Abstract
Heterotrimeric eukaryotic/archaeal translation initiation factor 2 (e/aIF2) binds initiator methionyl-tRNA and plays a key role in the selection of the start codon on messenger RNA. tRNA binding was extensively studied in the archaeal system. The γ subunit is able to bind tRNA, but the α subunit is required to reach high affinity whereas the β subunit has only a minor role. In Saccharomyces cerevisiae however, the available data suggest an opposite scenario with β having the most important contribution to tRNA-binding affinity. In order to overcome difficulties with purification of the yeast eIF2γ subunit, we designed chimeric eIF2 by assembling yeast α and β subunits to archaeal γ subunit. We show that the β subunit of yeast has indeed an important role, with the eukaryote-specific N- and C-terminal domains being necessary to obtain full tRNA-binding affinity. The α subunit apparently has a modest contribution. However, the positive effect of α on tRNA binding can be progressively increased upon shortening the acidic C-terminal extension. These results, together with small angle X-ray scattering experiments, support the idea that in yeast eIF2, the tRNA molecule is bound by the α subunit in a manner similar to that observed in the archaeal aIF2–GDPNP–tRNA complex.
INTRODUCTION
In eukaryotic and archaeal cells, the initiator tRNA carrier is the eukaryotic/archaeal translation initiation factor 2 (e/aIF2) heterotrimer. In its GTP-bound form, this factor specifically binds Met-tRNAiMet and handles it in the translation initiation complex. After start codon recognition, the factor, in its GDP-bound form, loses affinity for Met-tRNAiMet and eventually dissociates from the initiation complex. This leaves Met-tRNAiMet in the P-site of the small ribosomal subunit and allows the final steps of initiation to occur (1,2). In this process, specific binding of the initiator tRNA by e/aIF2 is crucial for accuracy. In both archaea and eukaryotes, the Kd values of the e/aIF2–GDPNP–Met-tRNAiMet complexes are in the nanomolar range (3–8).
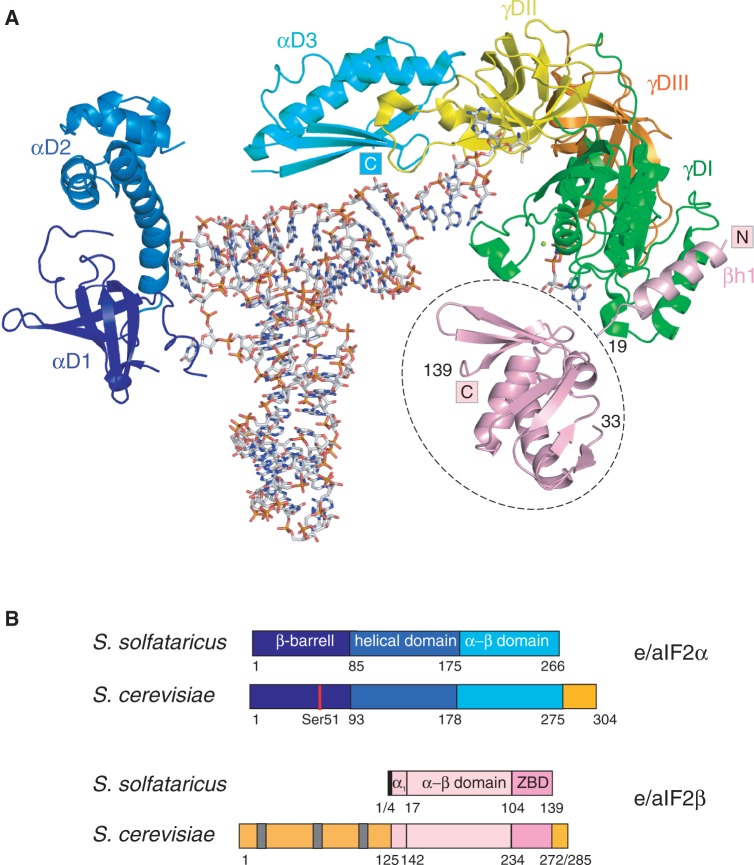
e/aIF2 results from the association of three subunits, α, β and γ. In archaea, the heterotrimer consists of a rigid central part, formed by the γ subunit, the C-terminal domain 3 of the α subunit and the N-terminal helix of the β subunit. Two mobile parts formed by domains 1 and 2 of the α subunit, and by the α–β and the zinc-binding domains of the β subunit are appended to the central core (Figure 1A) (6,9–12).
Figure 1.
Eukaryotic and archaeal e/aIF2. (A) Cartoon representation of Ss-aIF2 in complex with GDPNP and Met-tRNAfMet. The cartoon was drawn from PDB ID 3V11 (15). The color code is as follows: G-domain of γ (γDI, 1–210) in green, domain II (γDII, 211–327) in yellow, domain III (γDIII, 328–415) in orange, domain 1 of α in dark blue (αD1, 1–85), domain 2 of α in blue (αD2, 86–174), domain 3 of α in cyan (αD3, 175–266). The N-terminal α helix of the β subunit (3–19) anchored to γDI is colored in pink. Note that the position of the rest of the β subunit (residues 33–139, encircled with a dotted line) is only a tentative model, derived from SAXS data (15). The figure was drawn with PyMOL (http://www.pymol.org). (B) Schematic structural organizations of e/aIF2 α and β subunits. The colored boxes indicate the structural domains. Specific eukaryotic domains and extensions are colored in orange. Gray bars symbolize the K-boxes in the N-terminal domain of yeast eIF2β.
Until very recently, based on high structural resemblance of the γ subunit with the elongation factor EF1A as well as on site-directed mutagenesis studies, it was believed that the binding mode of the tRNA molecule on aIF2 was similar to that observed with the elongation factor (3,7,9,13,14). However, biochemical results and determination of the 5-Å crystal structure of archaeal Sulfolubus solfataricus IF2 (Ss-aIF2) bound to initiator methionyl-tRNA have broken this model (15,16). In the 3D structure of the aIF2–GDPNP–tRNA complex, the tRNA is bound by the α and γ subunits of aIF2 (Figure 1A). aIF2 approaches tRNA from the acceptor stem minor groove side, whereas EF1A approaches tRNA from the T-stem minor groove side. Despite this, thanks to a kinked conformation, the acceptor end of the tRNA fits in a channel on aIF2γ, which corresponds to the tRNA acceptor end-binding channel on EF1A. This model clearly explains why the isolated γ subunit of archaeal aIF2 is indeed able to bind initiator methionyl-tRNA but with a binding affinity highly reduced when compared with the tRNA-binding affinity for the complete aIF2 heterotrimer. Indeed, the α subunit provides the heterotrimer with almost its full tRNA-binding affinity, whereas the β subunit only slightly contributes to tRNA binding (5–7,15).
In contrast with archaeal aIF2, the construction of a Saccharomyces cerevisiae strain completely lacking eIF2α and allowing purification of an eIF2βγ heterodimer has shown that, in yeast, α would only slightly contribute to tRNA-binding affinity (no more than a factor of 5) (17), whereas β would have an important effect (18). From these data, an ‘eukaryotic behavior’, with a major role for the β subunit in the binding of the tRNA and a minor role for the α subunit would be opposed to an ‘archaeal behavior’ in which α has the major contribution. Hence, the possibility that the structural involvement of the peripheral subunits in the eukaryotic ternary initiation complex differs from that in the archaeal one cannot be completely excluded. Notably, eIF2 from the primitive eukaryote Encephalitozoon cuniculi displays an intermediate behavior with α and β subunits equally contributing to tRNA-binding affinity (19).
Functional dissection of S. cerevisiae eIF2 in vitro was impaired by the inability to produce the isolated γ subunit. However, very recently, we reported that a chimeric protein formed by assembling the γ subunit of S. solfataricus aIF2 with the α subunit of S. cerevisiae eIF2 was useful to study the role of the yeast eIF2α subunit (15). Therefore, chimeric proteins appear as an attractive tool for studying the role of the yeast peripheral subunits of eIF2 in tRNA binding. In the present study, using chimeric e/aIF2, we show that the β subunit of yeast has indeed an important role in tRNA-binding affinity. The N- and C-terminal domains of yeast eIF2β are necessary to obtain full tRNA-binding affinity, the C-terminal extension of β having the most important role. The effect of the β subunit on tRNA-binding affinity may be either direct or indirect. The intact α subunit apparently only slightly contributes to tRNA binding. However, shortening of the acidic C-terminal extension revealed a positive effect of the α subunit on tRNA binding (15). Here, we show that the negative effect of the C-terminal extension is directly related to the size of the acidic C-terminal tail. Moreover, the small-angle X-ray scattering (SAXS) diffusion curve obtained with the Ch-eIF2αΔCγ–GDPNP–met-tRNAfMet complex agrees well with the theoretical curve computed from the crystallographic structure of Ss-aIF2αγ bound to GDPND and met-tRNAfMet. Therefore, altogether, the results are compatible with the idea that, in yeast eIF2, the tRNA molecule is bound in an orientation similar to that observed in the archaeal aIF2–GDPNP–tRNA complex.
MATERIALS AND METHODS
Cloning, expression and production of yeast eIF2 α and β subunits and their variants
The genes encoding the α and β subunits of eIF2 from S. cerevisiae were amplified by PCR from genomic DNA and cloned in pET-derivative vectors. The gene coding for the α subunit was cloned between the NdeI and BamHI restriction sites of pET15b. The resulting plasmid called pET15bY-α led to the expression of an N-terminally tagged version of yeast α subunit in Escherichia coli. The gene coding for the β subunit was cloned between the NdeI and SacII restriction sites of pET3a to give pET3aY-β. This plasmid allows expression of an unmodified version of yeast eIF2β.
Deletions in the target genes were carried using the QuikChange Site-directed Mutagenesis method (Stratagen). The full sequence of all mutated genes was confirmed by resequencing. Deletion of the C-terminal extension of yeast eIF2α was achieved using pET15bY-α to give various truncated forms. pET15bY-αΔC274 produced a protein ending at residue T274 (15). pET15bY-αΔC283 produced a protein ending at residue L283, pET15bY-αΔC292 produced a protein ending at residue S292 and pET15bY-αΔC298 produced a protein ending at residue E298. The DNAs coding for yeast αD3 or yeast αD3ΔC274 were obtained by the QuikChange Site-directed Mutagenesis method with the introduction of a start codon at position 178.
Deletion of the C-terminal domain of yeast eIF2β was achieved using pET3aY-β to give pET3aY-βΔC. The construction allowed expression of a β subunit ending at residue I271. pET15bY-β allowed expression of an N-terminally tagged version of yeast eIF2β. pET15bY-βΔN was a derivative of pET15bY-β producing a β subunit deleted of its N-terminal domain (residues 1–125) His-tagged at its N-terminus. The doubly deleted mutant eIF2βΔNΔC was expressed from pET15bβΔNΔC, a derivative of pET15bβY-ΔN. The resulting protein corresponded to an N-terminally histidine-tagged version of yeast eIF2β comprised of the core domain of eIF2β, from residue E126 to I271.
Each subunit was overexpressed separately in E. coli BL21 Rosetta pLacI-Rare (Merck, Novagen). One-liter cultures were in 2xTY containing 50 µg/ml of ampicillin and 34 µg/ml of chloramphenicol. Expression was induced after an overnight culture at 37°C (OD650 ∼ 2.5) by adding 1 mM of IPTG. After induction, the cultures were continued for 5–6 h at 18°C.
Purification of chimeric eIF2
Overexpression of Ss-aIF2γ subunit was performed as described (6). Cultures of cells each overproducing one of the three subunits (250 ml for yeast α, 500 ml for yeast β and 500 ml for S. solfataricus aIF2γ) were harvested, mixed in 80 ml of buffer A (10 mM HEPES pH 7.5, 500 mM NaCl, 3 mM 2-mercaptoethanol, 0.1 mM PMSF and 0.1 mM benzamidine) and disrupted by sonication. After centrifugation, the supernatant was loaded onto a column (3 ml) containing Talon affinity resin (Clontech) equilibrated in the same buffer. The resin was first washed with 50 ml of buffer A and then with 50 ml of buffer A supplemented with 10 mM imidazole. The fractions containing the heterotrimer were finally recovered after elution with buffer A containing 125 mM imidazole. To remove the excess of the yeast α subunit, the affinity column eluate was diluted to 300 mM NaCl and then loaded onto a 5 ml S-Hiload column (10 mm × 5 cm; GE Healthcare) equilibrated in buffer B (10 mM HEPES pH 7.5, 300 mM NaCl, 10 mM 2-mercaptoethanol, 0.1 mM PMSF and 0.1 mM benzamidine). A gradient from 300 mM NaCl to 600 mM NaCl was used for elution (128 ml at a flow rate of 2 ml/min). After sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis, the fractions containing the heterotrimer were pooled and dialyzed against buffer B. Finally, the protein was concentrated by using Centricon 30 concentrators. Either GDPNP-Mg2+ (1 mM) or GDP-Mg2+ (1 mM) was added before storage at 4°C of the recovered heterotrimer (Figure 2, lane 1). The same procedure was applied to purify the Ch-eIF2αΔCβγ, Ch-eIF2βΔNγ and Ch-eIF2βΔNΔCγ (Figure 2, lanes 4, 6 and 5).
Figure 2.
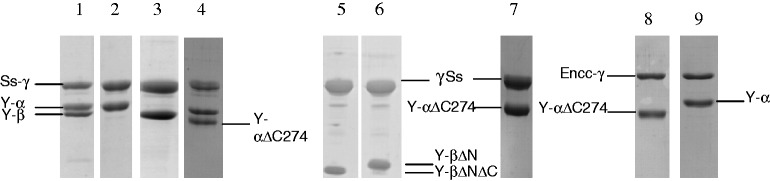
SDS–PAGE analysis of purified Ch-eIF2 variants. The 12.5% SDS–PAGE were stained with Coomassie Blue. Lane 1, Ch-eIF2 heterotrimer. Lane 2, Ch-eIF2αγ heterodimer. Lane 3, Ch-eIF2βγ heterodimer. Lane 4, Ch-eIF2(αΔC274)βγ. Positions of tagged yeast eIF2α (Y-α, 36·9 kDa), tagged yeast eIF2αΔC274 (Y-αΔC, 33·4 kDa), yeast eIF2β (Y-β, 31·4 kDa) and Ss-aIF2γ (Ss-γ, 45·6 kDa) are indicated. Lane 5, Ch-eIF2βΔNΔCγ heterodimer. Lane 6, Ch-eIF2βΔNγ heterodimer. Lane 7, Ch-eIF2αΔC274γ heterodimer. Positions of yeast eIF2βΔNΔC (Y-βΔNΔC), eIF2βΔN (Y-βΔN) and eIF2αΔC274 (Y-αΔC) are indicated. Lane 8, ChEncc-eIF2(αΔC274)γ heterodimer, Lane 9, ChEncc-eIF2αγ heterodimer. Positions of tagged yeast eIF2αΔC274 (Y-αΔC, 33·4 kDa), tagged yeast eIF2α (Y-α, 36·9 kDa) and E. cuniculi eIF2γ (Encc−γ, 48·9 kDa) are indicated.
Purification of Ch-eIF2βγ and Ch-eIF2βΔCγ
Cultures of cells each overproducing one of the two subunits (500 ml for yeast β or yeast βΔC and 500 ml for S. solfataricus aIF2γ) were harvested, mixed in 40 ml of buffer B (10 mM HEPES pH 7.5, 300 mM NaCl, 10 mM 2-mercaptoethanol, 0.1 mM PMSF and 0.1 mM benzamidine) and disrupted by sonication. After centrifugation, the supernatant was loaded onto a 15 ml S-Hiload column (16 mm × 20 cm; GE Healthcare) equilibrated in buffer B. A gradient from 300 mM NaCl to 600 mM NaCl was used for elution (120 ml at a flow rate of 2 ml/min). After SDS–PAGE analysis, the fractions containing the heterodimer were pooled and dialyzed against buffer B. To remove remaining nucleic acids, the recovered protein was then loaded onto a 15 ml Q-Hiload column (16 mm × 20 cm; GE Healthcare) equilibrated in buffer B. The heterodimer was recovered in the flow-through fraction and then concentrated by using Centricon 30 concentrators. Either GDPNP-Mg2+ (1 mM) or GDP-Mg2+ (1 mM) was added before storage at 4°C of the recovered heterodimer (see Figure 2, lane 3 for purified Ch-eIF2βγ).
Purification of Ch-eIF2αγ heterodimers and of its variants
The procedure used for the purification of Ch-eIF2αγ heterodimer was described previously (15) (Figure 2, lane 2). To purify the Ch-eIF2αΔCγ heterodimer variants (αΔC274, αΔC283, αΔC292 and αΔC298), the S-Hiload step was replaced by a molecular sieve using a Superdex 200 HR 10/30 column (GE Healthcare). The purified proteins (Figure 2, lane 7) were stored in buffer B in the presence of either 1 mM GDPNP-Mg2+ or 1 mM GDP-Mg2+.
The procedure used to purify Ch-eIF2αD3γ and Ch-eIF2αD3ΔCγ heterodimers was the same as that used for Ch-eIF2αΔCγ heterodimer variants, except that after the Talon column, size-exclusion chromatography on Superdex 75 HR 10/30 (GE Healthcare) was used to polish the preparation.
Ch-eIF2αΔCγ and Ch-eIF2αD3ΔCγ protein used to perform SAXS studies were purified as follows. αΔC and αD3ΔC were first purified using Talon affinity resin. The His-tag extensions of the recovered proteins were then removed during overnight dialysis in a buffer containing 10 mM HEPES pH 7.5, 200 mM NaCl, 3 mM 2-mercaptoethanol, 10 mM CaCl2 and Thrombin (0.25 U/mg of substrate protein) at 4°C. The γ subunit of S. solfataricus was purified as described (6). αΔC and/or αD3ΔC was assembled with Ss-aIF2γ and the heterodimers were finally purified using a last step of molecular sieving using a Superdex 200 HR 10/30 column (GE Healthcare) in buffer C (10 mM HEPES pH 7.5, 200 mM NaCl and 3 mM 2-mercaptoethanol).
Purification of Ch-Encc-eIF2αγ and of Ch-Encc-eIF2αΔCγ
BL21 Rosetta pLacI-Rare containing the pET28b+γtc-Encc plasmid was used to overexpress the γ subunit from E. cuniculi (19). The resulting protein carried a 6-histidine tag at its C-terminus. One-liter cultures were in 2xTY containing 25 µg/ml of kanamycin and 34 µg/ml of chloramphenicol. Expression was induced by adding 1 mM of IPTG when OD650 reached 0.8. After induction, the cultures were continued for 8–12 h at 18°C.
Cultures of cells each overproducing one of the two subunits (250 ml for yeast α, 500 ml for E. cuniculi eIF2γ) were harvested, mixed in 40 ml of buffer A and disrupted by sonication. After centrifugation, the supernatant was loaded onto a column (3 ml) containing Talon affinity resin (Clontech) equilibrated in the same buffer. The resin was first washed with 50 ml of buffer A and then with 50 ml of buffer A supplemented with 10 mM imidazole. The fractions containing the heterodimer were finally recovered after elution with buffer A containing 125 mM imidazole. To remove the excess of the yeast α subunit, the affinity column eluate was diluted to 250 mM NaCl and then loaded onto a 5 ml Q-Sepharose HP column (10 mm × 5 cm; GE Healthcare) equilibrated in buffer C (10 mM HEPES pH 7.5, 250 mM NaCl, 10 mM 2-mercaptoethanol, 0.1 mM PMSF and 0.1 mM benzamidine). The heterodimer flowed through the column whereas the excess yeast α subunit was retained. After SDS–PAGE analysis, the fractions containing the heterodimer were pooled and dialyzed against buffer B. Finally, the protein was concentrated by using Centricon 30 concentrators. Either GDPNP-Mg2+ (1 mM) or GDP-Mg2+ (1 mM) was added before storage at 4°C of the recovered heterodimers.
The same procedure was used to purify ChEncc-eIF2αΔCγ except that after the Talon column, the protein was loaded onto a Superdex 75 column (HR10/30, GE Healthcare) equilibrated in buffer B. The eluted heterodimer was loaded onto a 5 ml S-Sepharose HP column (10 mm × 5 cm, GE Healthcare) equilibrated in buffer D (10 mM HEPES pH 7.5, 200 mM NaCl, 10 mM 2-mercaptoethanol, 0.1 mM PMSF and 0.1 mM benzamidine). A gradient from 200 to 800 mM NaCl was used for elution (80 ml at a flow rate of 2 ml/min). The fractions containing the heterodimer were pooled and dialyzed against buffer B. Finally, the protein was concentrated by using Centricon 30 concentrators. Either GDPNP-Mg2+ (1 mM) or GDP-Mg2+ (1 mM) was added before storage at 4°C of the recovered heterodimer (Figure 2, lanes 8 and 9).
Protection assay
The protocol used was derived from (20). tRNAfMet was produced in E. coli from synthetic genes and purified as described (21,22). Endogenous tRNAiMet purified from S. cerevisiae was a generous gift of Dr Gérard Keith (Institut de Biologie Moléculaire et Cellulaire, Strasbourg, France). Full aminoacylation with [35S]-methionine (∼10 000 dpm/pmol; Perkin Elmer) was achieved using homogeneous E. coli M547 MetRS (23). tRNAfMetUAC, a derivative of tRNAfMet carrying a UAC (Val) anticodon was produced as described (21,22). Valylation with [14C]Val (563 dpm/pmol) was performed with homogeneous E. coli valyl-tRNA synthetase as described (7). Aminoacyl-tRNAs were precipitated with ethanol in the presence of 0.3 M NaAc pH 5.5 and stored at −20°C in 100% EtOH in small aliquots. Before use, aminoacylated tRNAs were redissolved in water and full aminoacylation was systematically controlled through measurement of radioactivity after precipitation in 5% trichloroacetic acid (TCA).
Protection by chimeric eIF2 (Ch-eIF2) variants of methionyl-tRNAfMet against spontaneous hydrolysis was assayed as follows. Reaction mixtures (150 µl) contained 20 mM HEPES–NaOH pH 8.0, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 0.2 mg/ml BSA (bovine serum albumin; ROCHE), 5% glycerol, 0.1% triton X-100, 1 mM GDPNP and 2 nM of E. coli methionyl-tRNAfMet. The concentrations of the proteins were determined from A280 measurements using extinction coefficients computed from the amino acid sequences. Concentrations of Ch-eIF2 variants were varied from 1 nM to 30 µM, using a range depending on the Kd value to be measured. The mixtures were incubated at 30°C. To determine the rate constants of deacylation, 20 µl aliquots were withdrawn at various times (usually, six aliquots from 5 to 60 min) and precipitated in 5% TCA in the presence of 80 µg of yeast RNA as carrier. In all cases, the deacylation curve as a function of time could be fitted with a single exponential. For each Kd measurement, a set of 8–10 experiments corresponding to 8–10 different protein concentrations was performed. The rate constants measured at variable protein concentrations were then fitted to simple binding curves (20) from which the dissociation constant of the studied protein–tRNA complexes and their associated standard errors could be deduced using the MC-Fit program (24). Each experiment was independently repeated at least twice, without significant variation of the results.
Small-angle X-ray scattering
SAXS experiments were conducted on the SWING beamline at the SOLEIL synchrotron as described (15). For these experiments, the His-tags of eIF2α3ΔC and of eIF2αΔC were removed before assembly with Ss-aIF2γ, as described above. A concentrated sample of Ch-eIF2(α3ΔC)γ (∼4 nmol in 40 μl) was injected onto a size-exclusion column (agilent Bio column SEC3; 300 Å) using an Agilent HPLC system and eluted directly into the SAXS flow-through capillary cell at a flow rate of 0.4 ml/min.
To collect data on Ch-eIF2(αΔC)γ–GDPNP–Met-tRNAfMet complex, the protein was mixed with a 1.15-fold molar excess of Met-tRNAfMet and injected as a concentrated sample (∼20 nmol in 250 µl) onto a size-exclusion column (Superdex 200 HR10/30, GE Healthcare), using an Agilent HPLC system and eluted directly into the SAXS flow-through capillary cell at a flow rate of 0.4 ml/min. The same procedure was used to collect data on Ss-aIF2–GDPNP–Met-tRNAfMet, Ss-aIF2–GDPNP–Met-tRNAfMet(A1–U72) and Ss-aIF2–GDPNP–Ss-Met-tRNAiMet complexes. tRNAfMet(A1–U72) and Ss-tRNAiMet were purified and aminoacylated as described (7,25).
For all experiments, the elution buffer consisted of 10 mM MOPS (pH 6.7), 200 mM NaCl and 5 mM MgCl2. SAXS data were collected continuously, with a frame duration of 1.5 s and a dead time between frames of 1.0 s. Selected frames corresponding to the main elution peak were averaged (26). A large number of frames were collected during the first minutes of the elution, and these were averaged to account for buffer scattering, which was subsequently subtracted from the signal during elution of the protein or protein–tRNA complex. Data reduction to absolute units, frame averaging and subtraction were done using FOXTROT (26). All subsequent data processing, analysis and modeling steps were carried out with PRIMUS and other programs of the ATSAS suite (27). Scattered intensity curves were calculated from the atomic coordinates of the crystallographic structure, using CRYSOL with 50 harmonics (28). This program was also used to fit the calculated curve to the experimental one, by adjusting the excluded volume, the averaged atomic radius and the contrast of the hydration layer surrounding the particle in solution.
RESULTS
Production and assembly of the Ch-eIF2 heterotrimer
The chimeric heterotrimer (Ch-eIF2) was formed by assembling the archaeal γ subunit of S. solfataricus aIF2 with the α and β subunits of S. cerevisiae eIF2. To facilitate purification of Ch-eIF2, we used an N-terminally tagged version of yeast eIF2α and native versions of yeast eIF2β and Ss-aIF2γ. The three subunits were produced independently in E. coli. The first step of purification of the assembled Ch-eIF2 heterotrimer was a metal affinity chromatography. This step showed that the archaeal γ subunit was able to interact with both yeast α and β subunits. To remove the excess of the yeast α subunit, we then used anion exchange chromatography. This purification step allowed to recover a correct stoechiometry between the three subunits, as judged by SDS–PAGE analysis (Figure 2, lane 1).
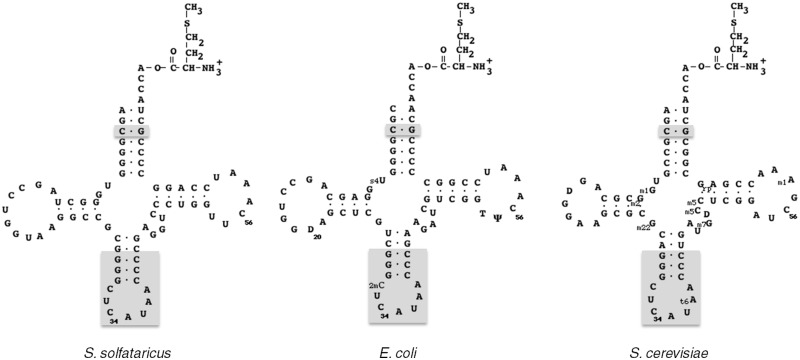
Binding of tRNAiMet onto e/aIF2 leads to protection of aminoacylated tRNA against spontaneous deacylation. Thus, dissociation constants of Met-tRNAiMet from an e/aIF2–GDPNP–Met-tRNAiMet complex can be estimated by following deacylation rates in the presence of various eIF2 concentrations at a fixed tRNA concentration (7,20). The sequence of the E. coli initiator tRNAfMet has a high homology with the one from the archaeal initiator tRNA (Figure 3). In particular, the acceptor stems of both tRNAs are identical, with the exception that the A1–U72 base pair in the archaeal tRNA is replaced by C1–A72 in the bacterial one. Moreover, it was previously shown that E. coli Met-tRNAfMet was as good a ligand of archaeal aIF2 as the cognate initiator Ss-tRNAiMet produced in E. coli (15). To further validate at a structural level, the use of E. coli initiator Met-tRNAfMet as a model ligand, we compared the SAXS curve of Ss-aIF2–GDPNP–Met-tRNAfMet with the corresponding ones with either Met-tRNAfMetA1–U72 variant or Met-tRNAiMet from S. solfataricus. The three SAXS curves were nicely superimposable (Supplementary Figure S1), showing that the three complexes were highly similar.
Figure 3.
Cloverleaf representations of initiator Met-tRNAs from S. solfataricus, E. coli and S. cerevisiae cytoplasm. Nucleotides denoted with gray boxes are universally conserved in initiator tRNAs. Post-transcriptional modifications are indicated in the cases of E. coli and S. cerevisiae.
Escherichia coli Met-tRNAfMet initiator tRNA was used as a model ligand to study tRNA binding by Ch-eIF2. Ch-eIF2 bound efficiently initiator methionyl-tRNA in the presence of GDPNP-Mg2+ with a Kd value of 87 ± 10 nM (Table 1, row 1). Moreover, in the presence of GDP instead of GDPNP, the Kd value was increased to 700 ± 300 nM. Therefore, GDP lowered the binding affinity of the initiator methionyl-tRNA by one order of magnitude, in keeping with the nucleotide effects observed with yeast eIF2 [factor of 20 (4)] or Ss-aIF2 [factor of 100, (6)]. Furthermore, we verified that Ch-eIF2 could still interact with authentic yeast Met-tRNAiMet (Kd = 27 ± 5 µM; Table 1, row 1; Figure 3). The binding affinity of both initiator tRNAs for the chimeric factor in the presence of GDPNP were reasonably high, when compared with the affinity for eIF2 from yeast [Kd = 9 nM, (4)] or for Ss-aIF2 [Kd = 1.5 nM, (6)]. The observed discrepancies between the Kd values measured with yeast eIF2, Ss-aIF2 or Ch-eIF2 could be in part due to differences in the assay conditions. Indeed, in the case of yeast eIF2 the temperature used in the assay was 26°C and in the case of Ss-aIF2, the temperature used in the assay was 51°C. Here, to avoid thermal effects on yeast α and β proteins, we performed the measurements at 30°C, a temperature sub-optimal for the archaeal γ subunit of Ch-eIF2 (5). Finally, it is known that archaeal and eukaryotic eIF2 strongly recognize the methionine moiety on initiator tRNAs (4,7,29,30). Therefore, as an additional control to validate the use of Ch-eIF2 as a tool, we verified that the chimeric heterotrimer had the same property. We used Val-tRNAfMet UAC in a standard protection assay. Instead of protection against deacylation, we observed a surprising 2-fold increase of the rate of deacylation in the presence of 7.4 µM Ch-eIF2. Possibly, low-affinity unspecific binding of the Val-tRNAfMet UAC on the factor may favor hydrolysis of the esterified amino acid. Nevertheless, this experiment allowed to conclude that the presence of the valyl group strongly affected specific aminoacyl-tRNA binding.
Table 1.
tRNA binding by Ch-eIF2
| Y-α | Y-β | Ss-γ | Kd (nM) | Kd (nM) | Kd (nM) | ||
|---|---|---|---|---|---|---|---|
| Met-tRNAfMet (GDPNP)a | Met-tRNAfMet (GDP)a | Met-tRNAiMet (GDPNP)b | |||||
| 1 | Ch-eIF2 | wt | wt | wt | 87 ± 10 | 700 ± 300 | 27 ± 5 |
| 2 | Ss-aIF2γ | – | – | wt | 55 000 ± 12 000 | n.d. | 27 000 ± 10 000 |
| 3 | Ch-eIF2βγ | wt | wt | 55 ± 20 | 180 ± 50 | 34 ± 6 | |
| 4 | Ch-eIF2(βΔN)γ | – | βΔN | wt | 231 ± 30 | 557 ± 125 | n.d. |
| 5 | Ch-eIF2(βΔC)γ | – | βΔC | wt | 532 ± 140 | 1670 ± 960 | n.d. |
| 6 | Ch-eIF2(βΔNΔC)γ | – | βΔNΔC | wt | 3500 ± 1400 | >9500 | n.d. |
| 7 | Ch-eIF2αγ | wt | – | wt | 9200 ± 2000 | >120 000 | 4200 ± 2200 |
| 8 | Ch-eIF2(αΔC274)γ | αΔC | – | wt | 73 ± 10 | 740 ± 70 | 36 ± 8 |
| 9 | Ch-eIF2(αΔC274)βγ | αΔC | wt | wt | 3 ± 1 | 26 ± 4 | n.d. |
| 10 | Ch-eIF2(αΔC283)γ | αΔC | – | wt | 950 ± 180 | n.d. | n.d. |
| 11 | Ch-eIF2(αΔC292)γ | αΔC | – | wt | 2600 ± 600 | n.d. | n.d. |
| 12 | Ch-eIF2(αΔC298)γ | αΔC | – | wt | 4200 ± 200 | n.d. | n.d. |
| 13 | Ch-eIF2(α3)γ | α3 | – | wt | >30 000 | n.d. | n.d. |
| 14 | Ch-eIF2(α3ΔC274)γ | α3ΔC | – | wt | 2900 ± 500 | >44 000 | n.d. |
Dissociation constants of Met-tRNAfMet from its complexes with the indicated versions of Ch-eIF2 were determined from protection experiments as described in ‘Materials and Methods’ section.
aMeasured with E. coli Met-tRNAfMet as a ligand.
bMeasured with S. cerevisiae Met-tRNAiMet as a ligand. n.d., not determined.
The yeast β subunit strongly influences tRNA binding on Ch-eIF2
At 30°C, only weak, though significant, GDPNP-dependent binding of initiator tRNA to Ss-aIF2γ occurs (Table 1, row 2). Therefore, the large difference between the Kd value measured with the Ss-γ subunit alone and that measured for the full Ch-eIF2 trimer showed that the α and/or the β subunits participate in tRNA binding (Table 1, rows 1 and 2). In order to examine the contribution of each peripheral subunit, Ch-eIF2αγ and Ch-eIF2βγ heterodimers were produced and purified (Figure 2, lanes 2 and 3). Using Ch-eIF2βγ, the Kd value of Met-tRNAfMet was 55 ± 10 nM (Table 1, row 3) whereas using Ch-eIF2αγ, the Kd value of Met-tRNAfMet was 9200 ± 2000 nM (Table 1, row 7). Hence, the β subunit of yeast eIF2 improved the binding affinity of Met-tRNAfMet by three orders of magnitude (Table 1, rows 2 and 3, Supplementary Figure S2A). The yeast α subunit also participated in the binding affinity of Met-tRNAfMet. Its contribution was however two orders of magnitude less important than that of the β subunit (Table 1, rows 2 and 7, Supplementary Figure S2A). Notably, in the absence of the α subunit, the affinity of Ch-eIF2βγ for Met-tRNAfMet was similar to that measured with the complete chimeric heterotrimer (compare rows 1 and 3, Table 1 and Supplementary Figure S2A). The same effects of the α and β subunits were observed using authentic yeast Met-tRNAiMet as a substrate (Table 1, rows 2, 3 and 7). This indicated that the association of the β subunit to γ was enough to retrieve almost the same binding affinity for tRNA as full Ch-eIF2. Ch-eIF2βγ remained specific of the methionine moiety of the tRNA ligand. Indeed, the presence of 10 µM Ch-eIF2βγ only decreased the deacylation rate of Val-tRNAfMet UAC by 20%, consistent with a dissociation constant of the order of several tens of micromolar. Notably, the behavior of the yeast peripheral subunits was opposite to that observed with archaeal α and β subunits (Supplementary Figure S2A).
N- and C-terminal extensions of yeast eIF2β are necessary for Met-tRNAfMet binding
The yeast β subunit is made of a conserved structural core corresponding to the archaeal version of the protein to which are appended eukaryote-specific domains, namely, an N-terminal domain and a short C-terminal extension (Figure 1B). To evaluate the roles of the yeast-eIF2β extensions in tRNA binding, three truncated versions of the subunit were produced. In eIF2βΔN, the first 125 residues were removed. In eIF2βΔC, residues 272–285 were removed. The two deletions were also performed simultaneously giving eIF2βΔNΔC.
The three heterodimers Ch-eIF2βΔNγ, Ch-eIF2βΔCγ and Ch-eIF2βΔNΔCγ were purified and Met-tRNAfMet-binding affinities were measured. As shown in Table 1, the N-terminal domain contributed by a factor of 4.2 to tRNA-binding affinity (rows 3 and 4) and the C-terminal extension of eIF2β contributed by a factor of 9.7 (rows 3 and 5). These results showed that both eukaryotic extensions participate in the binding of the tRNA molecule with the contribution of the short basic C-terminal extension being twice as much as that of the N-terminal domain. Moreover, with the double truncation (Ch-eIF2βΔNΔCγ), the dissociation constant fell down by a factor of 63 when compared with unmodified Ch-eIF2βγ (rows 3 and 6). Therefore, the positive effects of both extensions on the binding of the initiator tRNA were cumulative (Supplementary Figure S2B). Finally, using Ch-eIF2βΔNΔCγ, a Kd-value for Met- tRNAfMet binding of 3500 ± 1400 nM was measured. The affinity of the tRNA remained 15-fold higher than that with Ss-aIF2γ alone. Hence, the conserved core of yeast eIF2β still contains some features allowing it to participate in tRNA-binding affinity (Supplementary Figure S2B).
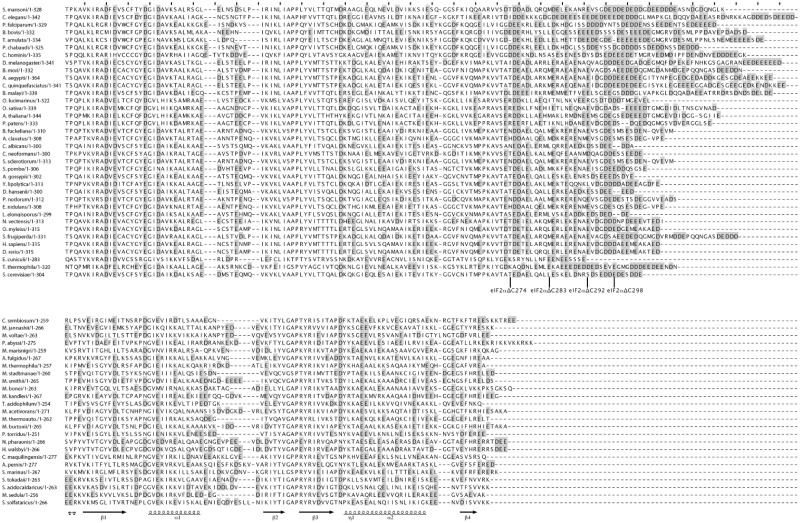
The acidic C-terminal extension of yeast eIF2α interferes with tRNA binding
The yeast α subunit of eIF2 comprises a conserved structural core corresponding to the archaeal version of the protein, with an additional highly acidic (pI = 3.14) C-terminal tail, containing 16 Asp or Glu out of 30 residues. This acidic C-terminal extension is characteristic of eukaryotic eIF2α (Figures 1B and 4). Previously, the C-terminal region corresponding to residues 275–304 was removed through site-directed mutagenesis of the yeast eIF2α gene (15). With the heterodimer Ch-eIF2αΔC274γ, a Kd-value for the initiator methionyl-tRNA of 73 ± 10 nM was measured (15). Therefore, removal of the C-terminal tail increased the binding affinity by more than two orders of magnitude (rows 7 and 8 in Table 1 and Supplementary Figure S2C). The same effect was observed using authentic yeast Met-tRNAiMet as a substrate (Kd = 36 ± 8 nM; Table 1). Hence, the C-terminal extension of the yeast α subunit is strongly unfavorable to the binding of the tRNA on Ch-eIF2αγ. Again, we verified that Ch-eIF2αΔC274γ remained specific of the methionine moiety of the tRNA ligand. Indeed, the presence of 10 µM Ch-eIF2αΔC274γ only decreased the deacylation rate of Val-tRNAfMet UAC by 37%, consistent with a dissociation constant in the 10 µM range.
In order to delineate the region responsible for the negative effect on tRNA binding, three deletions of various lengths in the C-terminal extension were performed. The three shortened proteins were named αΔC283, αΔC292 and αΔC298 (Figure 4). The corresponding chimeric heterodimers Ch-eIF2αΔCγ were purified and their tRNA-binding affinities were measured. As shown in Table 1 (rows 8 and 10–12), the tRNA-binding affinity increased when the size of the acidic C-terminal extension decreased, the highest tRNA-binding affinity being obtained upon deletion of the complete acidic C-terminal extension (Table 1 and Supplementary Figure S2C). In the context of the heterotrimer, complete removal of the acidic tail of α (αΔC274) increased the affinity by a factor of 30 (rows 1 and 9) leading to a heterotrimer even more efficient in tRNA binding than Ch-eIF2. These results strongly suggested that the apparent weak effect of yeast eIF2α on tRNA-binding results from a positive effect of the core part of the subunit, compensated by a negative effect of the acidic residues in its appended C-terminal tail.
Figure 4.
Alignment of e/aIF2α domain 3 sequences. Representative eukaryotic (upper block) and archaeal (lower block) sequences are shown. Aspartate and glutamate residues are boxed in gray. Secondary structures are drawn below the Ss-aIF2α sequence. The positions of the last residue in C-terminal truncated versions of eIF2α are indicated.
To confirm the effect of the C-terminal tail of yeast eIF2α, we examined it using another eIF2γ subunit. We therefore produced Ch-eIF2 using the central eukaryotic eIF2γ subunit from E. cuniculi (Encc-eIF2). Two chimeric proteins were produced and purified (Figure 2, lanes 8 and 9). ChEncc-eIF2αγ corresponds to the α subunit of S. cerevisiae bound to the γ subunit of E. cuniculi and ChEncc-eIF2αΔ274Cγ corresponds to the α subunit of S. cerevisiae truncated of its C-terminal tail bound to the γ subunit of E. cuniculi. Dissociation constants of Met-tRNAfMet from these proteins were determined using the same procedure as that used for yeast Ch-eIF2. Encc-eIF2γ bound Met-tRNA with a Kd value of 1700 ± 300 nM (19). Association of yeast eIF2α to Encc-eIF2γ only increased the affinity of Met-tRNA by a factor of 5.6 (Table 2, rows 1 and 2) whereas the increase of affinity was by a factor of 105 when eIF2αΔC was used in place of eIF2α (Table 2, rows 1 and 3).
Table 2.
Effects of the C-terminal acidic tail of yeast α subunit on tRNA binding in the context of Encephalitozoon cuniculi γ subunit
| Kd (nM) | Kd (nM) | ||||
|---|---|---|---|---|---|
| Y-α | Encc-γ | Met-tRNAfMet (GDPNP)a | Met-tRNAfMet (GDP)a | ||
| 1 | Encc-eIF2γb | – | wt | 1700 ± 300 | >24 000 |
| 2 | ChEncc-eIF2αγ | Wt | wt | 300 ± 50 | >16 000 |
| 3 | ChEncc-eIF2(αΔC)γ | αΔC | wt | 16 ± 1 | 3380 ± 550 |
Dissociation constants were determined from protection experiments as described in ‘Materials and Methods’ section.
aMeasured with E. coli Met-tRNAfMet as a ligand.
bData from (19).
Domains 1 and 2 of eIF2α contribute to tRNA-binding affinity
In the archaeal system, the αD3 domain is sufficient to confer on aIF2γ its full tRNA-binding affinity (6,7). Nevertheless, the 3D structure of aIF2 bound to the initiator tRNA revealed contacts of the αD12 domains with the tRNA molecule (15). To explain the apparent discrepancy between biochemical data and the observed contacts of αD12 with the tRNA in the structure, it was proposed that the gain in enthalpy resulting from the contacts of the αD12 domains with the tRNA just compensates for the entropic cost of the immobilization of αD12. We anticipated that these contributions may quantitatively vary depending on the α subunit studied.
We produced the αD3 domain of yeast eIF2α. Then, tRNA-binding affinity of Ch-eIF2(αD3)γ heterodimer was measured. Interestingly, the tRNA-binding affinity for Ch-eIF2(αD3)γ heterodimer was markedly lowered compared with that measured with the entire yeast α subunit bound to archaeal γ. Only slight protection of Met-tRNA was observed at a concentration of Ch-eIF2(αD3)γ heterodimer of 30 µM (compare rows 7 and 13 in Table 1). This result strongly suggested that domains 1 and 2 of yeast eIF2α contribute to tRNA-binding affinity. Consistently, although deletion of the C-terminal acidic tail from αD3 increased the binding affinity of the tRNA for the chimeric heterodimer by a factor of at least 10 (compare rows 13 and 14 in Table 1), the affinity remained much lower than that measured in the presence of the αD12 domains, with the Ch-eIF2αΔC274γ heterodimer (compare rows 14 and 8 in Table 1).
Structural studies in solution using SAXS
The region of contact between e/aIF2α and e/aIF2γ mainly involves two loops of the C-terminal domain of α (αD3) as well as an elongated loop and a β-strand of γ [Figure 1A (6)]. Moreover, the structure of the human eIF2α C-terminal domain is highly similar to the structure of archaeal aIF2α (6,31,32). Therefore, besides the C-terminal extension, the structure of αD3 is conserved between eukaryotes and archaea.
In a first step, we checked the structural similarity between the Ch-eIF2(α3ΔC)γ protein and the archaeal Ss-aIF2αD3γ protein. This was achieved by measuring the X-ray scattering curve of Ch-eIF2(α3ΔC)γ. This experimental curve was then compared with the theoretical X-ray scattering curve computed from the crystallographic structure of the archaeal αD3γ protein (from PDB ID 2AHO). The two curves showed a very good agreement (Supplementary Figure S3A, χ = 1.9). This result strongly argued in favor of a binding mode of yeast eIF2α3ΔC protein onto Ss-aIF2γ similar to that observed for the archaeal aIF2αD3 domain (6).
Finally, in order to evaluate the binding mode of the tRNA molecule by the yeast eIF2αΔC subunit, the scattering curve of Ch-eIF2αΔCγ bound to Met-tRNAfMet was measured after purification of the complex (see ‘Materials and Methods’ section). The SAXS diffusion curve obtained with the Ch-eIF2αΔCγ–GDPNP–Met-tRNAfMet complex agreed well with the theoretical curve computed from the crystallographic structure of Ss-aIF2αγ bound to Met-tRNAfMet (from PDB ID 3V11, Supplementary Figure S3B, χ = 2.8). This strongly argued in favor of a binding mode of the tRNA by yeast eIF2α similar to the binding mode observed for the archaeal Ss-aIF2α subunit.
DISCUSSION
In this study, we have constructed Ch-eIF2 formed by assembling yeast eIF2α and β subunits to archaeal Ss-aIF2γ core subunit. In such chimeric factors, the peripheral α and β subunits are likely to adopt a binding mode to the central γ subunit identical to that observed for archaeal aIF2 [Figure 1A (6,10–12)]. Indeed, the binding of the β subunit to the γ one involves an α-helix of e/aIF2β wedged between two α-helices of the G-domain of γ (10–12). The rest of the β subunit is mobile with respect to γ (10–12). Since the residues of the anchoring helix contacting the γ subunit are well conserved in all e/aIF2β species, it is most likely that yeast eIF2β is caught by Ss-aIF2γ in a similar manner. This idea is consistent with site-directed mutagenesis studies in the yeast system (33). The binding of yeast eIF2α to the archaeal γ subunit was probed by SAXS (Supplementary Figure S3A). The results clearly argued in favor of a binding mode of the yeast eIF2α subunit to the archaeal γ identical to that observed for the archaeal α subunit.
Consistently, the chimeric protein, Ch-eIF2, is indeed able to bind yeast initiator methionyl-tRNA with a Kd value (27 nM) similar to those measured with authentic versions of eIF2 (4,6,7,19). Moreover, we verified that the chimeric factor remained specific for the methionine moiety of its aminoacyl-tRNA ligand. These results validated the use of such chimeric proteins to study the influence of the α and β peripheral subunits on the tRNA-binding affinity. The role of each peripheral subunit in tRNA binding was studied in the context of heterodimers (Ch-eIF2αγ and Ch-eIF2βγ). We observe that yeast eIF2β is essential to retrieve full tRNA-binding affinity, whereas yeast eIF2α only weakly contributes to tRNA-binding affinity. These results are in keeping with the observations made using the yeast system (17,18) and shows that binding of yeast α and β subunits to archaeal γ subunit confers on Ch-eIF2 a eukaryotic behavior. The same effects of the yeast α and β subunits on tRNA binding were observed when E. coli Met-tRNAfMet was used as the ligand instead of authentic S. cerevisiae Met-tRNAiMet. The authentic tRNA was preferred by a factor of 2–3, whatever the combination of subunits (Table 1). This preference was also observed with the aIF2γ subunit alone, consistent with the idea that the stimulatory role of the A1–U72 pair of the eukaryotic initiator tRNA is mainly exerted through binding by the γ subunit (4,15,30,34). The E. coli initiator tRNA therefore appeared as a good substitute to further dissect the structure–function relationships of the yeast eIF2α and eIF2β subunits. Hence, the bacterial tRNA was used in the rest of the study since it can be obtained with a higher purity.
The two eukaryotic-specific extensions of eIF2β positively contribute to tRNA affinity. Removal of the two extensions makes a truncated eIF2β structurally homologous to its archaeal counterpart. However, in contrast to archaeal aIF2β, the core-restricted version of eIF2β retains a significant ability to increase tRNA-binding affinity (by a factor of at least 28). Therefore, the eIF2β core domain contains specific features involved in tRNA-binding affinity. Overall, each of the three parts of the β subunit contributes by one order of magnitude to tRNA-binding affinity. Furthermore, these contributions are additive, thereby suggesting that the two extensions independently cooperate to enhance tRNA affinity. Whether these contributions result from direct contacts of the β subunit with tRNA or from indirect effects, for instance on the conformation of the γ subunit, remains to be elucidated.
Interestingly, the near absence of effect of yeast eIF2α on tRNA affinity is only apparent. Indeed, a positive effect of the α subunit is compensated by a negative effect due to the appended eukaryotic acidic tail. The extent of the negative effect of the C-terminal tail increases with the size of the acidic peptide. The αΔC274 version of yeast eIF2α contributes by at least three orders of magnitude to tRNA-binding affinity. Within this subunit, domain 3 has apparently a minor role whereas domains 1 and 2 of eIF2α bring the major contribution to tRNA affinity. Nevertheless, SAXS experiments argue in favor of a binding mode for the tRNA molecule by Ch-eIF2αΔCγ identical to that observed within the authentic archaeal complex. The precise interactions of the domains of e/aIF2α with the tRNA molecule may explain the differences observed in the Kd measurements. In addition, at this stage, we cannot exclude that a part of the effects of the α subunit on tRNA binding may be indirect, through modulating the conformation of the γ subunit, for example at the level of the switch regions.
Finally, the present study, together with those of eIF2 from E. cuniculi (19) and archaeal aIF2 (6,7,14,35), shows that the roles of the peripheral subunits of e/aIF2 in tRNA binding vary from one organism to another. Despite this, the overall structure of the ternary initiation complex in solution is likely to remain similar in all organisms. The observed differences in the contributions of the peripheral subunits to tRNA binding might reflect species-specific adjustments in the mechanisms of handling of the initiator tRNA by e/aIF2 within the ribosomal initiation complex. For example, one may propose that the negative effect of the C-terminal tail of yeast eIF2α is cancelled on the small ribosomal subunit, thanks to an interaction with another partner of the ribosomal initiation complex. In agreement with this idea, the C-terminal tail of human eIF2α was shown to be mobile in solution (31). More generally, contacts of the α and the β subunits with other components of the initiation complex may contribute to modulate the affinity of e/aIF2 for the initiator tRNA along the translation initiation process (16). Such modulations would, in turn, contribute to start codon recognition, in agreement with the known importance of eIF2α and eIF2β for translation start specificity (36,37).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Centre National de la Recherche Scientifique; Ecole Polytechnique; Agence Nationale de la Recherche (ANR-06-PCVI-0018; MASTIC); Gaspard Monge PhD scholarship from Ecole Polytechnique (to M.N.). Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J. Perez for expert assistance during SAXS data collection on the SWING beamline of synchrotron SOLEIL and G. Keith (Institut de Biologie Moléculaire et Celulaire, Strasbourg) for the kind gift of yeast initiator tRNA.
REFERENCES
- 1.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson FL, Hannig EM. Ligand interactions with eukaryotic translation initiation factor 2: role of the gamma-subunit. EMBO J. 1996;15:6311–6320. [PMC free article] [PubMed] [Google Scholar]
- 4.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Pedulla N, Palermo R, Hasenohrl D, Blasi U, Cammarano P, Londei P. The archaeal eIF2 homologue: functional properties of an ancient translation initiation factor. Nucleic Acids Res. 2005;33:1804–1812. doi: 10.1093/nar/gki321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structural switch of the gamma subunit in an archaeal aIF2 alpha gamma heterodimer. Structure. 2006;14:119–128. doi: 10.1016/j.str.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Yatime L, Schmitt E, Blanquet S, Mechulam Y. Functional molecular mapping of archaeal translation initiation factor 2. J. Biol. Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- 8.Farruggio D, Chaudhuri J, Maitra U, RajBhandary UL. The A1 x U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol. Cell. Biol. 1996;16:4248–4256. doi: 10.1128/mcb.16.8.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt E, Blanquet S, Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokabe M, Yao M, Sakai N, Toya S, Tanaka I. Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region. Proc. Natl Acad. Sci. USA. 2006;103:13016–13021. doi: 10.1073/pnas.0604165103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stolboushkina E, Nikonov S, Nikulin A, Blasi U, Manstein DJ, Fedorov R, Garber M, Nikonov O. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the alpha- and beta-subunits. J. Mol. Biol. 2008;382:680–691. doi: 10.1016/j.jmb.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 12.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl Acad. Sci. USA. 2007;104:18445–18450. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BFC, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 14.Roll-Mecak A, Alone P, Cao C, Dever TE, Burley SK. X-ray structure of translation initiation factor eIF2gamma: implications for tRNA and eIF2alpha binding. J. Biol. Chem. 2004;279:10634–10642. doi: 10.1074/jbc.M310418200. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt E, Panvert M, Lazennec-Schurdevin C, Coureux PD, Perez J, Thompson A, Mechulam Y. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat. Struct. Mol. Biol. 2012;19:450–454. doi: 10.1038/nsmb.2259. [DOI] [PubMed] [Google Scholar]
- 16.Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE. Initiation factor eIF2gamma promotes eIF2-GTP-Met-tRNAi(Met) ternary complex binding to the 40S ribosome. Nat. Struct. Mol. Biol. 2011;18:1227–1234. doi: 10.1038/nsmb.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nika J, Rippel S, Hannig EM. Biochemical analysis of the eIF2bg complex reveals a structural function for eIF2a in catalyzed nucleotide exchange. J. Biol. Chem. 2001;276:1051–1060. doi: 10.1074/jbc.M007398200. [DOI] [PubMed] [Google Scholar]
- 18.Flynn A, Oldfield S, Proud CG. The role of the beta-subunit of initiation factor eIF-2 in initiation complex formation. Biochim. Biophys. Acta. 1993;1174:117–121. doi: 10.1016/0167-4781(93)90105-m. [DOI] [PubMed] [Google Scholar]
- 19.Naveau M, Lazennec-Schurdevin C, Panvert M, Mechulam Y, Schmitt E. tRNA binding properties of eukaryotic translation initiation factor 2 from Encephalitozoon cuniculi. Biochemistry. 2010;49:8680–8688. doi: 10.1021/bi1009166. [DOI] [PubMed] [Google Scholar]
- 20.Mechulam Y, Guillon L, Yatime L, Blanquet S, Schmitt E. Protection-based assays to measure aminoacyl-tRNA binding to translation initiation factors. Methods Enzymol. 2007;430:265–281. doi: 10.1016/S0076-6879(07)30011-6. [DOI] [PubMed] [Google Scholar]
- 21.Guillon JM, Meinnel T, Mechulam Y, Lazennec C, Blanquet S, Fayat S. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNAMetf formyltransferase. J. Mol. Biol. 1992;224:359–367. doi: 10.1016/0022-2836(92)91000-f. [DOI] [PubMed] [Google Scholar]
- 22.Meinnel T, Blanquet S. Maturation of pre-tRNAfMet by E. coli RNase P is specified by a guanosine of the 5′ flanking sequence. J. Biol. Chem. 1995;270:15906–15914. doi: 10.1074/jbc.270.26.15908. [DOI] [PubMed] [Google Scholar]
- 23.Mellot P, Mechulam Y, LeCorre D, Blanquet S, Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase:tRNA recognition. J. Mol. Biol. 1989;208:429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- 24.Dardel F. MC-Fit: Using Monte-Carlo methods to get accurate confidence limits on enzyme parameters. Comput. Applic. Biosci. 1994;10:273–275. doi: 10.1093/bioinformatics/10.3.273. [DOI] [PubMed] [Google Scholar]
- 25.Guillon L, Schmitt E, Blanquet S, Mechulam Y. Initiator tRNA binding by e/aIF5B, the eukaryotic/archaeal homologue of bacterial initiation factor IF2. Biochemistry. 2005;44:15594–15601. doi: 10.1021/bi051514j. [DOI] [PubMed] [Google Scholar]
- 26.David G, Perez J. combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchroron SOLEIL SWING beamline. J. Appl. Crystallogr. 2009;42:892–900. [Google Scholar]
- 27.Konarev PV, Volkov VV, Petoukhov MV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svergun DI, Barberato C, Koch MHJ. CRYSOL—a program to evaluate X-ray solution scaterring of biological macromoleucles from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 29.Wagner T, Gross M, Sigler PB. Isoleucyl initiator tRNA does not initiate eucaryotic protein synthesis. J. Biol. Chem. 1984;259:4706–4709. [PubMed] [Google Scholar]
- 30.Kapp LD, Kolitz SE, Lorsch JR. Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation. RNA. 2006;12:751–764. doi: 10.1261/rna.2263906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T, Marintchev A, Wagner G. Solution structure of human initiation factor eIF2alpha reveals homology to the elongation factor eEF1B. Structure Fold. Des. 2004;12:1693–1704. doi: 10.1016/j.str.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Yatime L, Schmitt E, Blanquet S, Mechulam Y. Structure-function relationships of the intact aIF2a subunit from the archaeon Pyrococcus abyssi. Biochemistry. 2005;44:8749–8756. doi: 10.1021/bi050373i. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto NN, Carnevalli LS, Castilho BA. Translation initiation at non-AUG codons mediated by weakened association of eukaryotic initiation factor (eIF) 2 subunits. Biochem. J. 2002;367:359–368. doi: 10.1042/BJ20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 2012;4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedulla N, Palermo R, Hasenohrl D, Blasi U, Cammarano P, Londei P. The archaeal eIF2 homologue: functional properties of an ancient translation initiation factor. Nucleic Acids Res. 2005;33:1804–1812. doi: 10.1093/nar/gki321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donahue TF, Cigan AM, Pabich EK, Valavicius BC. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 37.Cigan AM, Pabich EK, Feng L, Donahue TF. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc. Natl Acad. Sci. USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.