the top of each graph. The llr represents the statistical significance value of the difference in the expression profiles of each transgene in the inducible and in the knock-down condition compared with each control. The statistical significance is indicated by asterisk (llr > 0).
To assess whether the induction and/or the knock-down of E13 could influence the formation of radial glia cells and/or of some neuronal subtypes, we then analysed the expression of additional neuronal lineage specific markers (Figure 5). Vglut2, the vesicular glutamate transporter 2, which is essential for glutamate release from presynaptic vesicles in glutamatergic excitatory neurons (8), was specifically up-regulated by E13 overexpression in the inducible clones (Figure 5A). In contrast, both GAD65, a glutamate decarboxylase specific for GABAergic neurons (10), and Blbp, the brain-specific member of the lipid-binding protein family, which is required for the establishment of the radial glial fibre system in developing brain (11,12), were significantly down-regulated (Figure 5B and C).
Figure 5.
Expression profile of Vglut2, GAD65 and BLBP during neuronal differentiation. Expression analysis in two inducible non-tagged clones derived from parental ES cell line EB3 (left panels) and in two knock-down clones derived from parental ES cell line E14 (right panels) was evaluated by q-PCR: Vglut2 is a glutamatergic neuron marker (A), GAD65 is a GABAergic neuron marker (B), BLBP is a radial glia marker (C). The red bar represents the control, the green bar represents the expression profile of each transcript in the overexpressing and in the knock-down clones. For each expression graph, we reported on the x-axis the days of the differentiation protocol and on the y-axis the relative expression of the transcript expressed as 2-dCT. For each expression profile, the llr was calculated, and the value was reported on the top of each graph. The llr represents the statistical significance value of the difference in the expression profiles of each transgene in the inducible and in the knock-down condition compared with each control. The statistical significance is indicated by asterisk (llr > 0).
In addition, we also analysed the expression of Chat (choline acetyltransferase), a marker of mammalian cholinergic system (52), as well as that of the rate-limiting enzyme in dopamine biosynthesis, the tyrosine hydroxylase TH (53), and of the serotonin biosynthetic enzyme tryptophan hydroxylase Tph2 (54). The expression of Chat was significantly, albeit transiently, affected by the knock-down of E13, whereas the expression of TH and of Tph2 did not change significantly following E13 induction or knock-down (Supplementary Figure S9).
To verify whether the aforementioned variations in the expression of the glutamatergic, GABAergic and radial glial markers also resulted in a variation in the number of cells belonging to those specific subpopulations, we performed immunofluorescence experiments with anti-Vglut2, anti-GAD65 and anti-Blbp antibodies during differentiation of E13-inducible clones and knock-down clones (Supplementary Figure S10). We found that the number of Blbp-positive cells and of GAD65-positive cells in induced clones was significantly lower (negative binomial P = 0.02 and P = 0.009, respectively) than in uninduced control clones (Supplementary Figure S10B). These data suggest that the down-regulation of the GAD65 and Blbp transcripts (Figure 5B and C) following E13 overexpression is because of the reduced number of GAD65-positive and Blbp-positive cells.
Expression of E13 in brain
The aforementioned results prompted us to study the expression pattern of E13 in the central nervous system. Towards this goal, we performed RNA ISH analysis with an E13 probe in mouse brain starting at embryonic Day 12.0 (E12.0), (Figures 6 and 7). At this stage, the first post-mitotic excitatory neurons are clearly visible at the pial border of the dorsal pallium (55,56), while the interneuronal population emerging in the ganglionic eminences of the ventral pallium is approaching its tangential migration (57–59).
Figure 6.
Expression of E13 in developing mouse brain. (A) Expression of E13 during mouse brain development evaluated by RNA ISH at embryonic Day 12.0 (E12.0). High levels of E13 are detected in the dorsal pallium from the ventricular to the pial surface, along the entire rostro-caudal (R-C) axis. In the ventral pallium, E13 expression is restricted to the ventricular zone of the ganglionic eminences (dashed lines). Scale bar: 500 µm. (B) High magnification of E12.0 embryonic brain at the rostro-caudal level indicated by the asterisk in (A). A graded expression of E13 is clearly visible in all the cortical primordium. Double ISH/IHC shows that although at low levels of expression. E13 levels gradually increase and co-localize with the Tbr2+ intermediate progenitor cells of the subventricular zone, which are about to undergo the final neurogenic division. The maximum rate of E13 is evident in the post-mitotic neurons of the preplate that are positive to NeuN staining (arrows and arrow heads in the higher magnification and insets). Please note that in the ganglionic eminence, E13 is detected only in the ventricular zone. Scale bars: 100 µm, 50 µm in high magnifications. ctx, cortex; hp, hippocampus; vz, ventricular zone; svz, subventricular zone; ppl, preplate; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; CGE, caudal ganglionic eminence; mz, mantle zone; Th, thalamus; Hyp, hypothalamus.
Figure 7.
E13 is expressed in cortical glutamatergic neurons and in the radial glia stem cells-like of the SGZ neurogenesis niche in the adult brain. (A) RNA ISH on P21 mouse brain reveals a strong signal of E13 in the cerebral cortex, in the hippocampus and in the amygdaloid nuclei (arrowheads). Scale bar: 500 µm. (B) A more detailed analysis of E13 expression in primary somatosensory cortex indicates a widespread signal of E13 in all the cortical layers. Co-localization of the vesicular glutamate transporter 1, Vglut1, is detected in E13+ cortical neurons (high magnification and arrow in insets). No co-localization of E13 with Blbp and GFAP is found (high magnification and arrow in insets). Scale bar: 100 µm. (C) In the hippocampus, E13 expression is present in both the glutamatergic main neurons: CA pyramidal cells and the DG granules. Interestingly in the DG, E13 shows a gradient similar to that found in the developing cortex, namely higher in the more mature granule cells. Scale bar: 50 µm. (D) By adjacent ISH for E13 and GAD67 in P21 brains, at the hippocampal level [dotted box in (A)], E13 appears also in the in the GABAergic interneurons populating the oriens and radiatum strata of the hippocampus (arrows). so, stratum oriens; Pyr, pyramidal layer; sr, stratum radiatum; Rs, Retro splenial cortex; Ss1, primary somatosensory cortex; Ss2, secondary somatosensory cortex; Pir, Piriform cortex; BA, basal amygdaloid nuclei; Hp, hippocampal formation; CA1, Cornus Ammonis field 1 of hippocampus proper; Iml, inner molecular layer; GCsL, granule cells layer; sgz, subgranular zone; po, polymorphic layer.
E13 mRNA was strongly expressed in the developing cerebral cortex, from which glutamatergic neurons and glial cells derive (Figure 6A). At higher magnification, a graded expression of E13 in the developing cortex (Figure 6B) is clearly visible, from the ventricular (low) to the pial (high) surface. By double ISH/IHC, E13 appears to be strongly expressed in the post-mitotic neuronal population of the pre-plate, as marked by the neuronal nuclei staining [NeuN, Rbfox3 (60)] (Figure 6B). E13 levels gradually increase in the subventricular zone, co-localizing with the intermediate progenitor cells, which are positive for the neuronal marker Tbr2 (Figure 6B) (61–63).
E13 expression in adult brain was localized to cortical glutamatergic neurons by RNA ISH in P21 mouse brain, and appeared to be particularly high in the cerebral cortex, especially in cingulate (not shown), somatosensory and piriform cortex, in the hippocampal formation, and in the amygdaloid nuclei (Figure 7A). A detailed analysis of E13 in the cortical domains indicated a large representation in all the cortical layers, showing high co-localization with the vesicular glutamate transporter 1, Vglut1, a marker of adult glutamatergic cells, whereas no co-localization is apparent in Blbp-positive (11,12), or GFAP-positive mature astroglial cells, in the cortex (64,65) (Figure 7B).
E13 is also highly represented in the hippocampal formation, in both the glutamatergic principal neuronal populations, the pyramidal neurons of the Cornus Ammonis 1 (CA1) to the CA3 fields of the hippocampus proper and the granule cells of the dentate gyrus (DG) (Figure 7C and D). Interestingly, in the DG, one of the two brain niches where neurogenesis occurs throughout adult life (66–70), E13, shows a gradient similar to that observed in the developing cortex, with high levels in the mature granule cells, as confirmed by co-localization with the calcium binding protein, Calbindin (Figure 7C) (71).
To evaluate the expression of E13 in neural populations whose transcriptome profiles, in vitro, do not appear to be influenced by E13 manipulation, we examined the TH-positive population that correspond to dopaminergic neurons (53). We followed this population history, from the embryonic brain development to the adulthood, and we found no co-localization with E13 neither at E12.0 (Supplementary Figure S11A), nor at P21 (Supplementary Figure S11B).
To further assess in which neuronal subtype E13 gene was significantly expressed, we analysed 36 GEPs collected from 12 different neuronal cell types in the adult mouse forebrain (GSE2882) (72). As shown in Supplementary Figure S12, the expression of E13 varies significantly across the 12 different neuronal cell types (analysis of variance P = 3 × 10−6), with higher expression in the five glutamatergic pyramidal neuron populations, as compared with the six GABAergic populations of interneurons.
DISCUSSION
Regulatory interactions among genes can be ‘reverse engineered’ by considering pairs of genes and checking whether they are co-expressed across different experimental conditions (‘co-expression’ networks). Reverse engineering is a powerful tool to generate hypotheses on gene function (73). In this work, we have produced a collection of GEPs measured in mouse ES cells from our previous study (7) together with a new collection of microarrays and used a systems biology reverse engineering approach to gain initial insight into the functional role of a previously uncharacterized gene, E130012A19Rik. We applied the ARACNe reverse engineering algorithm, which is based on computing a pairwise MI between two genes, as the nature of the GEPs we collected prevented use of more sophisticated strategies (such as those based on time series GEPs). We observe that there is a plethora of reverse engineering algorithms available, and new and improved methods are being developed (32). Therefore, different reverse engineering methods could reveal different aspects of the network, and thus either confirm or reveal additional roles of E13, as well as of other genes with unknown functions.
Interestingly, E13 was the uncharacterized ES-specific ‘hub’ gene with the highest number of connections in the inferred network. Hub genes have been found to be master regulators of specific transcriptional programs both in normal and pathogenic conditions (74,75). Here, by transcriptomics, immuno-based affinity purification experiments and ISH, we indeed identified a role of E13 in neuronal subpopulation specification.
The capability of neurons to adopt the correct neurotransmitter phenotype during early development is the critical point for the proper functioning of the vertebrate adult nervous system. A multiple array of neuronal types have to arise from a field of undifferentiated progenitors; how a cell acquires a given neurotransmitter phenotype is a central issue in developmental neurobiology (76,77). This is particularly true for glutamatergic and γ-aminobutyric acid GABAergic neurons, which are the most abundant excitatory and inhibitory neurons, respectively, in the vertebrate’s central nervous system. It is likely that only a proportion of the factors required for neuronal identity have so far been identified, and the precise way in which such factors interact to specify the timing and terminal differentiation of particular neuronal subpopulations is not yet defined (78).
In addition, the emerging role that epigenetic control plays in brain development implies that the interplay of TFs and epigenetic modifiers, including histone modifications, DNA methylation and microRNAs, is essential for the acquisition of specific cell fates (79). Several chromatin-modifying complexes regulate the renewal or differentiation of a range of neural stem cells, but the Polycomb repressor complexes (PRCs) are of particular interest in this context. This is particularly true for some members of PRC2, such as EzH2, whose alterated expression is able to change the competence of cortical progenitors to generate neurons of different cortical layers, orchestrating the switch from neurogenesis to gliogenesis (80,81). Members of this Polycomb complex are also required for the subgranular zone (SGZ)-hippocampal adult neurogenesis (82,83) and impact excitatory synaptic plasticity (84,85).
Here, we show that E13 may be a component of the genetic and epigenetic networks controlling neuronal specification. Indeed, induction of the expression of E13 in stable ES cell lines results in the significant up-regulation of markers of glutamatergic excitatory neurons, and a strong down-regulation of the GABAergic neurons and radial glia markers. Our study further suggest that the effects of E13 on neuronal differentiation may be exerted via epigenetic mechanisms; the results we obtained by immune-based affinity purification show that E13 interacts with Eed (EED gene) and Suz12 (suppressor of zeste 12 homologue), which are both components of the PRC-EED-EZH2 Polycomb chromatin modelling complex (48), as well as the retinoblastoma binding protein Rbbp4, a core histone-binding protein (86).
Taken together, the interaction of E13 with these proteins suggest it as member of the epigenetic regulation machinery of the neuronal subtypes and glia commitment (Figure 8). Further studies, including mouse models and chromatin IP, are needed to clarify the specific mechanisms by which E13 exerts its function on neuron specification.
Figure 8.
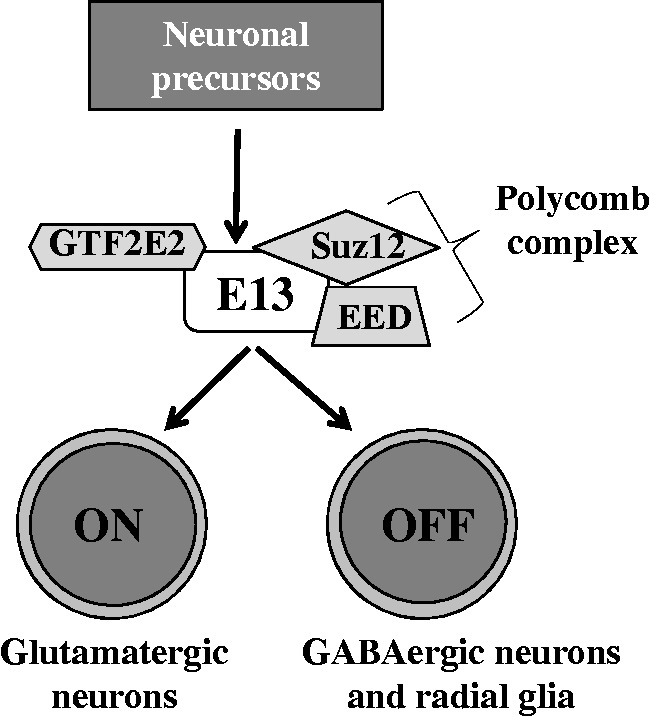
Proposed mechanism of action of E13 in regulating neuronal cell differentiation. E13 may interact with proteins of PRCs to regulate neuronal specification by modulating the transcriptional program of differentiating cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–17, Supplementary Figures 1–12 and Supplementary Files 1–3.
FUNDING
FP7 European Union grant ‘Aneuploidy’ [037627]; Italian Telethon Foundation Grant [TGM11SB1]; Italian Institute of Technology NoBrain Project. Funding for open access charge: Italian Telethon Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nicoletta D'Alessio for technical assistance in the generation of E13 transgenic mouse ES cell lines. They also thank Dr Hitoshi Niwa for providing the recombinant plasmid pPthC-Oct-3/4 and the cell line EBRTcH3 (EB3). They thank Marchesa Bilio for technical assistance in the IHC and ISH. They thank Ms Grit Nebrich and Mr Oliver Klein for their assistance of mass spectrometer measurements. They thank Margherita Mutarelli for the bioinformatics analysis on the E13 sequence.
REFERENCES
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AG. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 4.Suda Y, Suzuki M, Ikawa Y, Aizawa S. Mouse embryonic stem cells exhibit indefinite proliferative potential. J.Cell. Physiol. 1987;133:197–201. doi: 10.1002/jcp.1041330127. [DOI] [PubMed] [Google Scholar]
- 5.Palmqvist L, Glover CH, Hsu L, Lu M, Bossen B, Piret JM, Humphries RK, Helgason CD. Correlation of murine embryonic stem cell gene expression profiles with functional measures of pluripotency. Stem Cells (Dayton, Ohio) 2005;23:663–680. doi: 10.1634/stemcells.2004-0157. [DOI] [PubMed] [Google Scholar]
- 6.Macarthur BD, Ma'ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat. Rev. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Cegli R, Romito A, Iacobacci S, Mao L, Lauria M, Fedele AO, Klose J, Borel C, Descombes P, Antonarakis SE, et al. A mouse embryonic stem cell bank for inducible overexpression of human chromosome 21 genes. Genome Biol. 2010;11:R64. doi: 10.1186/gb-2010-11-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai L, Xu H, Collins JF, Ghishan FK. Molecular and functional analysis of a novel neuronal vesicular glutamate transporter. J. Biol. Chem. 2001;276:36764–36769. doi: 10.1074/jbc.M104578200. [DOI] [PubMed] [Google Scholar]
- 9.Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- 10.Westmoreland JJ, Hancock CR, Condie BG. Neuronal development of embryonic stem cells: a model of GABAergic neuron differentiation. Biochem. Biophys Res Commun. 2001;284:674–680. doi: 10.1006/bbrc.2001.5031. [DOI] [PubMed] [Google Scholar]
- 11.Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 12.Domowicz MS, Sanders TA, Ragsdale CW, Schwartz NB. Aggrecan is expressed by embryonic brain glia and regulates astrocyte development. Dev. Biol. 2008;315:114–124. doi: 10.1016/j.ydbio.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, III, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 16.Kohane IS, Kho AT, Butte AJ. Microarrays for an Integrative Genomics. Cambridge, MA: MIT Press; 2003. [Google Scholar]
- 17.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 18.Masui S, Shimosato D, Toyooka Y, Yagi R, Takahashi K, Niwa H. An efficient system to establish multiple embryonic stem cell lines carrying an inducible expression unit. Nucleic Acids Res. 2005;33:e43. doi: 10.1093/nar/gni043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 20.Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 21.Fisher RA. Statistical Methods for Research Workers. Edinburgh: Oliver & Boyd; 1925. [Google Scholar]
- 22.Fico A, Manganelli G, Simeone M, Guido S, Minchiotti G, Filosa S. High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cells Dev. 2008;17:573–584. doi: 10.1089/scd.2007.0130. [DOI] [PubMed] [Google Scholar]
- 23.Kalaitzis AA, Lawrence ND. A simple approach to ranking differentially expressed gene expression time courses through Gaussian process regression. BMC Bioinformatics. 2011;12:180. doi: 10.1186/1471-2105-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 25.Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiveron MC, Hirsch MR, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. J. Neurosci. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell. Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 29.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol. Syst. Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Kellis M, Collins JJ, Stolovitzky G. Wisdom of crowds for robust gene network inference. Nat. Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregoretti F, Belcastro V, Di Bernardo D, Oliva G. A parallel implementation of the network identification by multiple regression (NIR) algorithm to reverse-engineer regulatory gene networks. PLoS One. 2010;5:e10179. doi: 10.1371/journal.pone.0010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl Acad. Sci. USA. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belcastro V, Siciliano V, Gregoretti F, Mithbaokar P, Dharmalingam G, Berlingieri S, Iorio F, Oliva G, Polishchuck R, Brunetti-Pierri N, et al. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic Acids Res. 2011;39:8677–8688. doi: 10.1093/nar/gkr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka TS, Lopez de Silanes I, Sharova LV, Akutsu H, Yoshikawa T, Amano H, Yamanaka S, Gorospe M, Ko MS. Esg1, expressed exclusively in preimplantation embryos, germline, and embryonic stem cells, is a putative RNA-binding protein with broad RNA targets. Dev. Growth Differ. 2006;48:381–390. doi: 10.1111/j.1440-169X.2006.00875.x. [DOI] [PubMed] [Google Scholar]
- 39.Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng HH, Stanton LW. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol. Biol. Cell. 2007;18:1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin CH, Yang CH, Chen YR. UTF1 deficiency promotes retinoic acid-induced neuronal differentiation in P19 embryonal carcinoma cells. Int. J Biochem. Cell Biol. 2012;44:350–357. doi: 10.1016/j.biocel.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Kooistra SM, Thummer RP, Eggen BJ. Characterization of human UTF1, a chromatin-associated protein with repressor activity expressed in pluripotent cells. Stem Cell Res. 2009;2:211–218. doi: 10.1016/j.scr.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Belcastro V, Siciliano V, Gregoretti F, Mithbaokar P, Dharmalingam G, Berlingieri S, Iorio F, Oliva G, Polishchuck R, Brunetti-Pierri N, et al. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic Acids Res. 2011;39:8677–8688. doi: 10.1093/nar/gkr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 45.Macarthur BD, Ma'ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat. Rev. Mol. Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul FE, Hosp F, Selbach M. Analyzing protein-protein interactions by quantitative mass spectrometry. Methods (San Diego, Calif.) 2011;54:387–395. doi: 10.1016/j.ymeth.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Zheng XM, Moncollin V, Egly JM, Chambon P. A general transcription factor forms a stable complex with RNA polymerase B (II) Cell. 1987;50:361–368. doi: 10.1016/0092-8674(87)90490-9. [DOI] [PubMed] [Google Scholar]
- 48.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Connor MD, Wederell E, Robertson G, Delaney A, Morozova O, Poon SS, Yap D, Fee J, Zhao Y, McDonald H, et al. Retinoblastoma-binding proteins 4 and 9 are important for human pluripotent stem cell maintenance. Exp. Hematol. 2011;39:866–879. doi: 10.1016/j.exphem.2011.05.008. e861. [DOI] [PubMed] [Google Scholar]
- 50.Michalczyk K, Ziman M. Nestin structure and predicted function in cellular cytoskeletal organisation. Histol. Histopathol. 2005;20:665–671. doi: 10.14670/HH-20.665. [DOI] [PubMed] [Google Scholar]
- 51.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 52.Trifonov S, Houtani T, Hamada S, Kase M, Maruyama M, Sugimoto T. In situ hybridization study of the distribution of choline acetyltransferase mRNA and its splice variants in the mouse brain and spinal cord. Neuroscience. 2009;159:344–357. doi: 10.1016/j.neuroscience.2008.12.054. [DOI] [PubMed] [Google Scholar]
- 53.Barberi T, Klivenyi P, Calingasan NY, Lee H, Kawamata H, Loonam K, Perrier AL, Bruses J, Rubio ME, Topf N, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat. Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 54.Gutknecht L, Kriegebaum C, Waider J, Schmitt A, Lesch KP. Spatio-temporal expression of tryptophan hydroxylase isoforms in murine and human brain: convergent data from Tph2 knockout mice. Eur. Neuropsychopharmacol. 2009;19:266–282. doi: 10.1016/j.euroneuro.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev. Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 56.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat. Rev. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 57.Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat. Rev. Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 58.Marin O, Rubenstein JL. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 59.Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat. Neurosci. 2001;4(Suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- 60.Dredge BK, Jensen KB. NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl Acad. Sci. USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development (Cambridge, England) 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 63.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 64.Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J. Neurosci. 2009;29:7256–7270. doi: 10.1523/JNEUROSCI.5653-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bannerman P, Hahn A, Soulika A, Gallo V, Pleasure D. Astrogliosis in EAE spinal cord: derivation from radial glia, and relationships to oligodendroglia. Glia. 2007;55:57–64. doi: 10.1002/glia.20437. [DOI] [PubMed] [Google Scholar]
- 66.Wang S, Scott BW, Wojtowicz JM. Heterogenous properties of dentate granule neurons in the adult rat. J. Neurobiol. 2000;42:248–257. [PubMed] [Google Scholar]
- 67.Cameron HA, McKay RD. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J. Comp. Neurol. 2001;435:406–417. doi: 10.1002/cne.1040. [DOI] [PubMed] [Google Scholar]
- 68.Cameron HA, McKay R. Stem cells and neurogenesis in the adult brain. Curr. Opin. Neurobiol. 1998;8:677–680. doi: 10.1016/s0959-4388(98)80099-8. [DOI] [PubMed] [Google Scholar]
- 69.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J. Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 1965;124:319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 71.Baimbridge KG. Calcium-binding proteins in the dentate gyrus. Epilepsy Res. 1992;7:211–220. [PubMed] [Google Scholar]
- 72.Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 73.Veiga DF, Dutta B, Balazsi G. Network inference and network response identification: moving genome-scale data to the next level of biological discovery. Mol. Biosyst. 2010;6:469–480. doi: 10.1039/b916989j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 75.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howard MJ. Mechanisms and perspectives on differentiation of autonomic neurons. Dev. Biol. 2005;277:271–286. doi: 10.1016/j.ydbio.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 77.Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi H, Liu FC. Genetic patterning of the mammalian telencephalon by morphogenetic molecules and transcription factors. Birth Defects Res. C Embryo Today. 2006;78:256–266. doi: 10.1002/bdrc.20077. [DOI] [PubMed] [Google Scholar]
- 79.Stolp H, Neuhaus A, Sundramoorthi R, Molnar Z. The long and the short of it: gene and environment interactions during early cortical development and consequences for long-term neurological disease. Front. Psychiatry. 2012;3:50. doi: 10.3389/fpsyt.2012.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl Acad. Sci. USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 82.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun J, Ming GL, Song H. Epigenetic regulation of neurogenesis in the adult mammalian brain. Eur. J. Neurosci. 2011;33:1087–1093. doi: 10.1111/j.1460-9568.2011.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Graff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol. Rev. 2011;91:603–649. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- 85.Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J. Biol. Chem. 2007;282:9962–9972. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- 86.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









